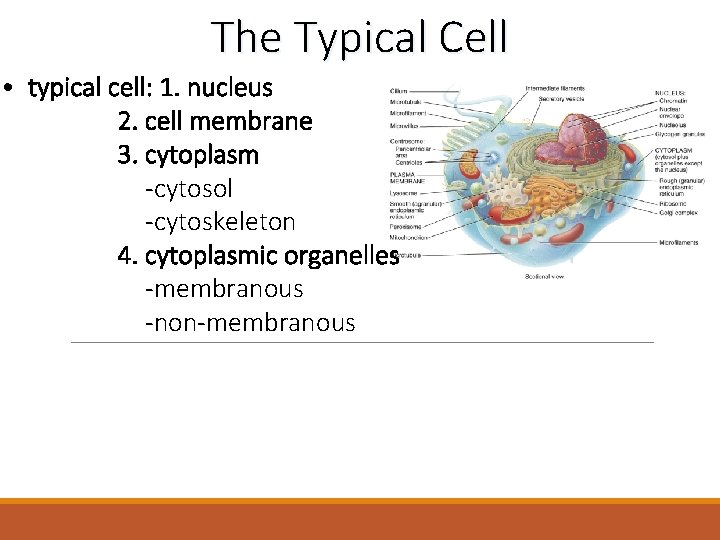
The Typical Cell typical cell 1 nucleus 2











- Slides: 72

The Typical Cell • typical cell: 1. nucleus 2. cell membrane 3. cytoplasm -cytosol -cytoskeleton 4. cytoplasmic organelles -membranous -non-membranous

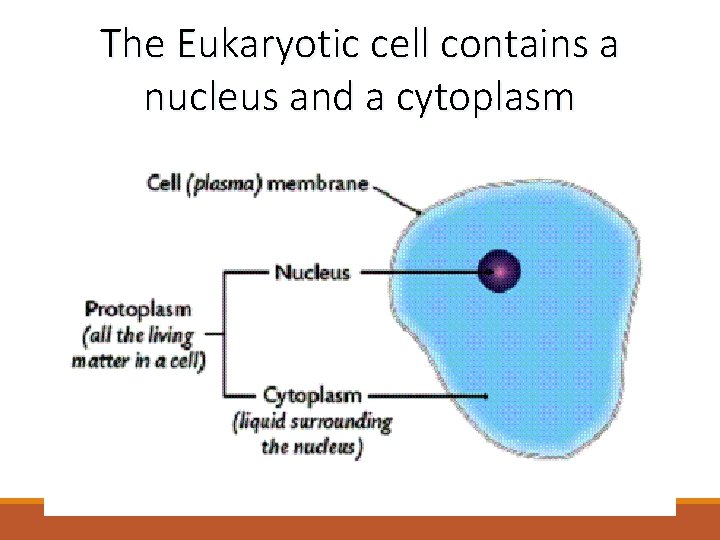
The Eukaryotic cell contains a nucleus and a cytoplasm

Cytoplasm • • semi-fluid-like jelly within the cell division into three subdivisions: • 1. fluid component: cytosol • 2. supportive framework of proteins: cytoskeleton • 3. organelles – membranous and nonmembranous


The Cytosol • eukaryotic cells – cytosol is part of the cytoplasm • fluid surrounding the organelles ◦ about 55% of the cell’s volume ◦ mostly water - 70 -90% ◦ PLUS ◦ ions ◦ dissolved nutrients – e. g. glucose ◦ soluble and insoluble proteins ◦ waste products ◦ macromolecules and their components - amino acids, fatty acids ◦ ATP

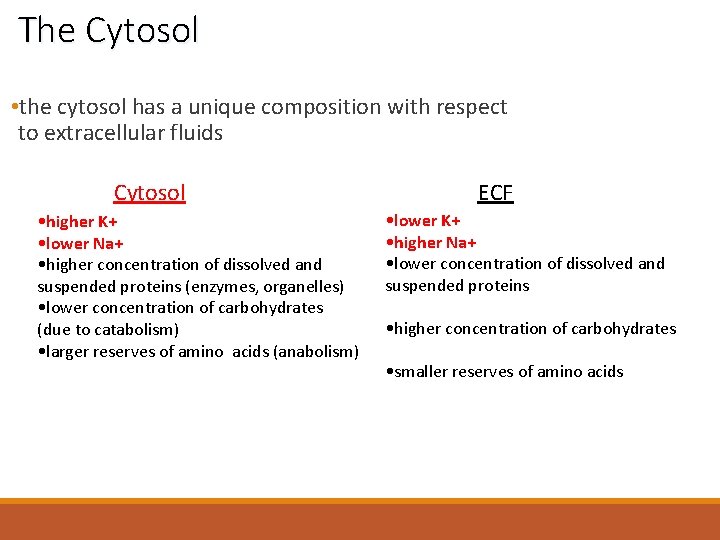
The Cytosol • the cytosol has a unique composition with respect to extracellular fluids Cytosol • higher K+ • lower Na+ • higher concentration of dissolved and suspended proteins (enzymes, organelles) • lower concentration of carbohydrates (due to catabolism) • larger reserves of amino acids (anabolism) ECF • lower K+ • higher Na+ • lower concentration of dissolved and suspended proteins • higher concentration of carbohydrates • smaller reserves of amino acids

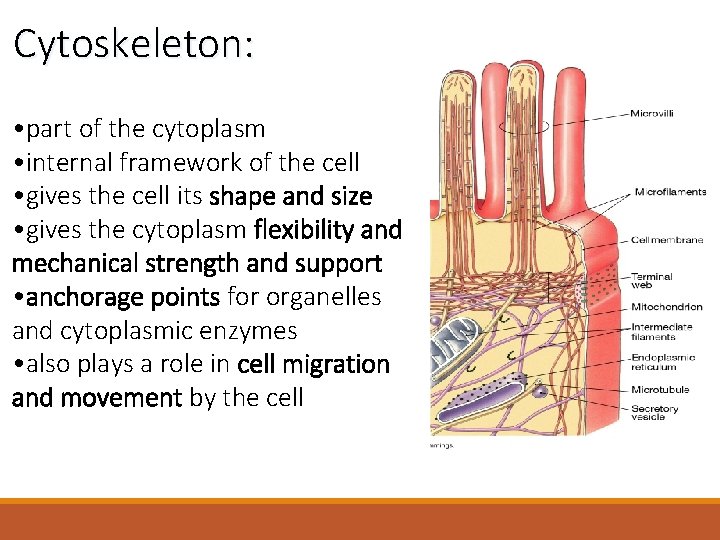
Cytoskeleton: • part of the cytoplasm • internal framework of the cell • gives the cell its shape and size • gives the cytoplasm flexibility and mechanical strength and support • anchorage points for organelles and cytoplasmic enzymes • also plays a role in cell migration and movement by the cell

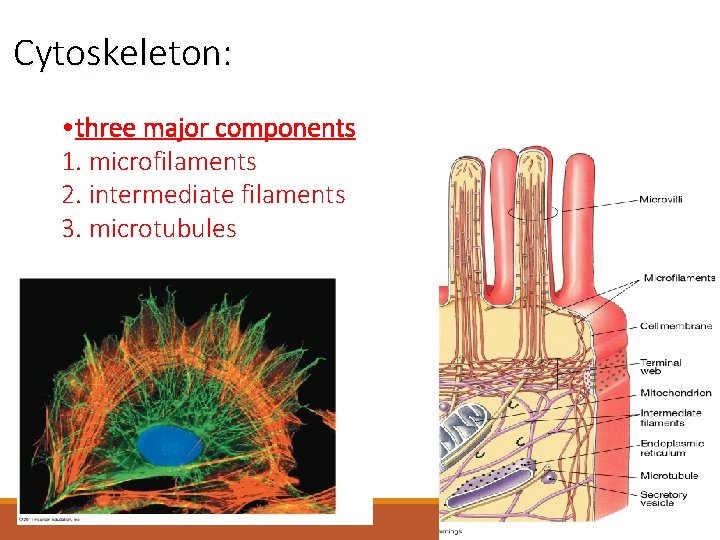
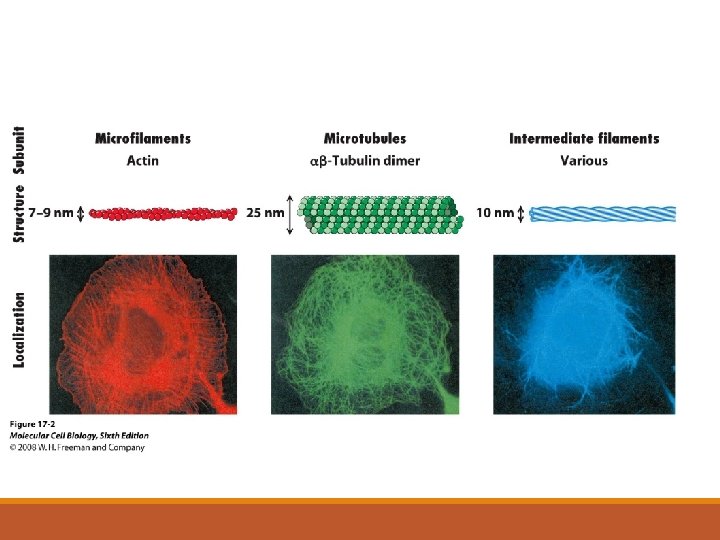
Cytoskeleton: • three major components 1. microfilaments 2. intermediate filaments 3. microtubules

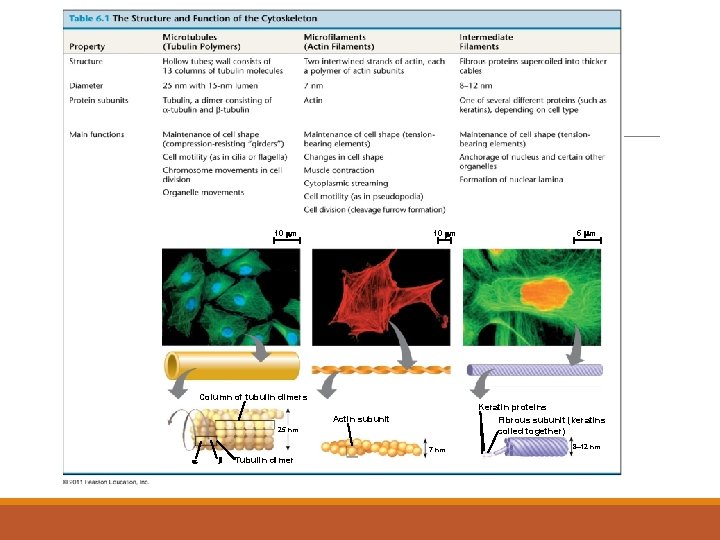
10 m 5 m Column of tubulin dimers Keratin proteins Fibrous subunit (keratins coiled together) Actin subunit 25 nm 7 nm Tubulin dimer 8 12 nm

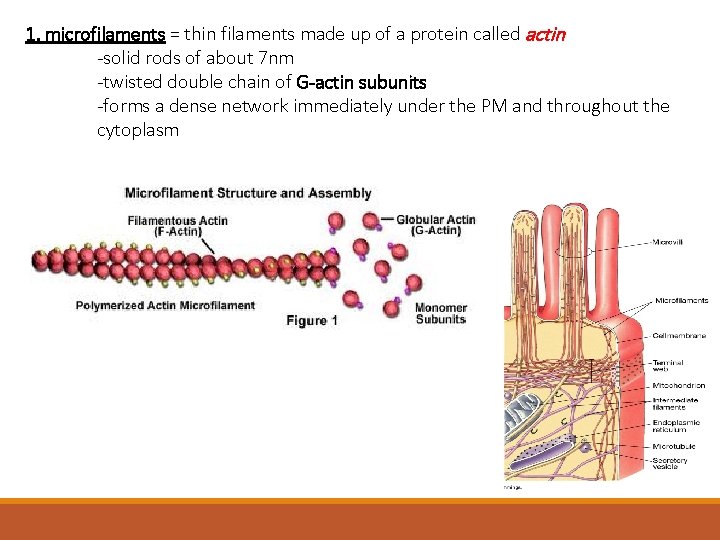
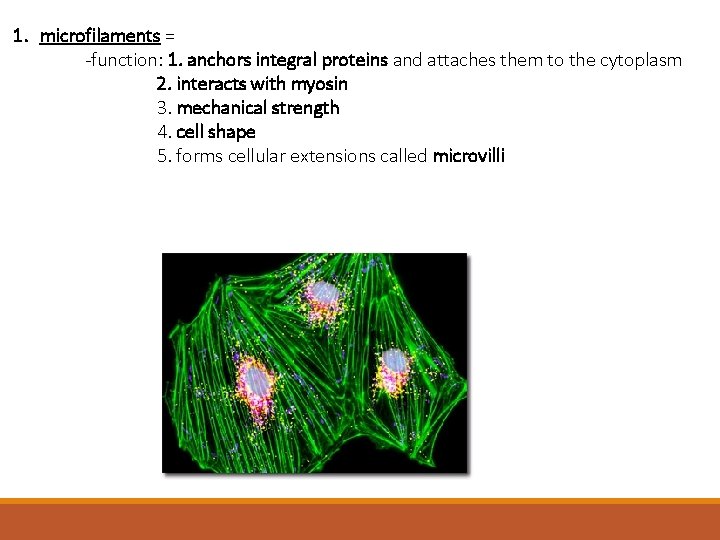
1. microfilaments = thin filaments made up of a protein called actin -solid rods of about 7 nm -twisted double chain of G-actin subunits -forms a dense network immediately under the PM and throughout the cytoplasm

1. microfilaments = -function: 1. anchors integral proteins and attaches them to the cytoplasm 2. interacts with myosin 3. mechanical strength 4. cell shape 5. forms cellular extensions called microvilli

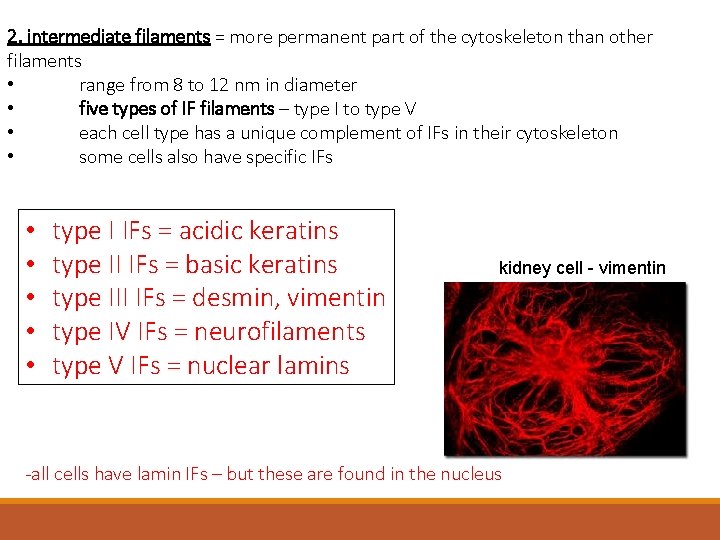
2. intermediate filaments = more permanent part of the cytoskeleton than other filaments • range from 8 to 12 nm in diameter • five types of IF filaments – type I to type V • each cell type has a unique complement of IFs in their cytoskeleton • some cells also have specific IFs • • • type I IFs = acidic keratins type II IFs = basic keratins type III IFs = desmin, vimentin type IV IFs = neurofilaments type V IFs = nuclear lamins kidney cell - vimentin -all cells have lamin IFs – but these are found in the nucleus

2. intermediate filaments = function: 1. give strength to the cytoskeleton 2. support cell shape 3. anchors & stabilizes organelles 4. transports materials

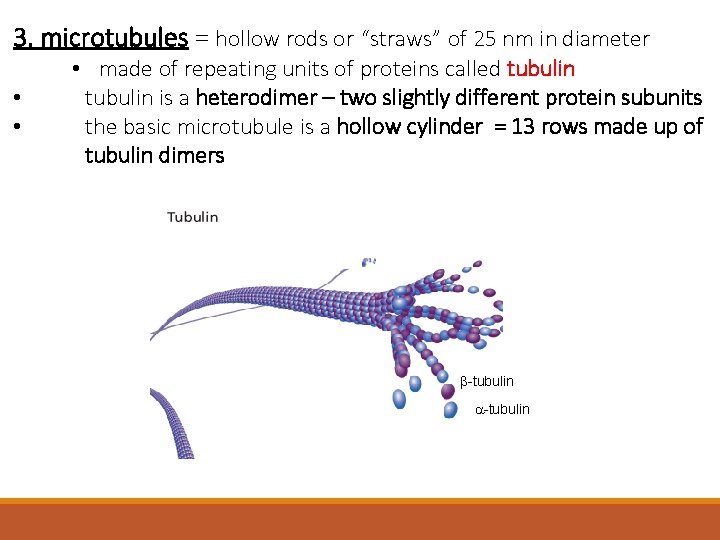
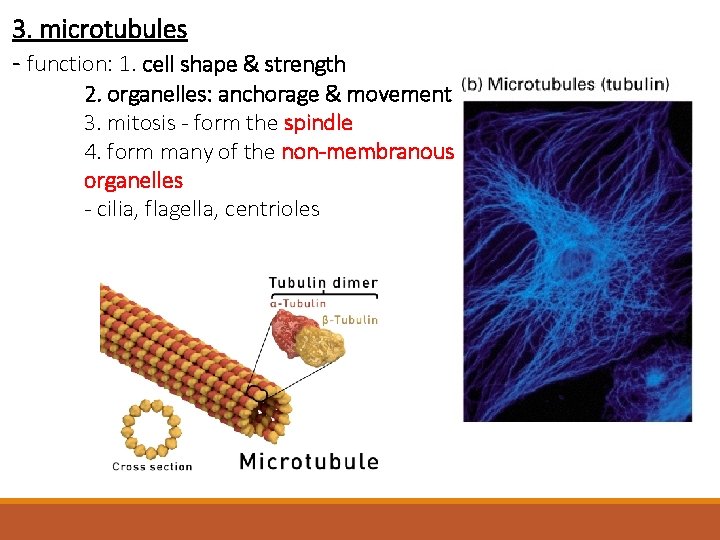
3. microtubules = hollow rods or “straws” of 25 nm in diameter • • • made of repeating units of proteins called tubulin is a heterodimer – two slightly different protein subunits the basic microtubule is a hollow cylinder = 13 rows made up of tubulin dimers b-tubulin a-tubulin

3. microtubules - function: 1. cell shape & strength 2. organelles: anchorage & movement 3. mitosis - form the spindle 4. form many of the non-membranous organelles - cilia, flagella, centrioles

3. microtubules • in animal cells – microtubule assembly occurs in the MTOC (microtubule organizing center or centrosome) -area of proteins located near the nucleus -within the MTOC : 1. tubulin dimers and other proteins 2. assembling MTs 3. modified MTs - called a centriole


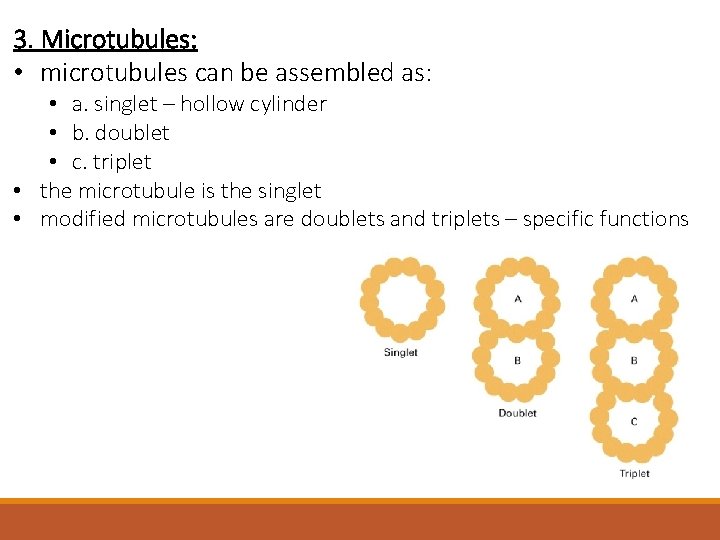
3. Microtubules: • microtubules can be assembled as: • a. singlet – hollow cylinder • b. doublet • c. triplet • the microtubule is the singlet • modified microtubules are doublets and triplets – specific functions

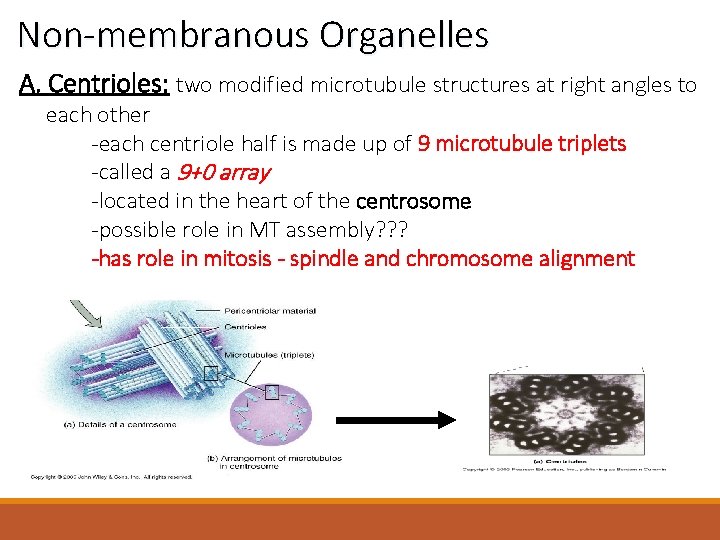
Non-membranous Organelles A. Centrioles: two modified microtubule structures at right angles to each other -each centriole half is made up of 9 microtubule triplets -called a 9+0 array -located in the heart of the centrosome -possible role in MT assembly? ? ? -has role in mitosis - spindle and chromosome alignment

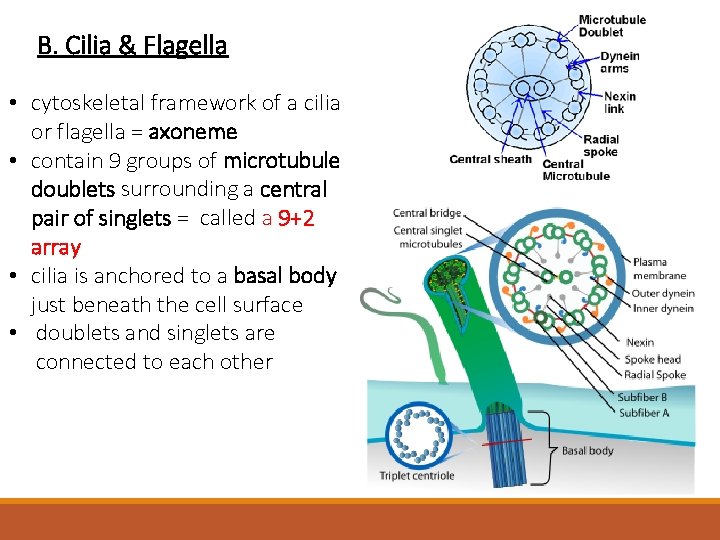
B. Cilia & Flagella • cilia = projections off of the plasma membrane of eukaryotic cells • exposed area is covered with plasma membrane - BUT NOT A MEMBRANOUS ORGANELLE • about 0. 25 um in diameter and only 20 um long • beat rhythmically to transport material • found in linings of several major organs covered with mucus where they function in cleaning Trachea

B. Cilia & Flagella • cytoskeletal framework of a cilia or flagella = axoneme • contain 9 groups of microtubule doublets surrounding a central pair of singlets = called a 9+2 array • cilia is anchored to a basal body just beneath the cell surface • doublets and singlets are connected to each other

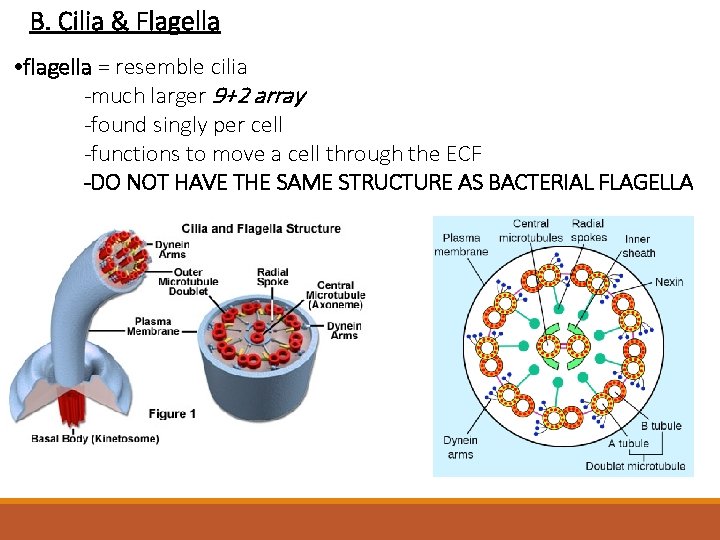
B. Cilia & Flagella • flagella = resemble cilia -much larger 9+2 array -found singly per cell -functions to move a cell through the ECF -DO NOT HAVE THE SAME STRUCTURE AS BACTERIAL FLAGELLA

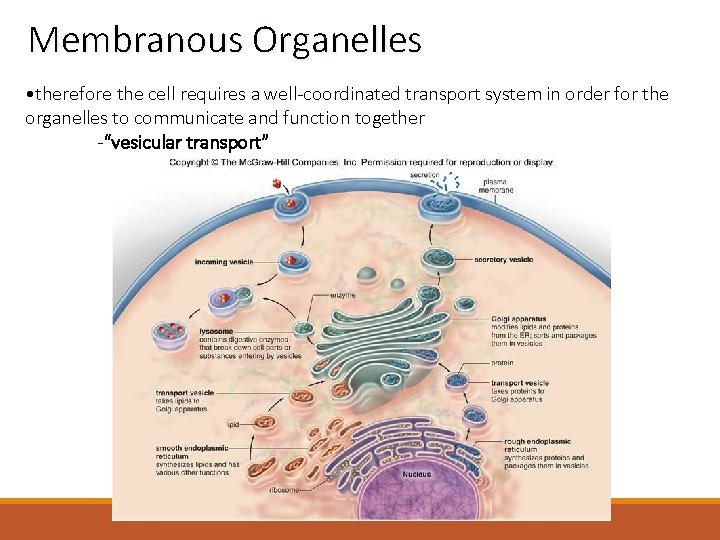
Membranous Organelles • completely surrounded by a phospholipid bilayer similar to the plasma membrane surrounding the cell • allows for isolation of each individual organelle - so that the interior of each organelle does not mix with the cytosol -known as compartmentalization • BUT THAT’S A PROBLEM • cellular compartments must “talk” to each other

Membranous Organelles • therefore the cell requires a well-coordinated transport system in order for the organelles to communicate and function together -“vesicular transport”

Membranous Organelles • major functions of membranous organelles • 1. protein synthesis & secretion – ER and Golgi • 2. energy production – mitochondria • 3. waste management – lysosomes and peroxisomes

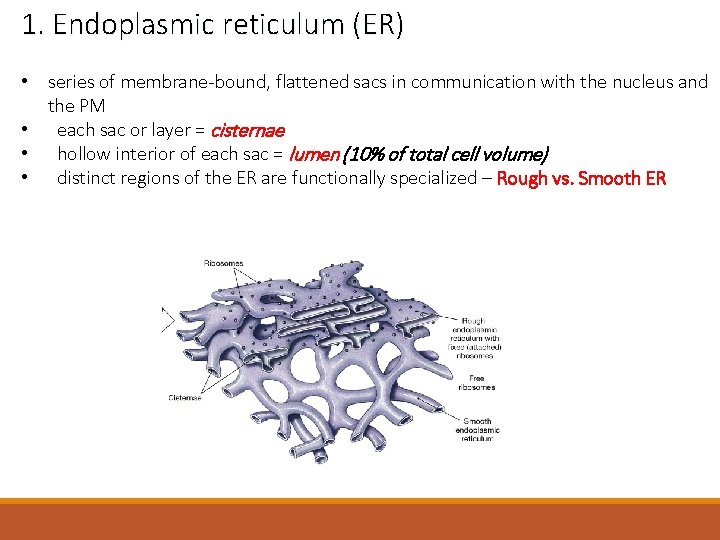
1. Endoplasmic reticulum (ER) • series of membrane-bound, flattened sacs in communication with the nucleus and the PM • each sac or layer = cisternae • hollow interior of each sac = lumen (10% of total cell volume) • distinct regions of the ER are functionally specialized – Rough vs. Smooth ER

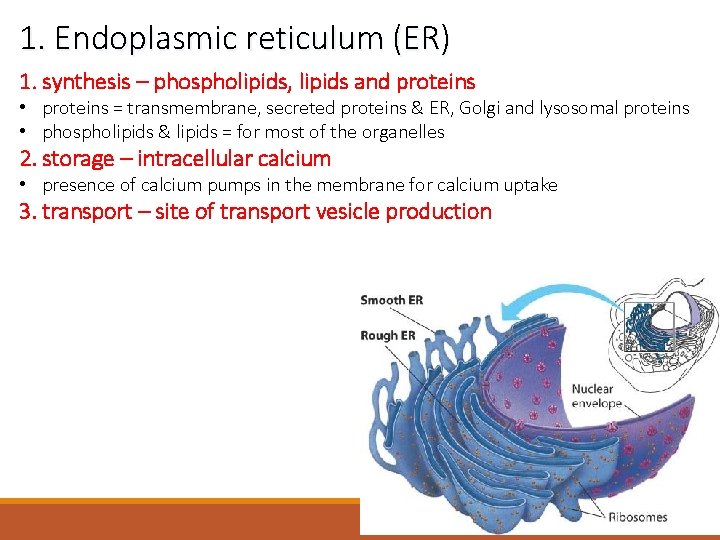
1. Endoplasmic reticulum (ER) 1. synthesis – phospholipids, lipids and proteins • proteins = transmembrane, secreted proteins & ER, Golgi and lysosomal proteins • phospholipids & lipids = for most of the organelles 2. storage – intracellular calcium • presence of calcium pumps in the membrane for calcium uptake 3. transport – site of transport vesicle production

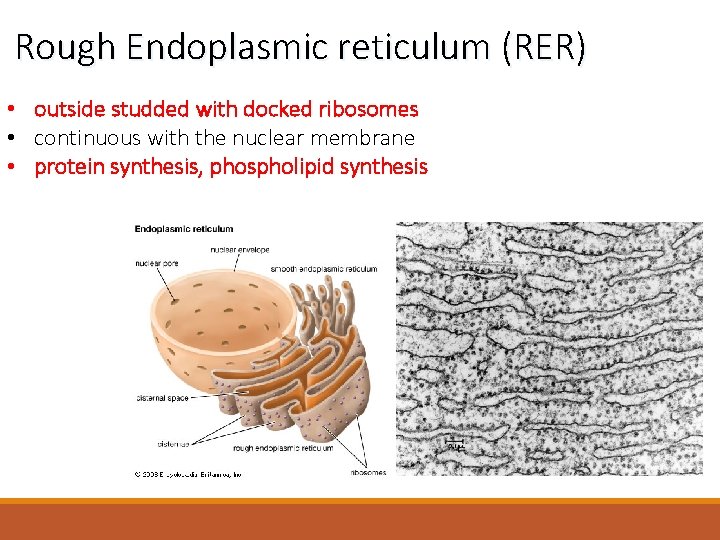
Rough Endoplasmic reticulum (RER) • outside studded with docked ribosomes • continuous with the nuclear membrane • protein synthesis, phospholipid synthesis

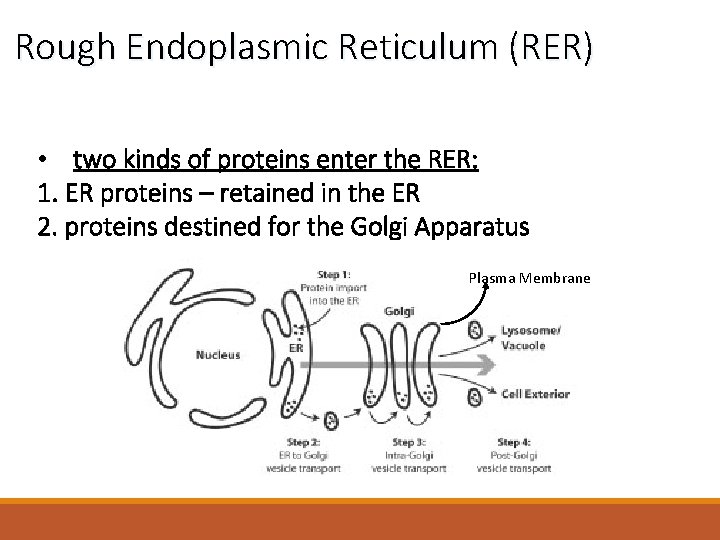
Rough Endoplasmic Reticulum (RER) • two kinds of proteins enter the RER: 1. ER proteins – retained in the ER 2. proteins destined for the Golgi Apparatus Plasma Membrane

Modifications in the RER 1. folding of the peptide chain 2. formation of disulfide bonds 3. addition and processing of carbohydrates = glycosylation 4. breakage of specific peptide bonds – proteolytic cleavage 5. assembly into globular proteins (more than one chain) http: //sumanasinc. com/webcontent/animations/content/proteinsecretion_mb. html

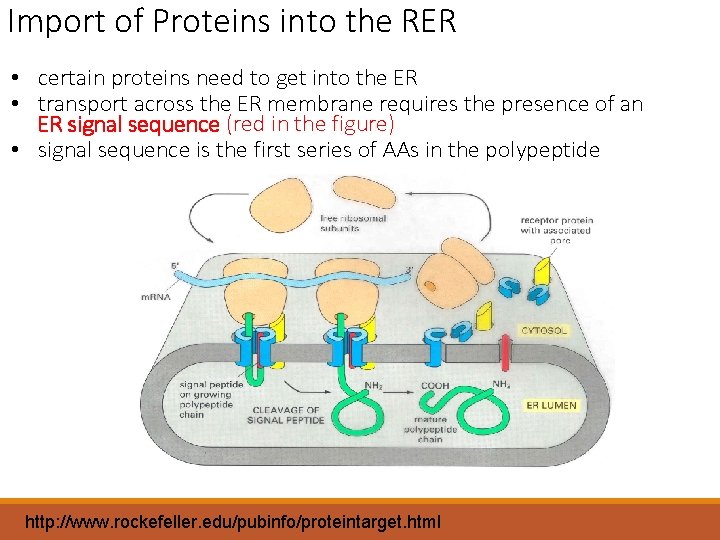
Import of Proteins into the RER • certain proteins need to get into the ER • transport across the ER membrane requires the presence of an ER signal sequence (red in the figure) • signal sequence is the first series of AAs in the polypeptide http: //www. rockefeller. edu/pubinfo/proteintarget. html

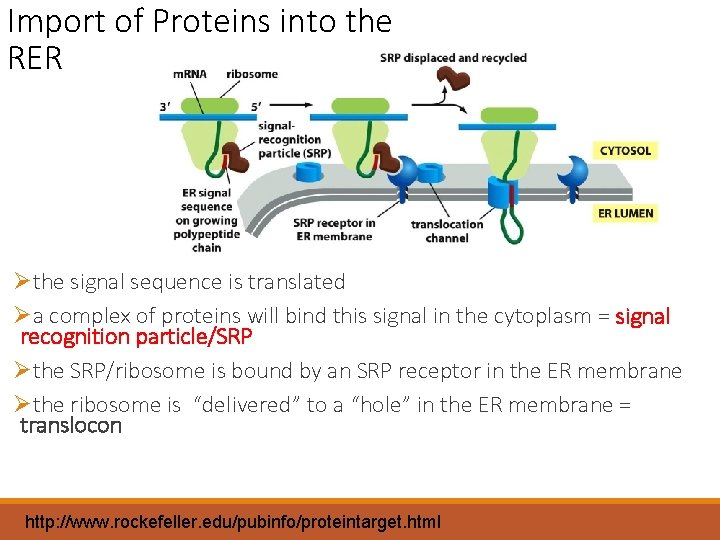
Import of Proteins into the RER Øthe signal sequence is translated Øa complex of proteins will bind this signal in the cytoplasm = signal recognition particle/SRP Øthe SRP/ribosome is bound by an SRP receptor in the ER membrane Øthe ribosome is “delivered” to a “hole” in the ER membrane = translocon http: //www. rockefeller. edu/pubinfo/proteintarget. html

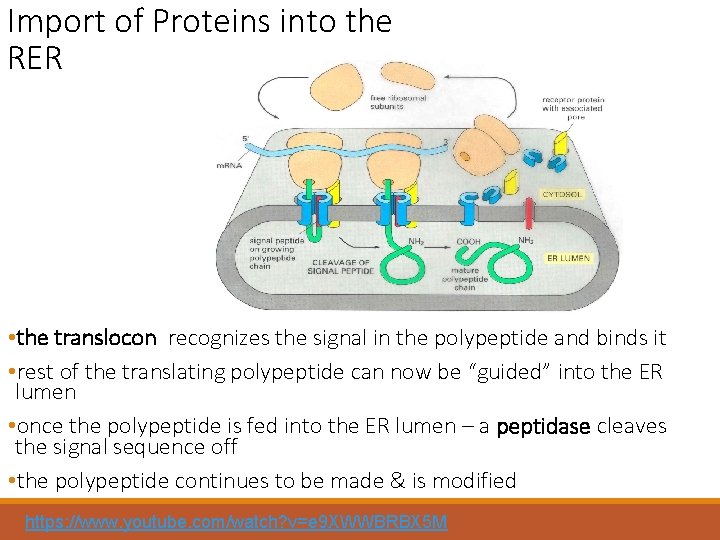
Import of Proteins into the RER • the translocon recognizes the signal in the polypeptide and binds it • rest of the translating polypeptide can now be “guided” into the ER lumen • once the polypeptide is fed into the ER lumen – a peptidase cleaves the signal sequence off • the polypeptide continues to be made & is modified https: //www. youtube. com/watch? v=e 9 XWWBRBX 5 M

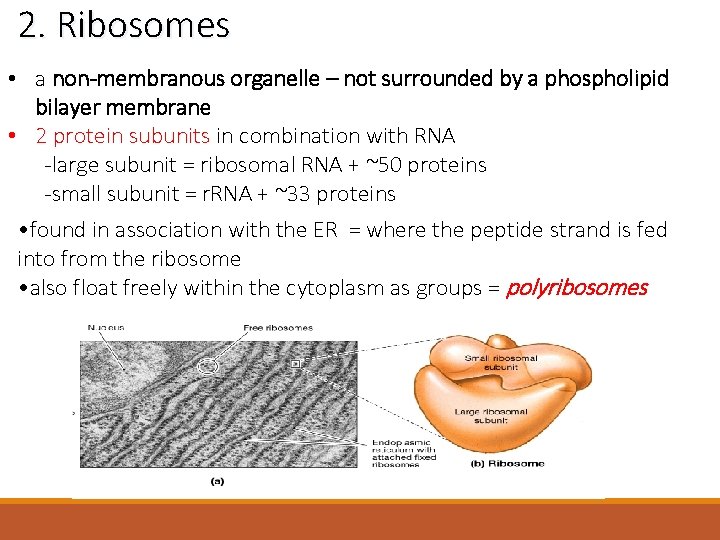
2. Ribosomes • a non-membranous organelle – not surrounded by a phospholipid bilayer membrane • 2 protein subunits in combination with RNA -large subunit = ribosomal RNA + ~50 proteins -small subunit = r. RNA + ~33 proteins • found in association with the ER = where the peptide strand is fed into from the ribosome • also float freely within the cytoplasm as groups = polyribosomes

Smooth Endoplasmic Reticulum (SER) • extends from the RER • free of ribosomes • many enzymes found on the surface of the SER 1. lipid synthesis for membranes 2. steroid biosynthesis 3. detoxification 4. transport vesicle formation 5. cleaves glucose

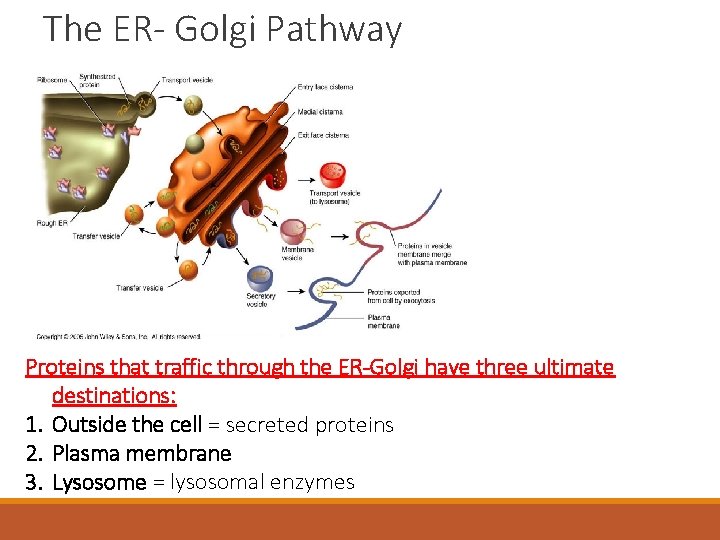
The ER- Golgi Pathway Proteins that traffic through the ER-Golgi have three ultimate destinations: 1. Outside the cell = secreted proteins 2. Plasma membrane 3. Lysosome = lysosomal enzymes

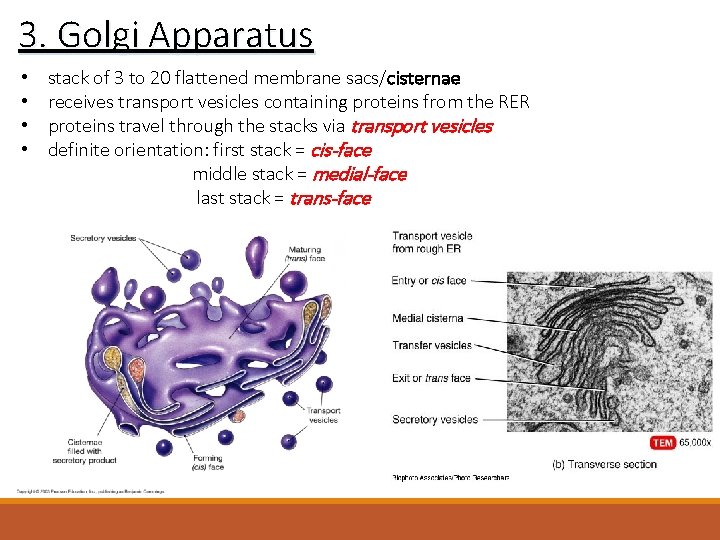
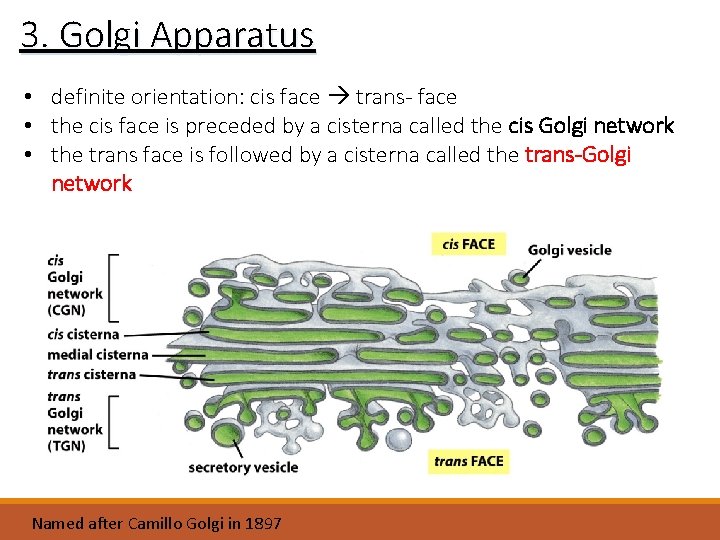
3. Golgi Apparatus • • stack of 3 to 20 flattened membrane sacs/cisternae receives transport vesicles containing proteins from the RER proteins travel through the stacks via transport vesicles definite orientation: first stack = cis-face middle stack = medial-face last stack = trans-face

3. Golgi Apparatus • definite orientation: cis face trans- face • the cis face is preceded by a cisterna called the cis Golgi network • the trans face is followed by a cisterna called the trans-Golgi network Named after Camillo Golgi in 1897

3. Golgi Apparatus • site of final protein modification and packaging of the finished protein • functions: • 1. major site of carbohydrate addition to proteins = glycosylation • 2. site for phosphate addition to proteins = phosphorylation • 3. production of sugars • 4. formation of the lysosome • 5. packaging of proteins and transport to their final destination • Golgi acts as a sorting station for transport vesicles

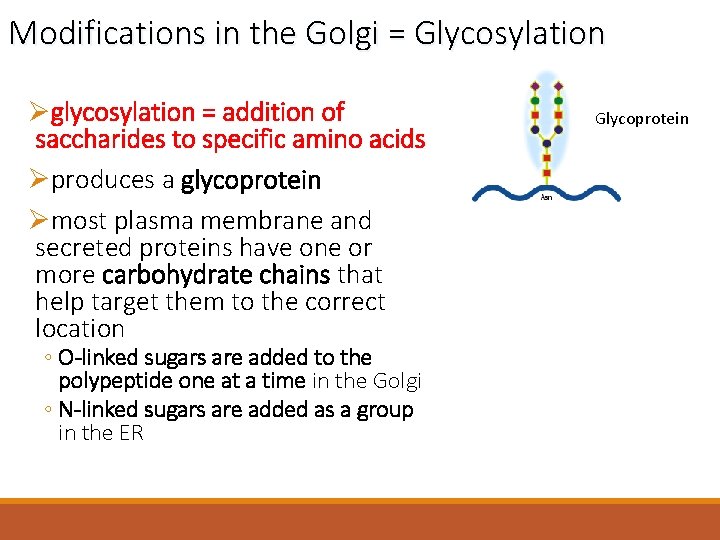
Modifications in the Golgi = Glycosylation Øglycosylation = addition of saccharides to specific amino acids Øproduces a glycoprotein Ømost plasma membrane and secreted proteins have one or more carbohydrate chains that help target them to the correct location ◦ O-linked sugars are added to the polypeptide one at a time in the Golgi ◦ N-linked sugars are added as a group in the ER Glycoprotein

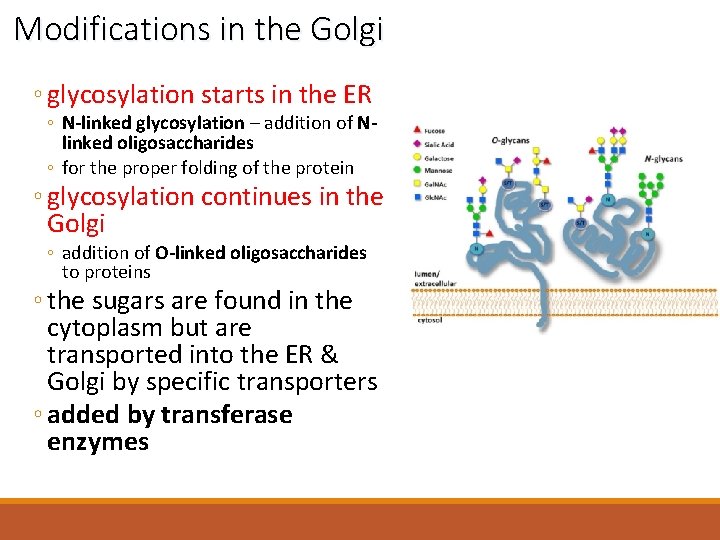
Modifications in the Golgi ◦ glycosylation starts in the ER ◦ N-linked glycosylation – addition of Nlinked oligosaccharides ◦ for the proper folding of the protein ◦ glycosylation continues in the Golgi ◦ addition of O-linked oligosaccharides to proteins ◦ the sugars are found in the cytoplasm but are transported into the ER & Golgi by specific transporters ◦ added by transferase enzymes

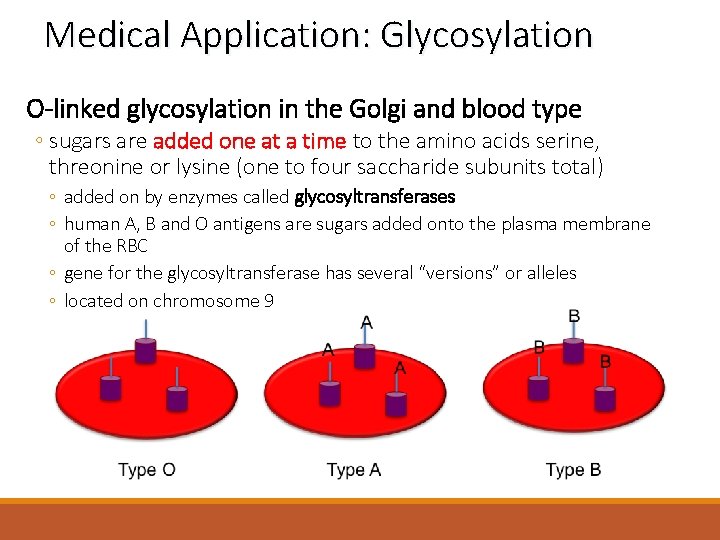
Medical Application: Glycosylation O-linked glycosylation in the Golgi and blood type ◦ sugars are added one at a time to the amino acids serine, threonine or lysine (one to four saccharide subunits total) ◦ added on by enzymes called glycosyltransferases ◦ human A, B and O antigens are sugars added onto the plasma membrane of the RBC ◦ gene for the glycosyltransferase has several “versions” or alleles ◦ located on chromosome 9

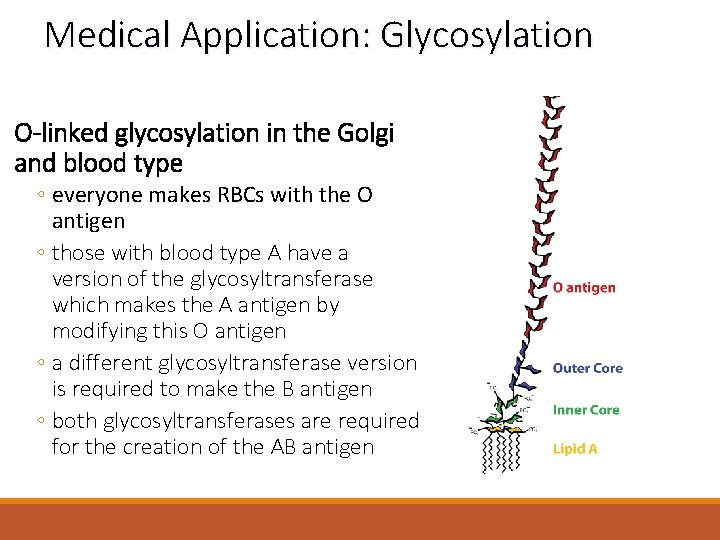
Medical Application: Glycosylation O-linked glycosylation in the Golgi and blood type ◦ everyone makes RBCs with the O antigen ◦ those with blood type A have a version of the glycosyltransferase which makes the A antigen by modifying this O antigen ◦ a different glycosyltransferase version is required to make the B antigen ◦ both glycosyltransferases are required for the creation of the AB antigen

Modifications in the Golgi: Protein Trimming Øsome PM proteins and most secretory proteins are synthesized as larger, inactive pro-proteins that will require additional processing to become active ◦ e. g. albumin, insulin, glucagon Øthis processing occurs very late in maturation ◦ processing is catalyzed by protein-specific enzymes called proteases ◦ occurs in secretory vesicles that bud from the trans-Golgi face

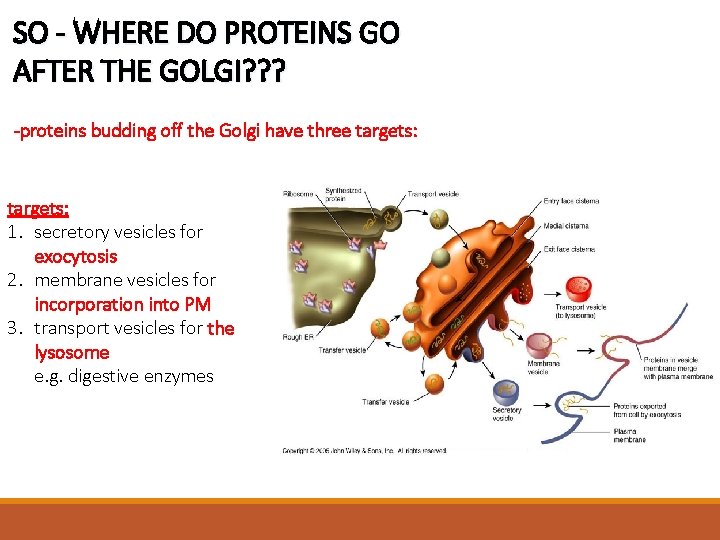
SO - WHERE DO PROTEINS GO AFTER THE GOLGI? ? ? -proteins budding off the Golgi have three targets: 1. secretory vesicles for exocytosis 2. membrane vesicles for incorporation into PM 3. transport vesicles for the lysosome e. g. digestive enzymes

WHAT IF YOU AREN’T ONE OF THESE PROTEINS? ? ØER proteins stay in the ER ◦ never traffic to the Golgi ◦ these ER proteins will have a retention signal ØRibosomal proteins ◦ translation of ribosomal proteins are done in the cytoplasm by polyribosomes ◦ imported into the nucleus for assembly with r. RNA into large and small subunits Ømitochondrial proteins ◦ the mitochondria has its own DNA, transcribes its own m. RNA and has its own ribosomes for translation

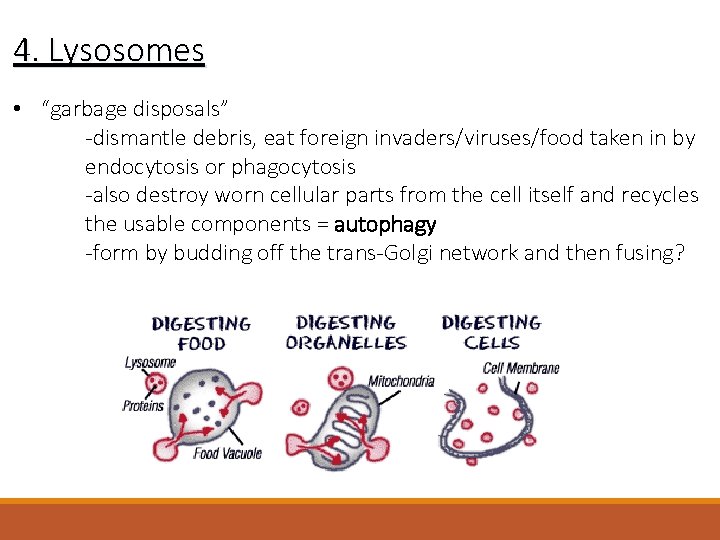
4. Lysosomes • “garbage disposals” -dismantle debris, eat foreign invaders/viruses/food taken in by endocytosis or phagocytosis -also destroy worn cellular parts from the cell itself and recycles the usable components = autophagy -form by budding off the trans-Golgi network and then fusing?

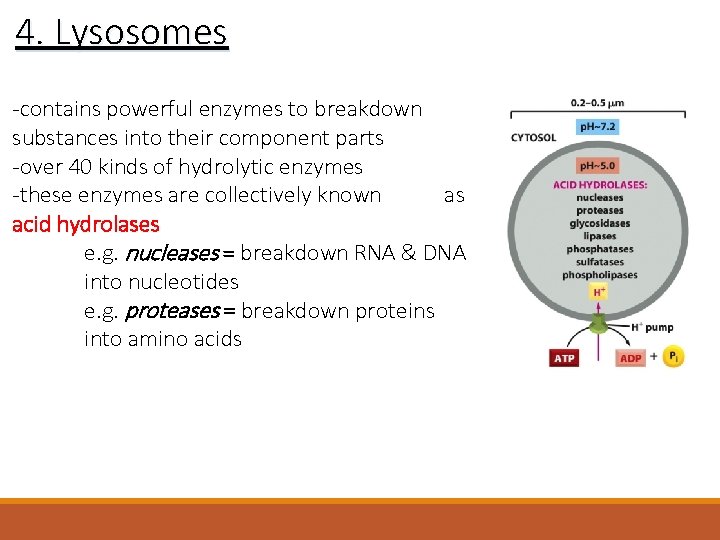
4. Lysosomes -contains powerful enzymes to breakdown substances into their component parts -over 40 kinds of hydrolytic enzymes -these enzymes are collectively known as acid hydrolases e. g. nucleases = breakdown RNA & DNA into nucleotides e. g. proteases = breakdown proteins into amino acids

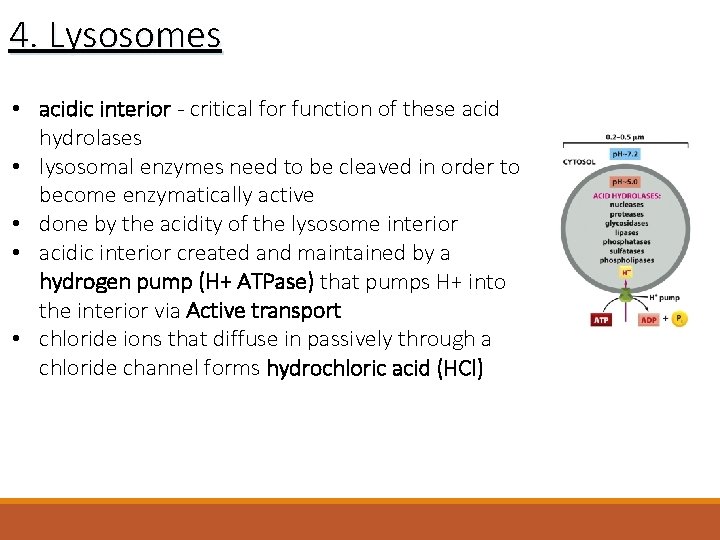
4. Lysosomes • acidic interior - critical for function of these acid hydrolases • lysosomal enzymes need to be cleaved in order to become enzymatically active • done by the acidity of the lysosome interior • acidic interior created and maintained by a hydrogen pump (H+ ATPase) that pumps H+ into the interior via Active transport • chloride ions that diffuse in passively through a chloride channel forms hydrochloric acid (HCl)

5. Peroxisomes: • found in all cells but abundant in liver and kidney cells • may arise from pre-existing peroxisomes or may bud from the ER

5. Peroxisomes: • major function is oxidation (breakdown) of long chain fatty acids (beta-oxidation) • results in the conversion of the fatty acid into acetyl co. A Kreb’s cycle • beta-oxidation is done by oxidases = enzymes that use oxygen to oxidize substances • generates hydrogen peroxide (H 2 O 2)

5. Peroxisomes: • PROBLEM#1: H 2 O 2 is very corrosive -therefore peroxisomes also contain an enzyme called catalase to break this peroxide down into water and oxygen • PROBLEM #2: the electron transport chain in mitochondria produces superoxide radicals (O 2 -) as a normal consequence of electrons “leaking” (from complex I) -peroxisomes also contain anti-oxidant enzymes to break down other dangerous oxidative chemicals made by the cell during metabolism -other functions of peroxisomes: 1. synthesis of bile acids 2. breakdown of alcohol by liver cells

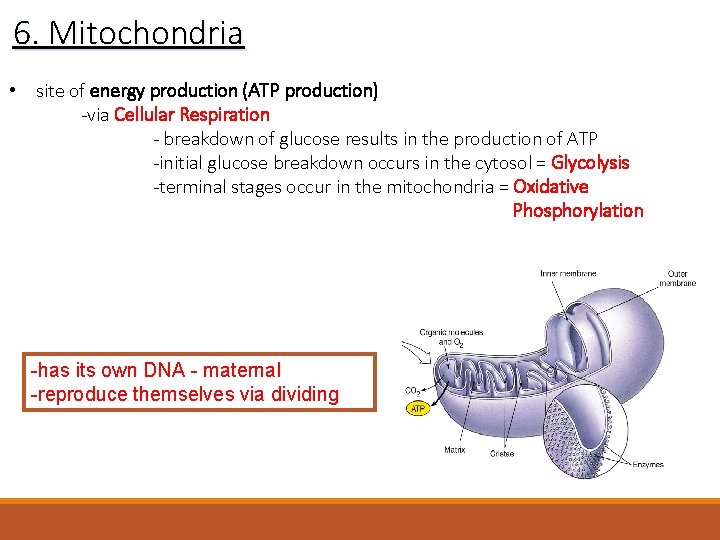
6. Mitochondria • site of energy production (ATP production) -via Cellular Respiration - breakdown of glucose results in the production of ATP -initial glucose breakdown occurs in the cytosol = Glycolysis -terminal stages occur in the mitochondria = Oxidative Phosphorylation -has its own DNA - maternal -reproduce themselves via dividing

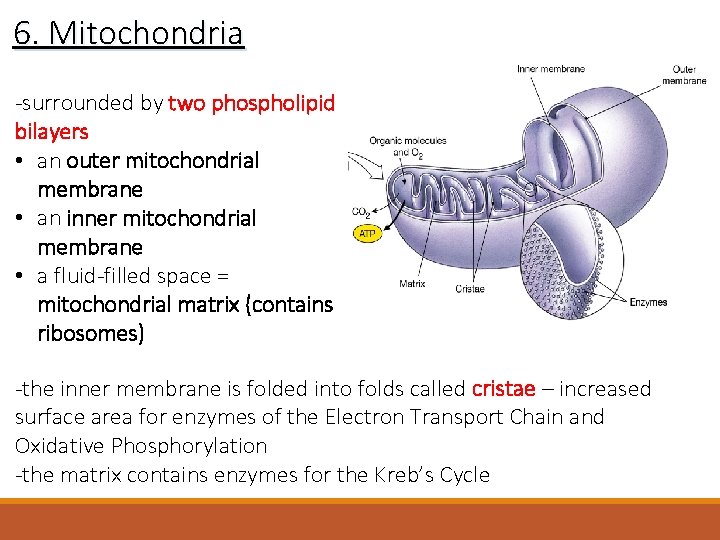
6. Mitochondria -surrounded by two phospholipid bilayers • an outer mitochondrial membrane • an inner mitochondrial membrane • a fluid-filled space = mitochondrial matrix (contains ribosomes) -the inner membrane is folded into folds called cristae – increased surface area for enzymes of the Electron Transport Chain and Oxidative Phosphorylation -the matrix contains enzymes for the Kreb’s Cycle

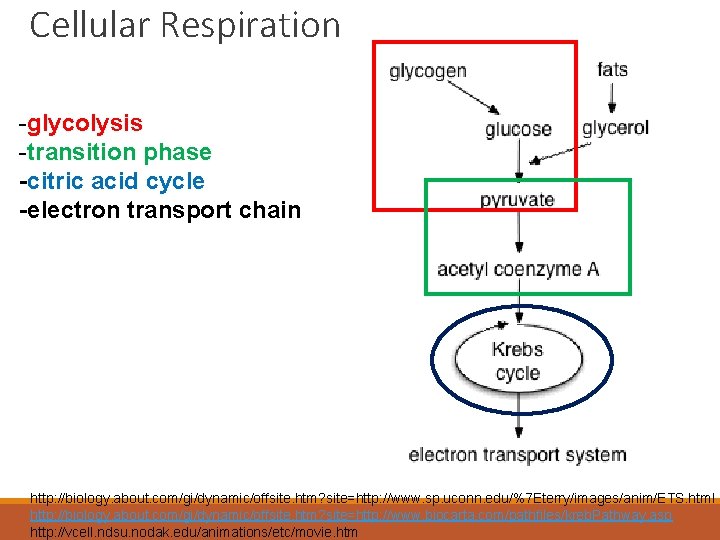
Cellular Respiration -glycolysis -transition phase -citric acid cycle -electron transport chain http: //biology. about. com/gi/dynamic/offsite. htm? site=http: //www. sp. uconn. edu/%7 Eterry/images/anim/ETS. html http: //biology. about. com/gi/dynamic/offsite. htm? site=http: //www. biocarta. com/pathfiles/kreb. Pathway. asp http: //vcell. ndsu. nodak. edu/animations/etc/movie. htm

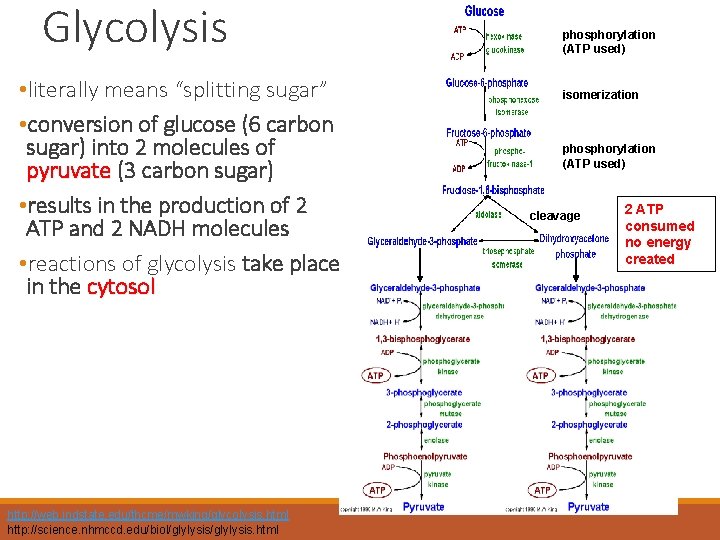
Glycolysis • literally means “splitting sugar” • conversion of glucose (6 carbon sugar) into 2 molecules of pyruvate (3 carbon sugar) • results in the production of 2 ATP and 2 NADH molecules • reactions of glycolysis take place in the cytosol http: //web. indstate. edu/thcme/mwking/glycolysis. html http: //science. nhmccd. edu/biol/glylysis. html phosphorylation (ATP used) isomerization phosphorylation (ATP used) cleavage 2 ATP consumed no energy created

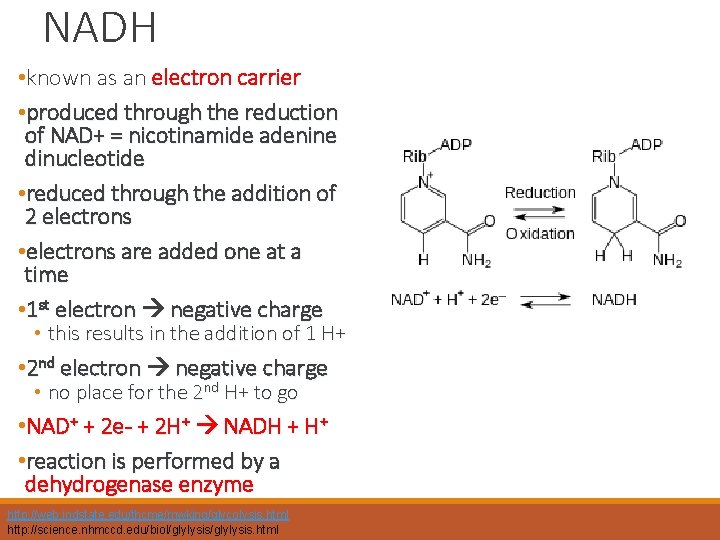
NADH • known as an electron carrier • produced through the reduction of NAD+ = nicotinamide adenine dinucleotide • reduced through the addition of 2 electrons • electrons are added one at a time • 1 st electron negative charge • this results in the addition of 1 H+ • 2 nd electron negative charge • no place for the 2 nd H+ to go • NAD+ + 2 e- + 2 H+ NADH + H+ • reaction is performed by a dehydrogenase enzyme http: //web. indstate. edu/thcme/mwking/glycolysis. html http: //science. nhmccd. edu/biol/glylysis. html

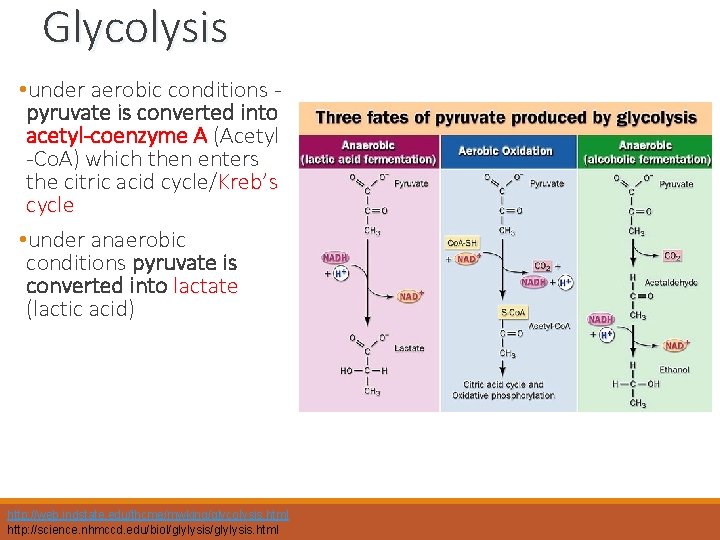
Glycolysis • under aerobic conditions pyruvate is converted into acetyl-coenzyme A (Acetyl -Co. A) which then enters the citric acid cycle/Kreb’s cycle • under anaerobic conditions pyruvate is converted into lactate (lactic acid) http: //web. indstate. edu/thcme/mwking/glycolysis. html http: //science. nhmccd. edu/biol/glylysis. html

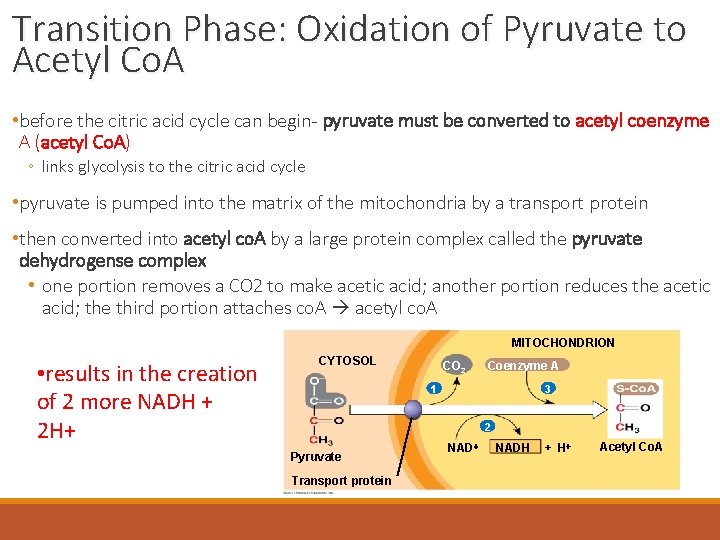
Transition Phase: Oxidation of Pyruvate to Acetyl Co. A • before the citric acid cycle can begin- pyruvate must be converted to acetyl coenzyme A (acetyl Co. A) ◦ links glycolysis to the citric acid cycle • pyruvate is pumped into the matrix of the mitochondria by a transport protein • then converted into acetyl co. A by a large protein complex called the pyruvate dehydrogense complex • one portion removes a CO 2 to make acetic acid; another portion reduces the acetic acid; the third portion attaches co. A acetyl co. A MITOCHONDRION • results in the creation of 2 more NADH + 2 H+ CYTOSOL CO 2 Coenzyme A 3 1 2 Pyruvate Transport protein NADH + H Acetyl Co. A

Pyruvate processing • pyruvate can be processed in many ways other than acetyl co. A • pyruvate can also be processed to form: ◦ 1. lactate – by animal cells in the absence of oxygen ◦ also produces NAD+ ◦ 2. ethanol – by yeast and bacteria ◦ pyruvate acetylaldehyde ethanol + NAD+ ◦ catalyzed by alcohol dehydrogenase ◦ chemically a dehydrogenase removes 2 electrons and 2 H+ from one substrate and adds it to another ◦ biologically, the dehydrogenase removes 2 electrons and 2 H+ and adds it to NAD+ NADH + H+

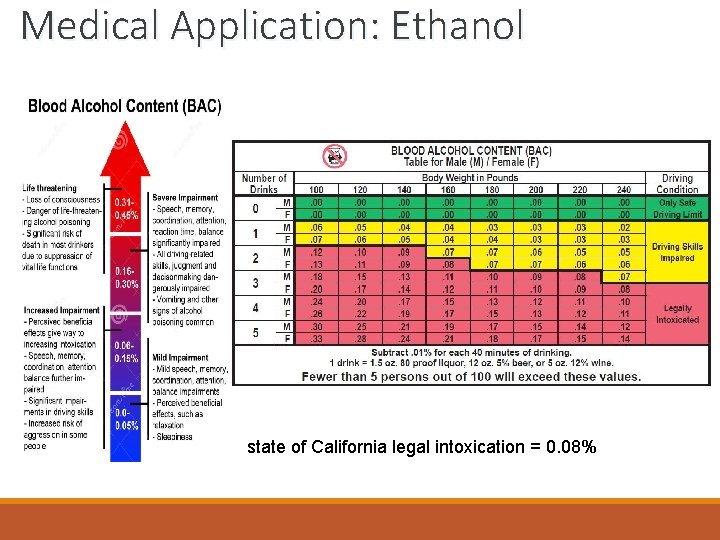
Medical Application: Ethanol ◦ in animals: ethanol aldehyde ◦ also catalyzed by alcohol dehydrogenase (ADH) ◦ ◦ there at least 7 genes for synthesis of ADH there are 5 classes of ADH genes breakdown of alcohol is done by ADH in the liver and the lining of the stomach this ADH is made of 3 subunits – encoded for by three genes: ADH 1, ADH 2, ADH 3 ◦ ADH 2 and ADH 3 genes show polymorphism – different forms and activities found in different populations ◦ expression of ADH differs according to sex and age ◦ increases in women as they age; decreases in men as they age ◦ unprocessed alcohol has multiple targets ◦ the overall effect is to slow the functional processes of the brain cell ◦ inhibits production of GABA - commonly known as the brain's "brake" mechanism. ◦ toxic effects = drunk

Medical Application: Ethanol state of California legal intoxication = 0. 08%

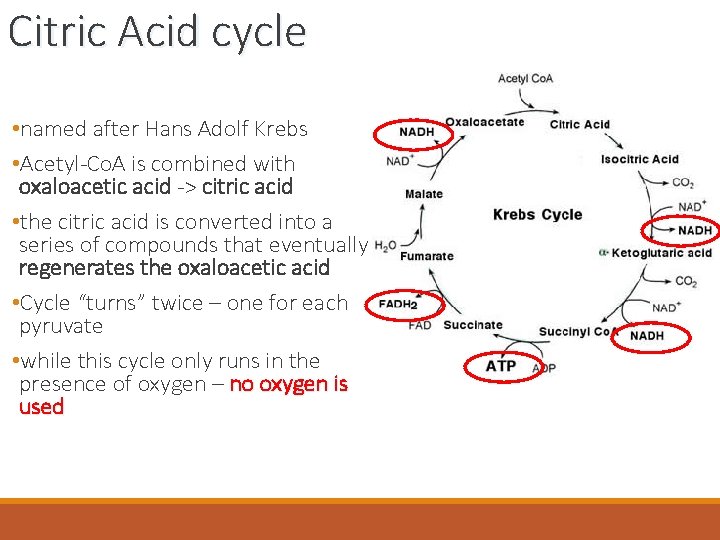
Citric Acid cycle • named after Hans Adolf Krebs • Acetyl-Co. A is combined with oxaloacetic acid -> citric acid • the citric acid is converted into a series of compounds that eventually regenerates the oxaloacetic acid • Cycle “turns” twice – one for each pyruvate • while this cycle only runs in the presence of oxygen – no oxygen is used

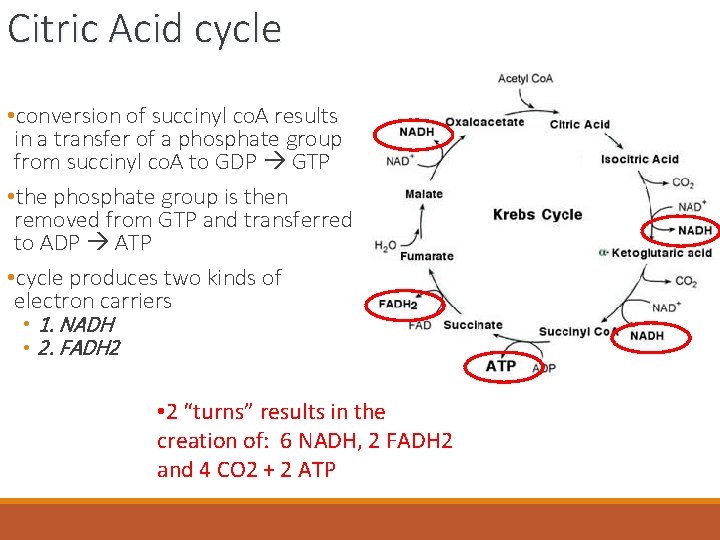
Citric Acid cycle • conversion of succinyl co. A results in a transfer of a phosphate group from succinyl co. A to GDP GTP • the phosphate group is then removed from GTP and transferred to ADP ATP • cycle produces two kinds of electron carriers • 1. NADH • 2. FADH 2 • 2 “turns” results in the creation of: 6 NADH, 2 FADH 2 and 4 CO 2 + 2 ATP

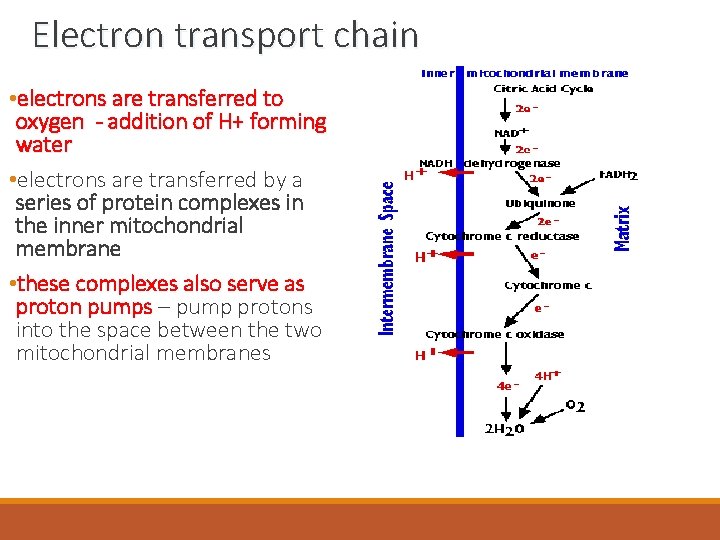
Electron transport chain • electrons are transferred to oxygen - addition of H+ forming water • electrons are transferred by a series of protein complexes in the inner mitochondrial membrane • these complexes also serve as proton pumps – pump protons into the space between the two mitochondrial membranes cytochrome c oxidase

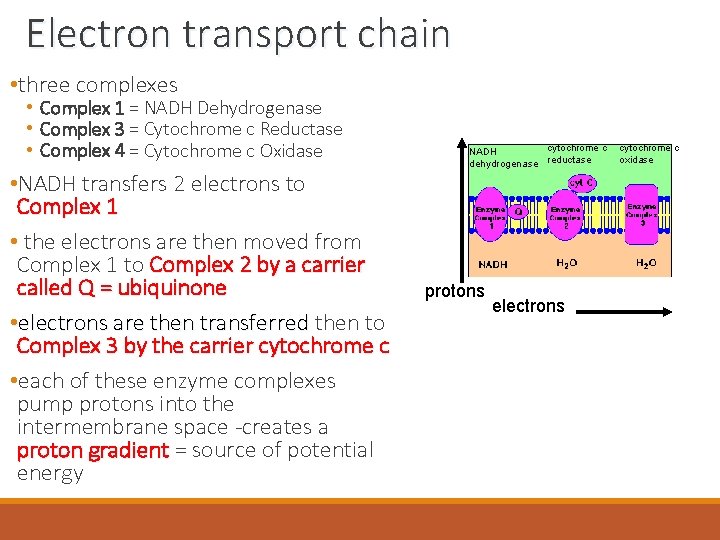
Electron transport chain • three complexes • Complex 1 = NADH Dehydrogenase • Complex 3 = Cytochrome c Reductase • Complex 4 = Cytochrome c Oxidase • NADH transfers 2 electrons to Complex 1 • the electrons are then moved from Complex 1 to Complex 2 by a carrier called Q = ubiquinone • electrons are then transferred then to Complex 3 by the carrier cytochrome c • each of these enzyme complexes pump protons into the intermembrane space -creates a proton gradient = source of potential energy cytochrome c NADH dehydrogenase reductase protons electrons cytochrome c oxidase

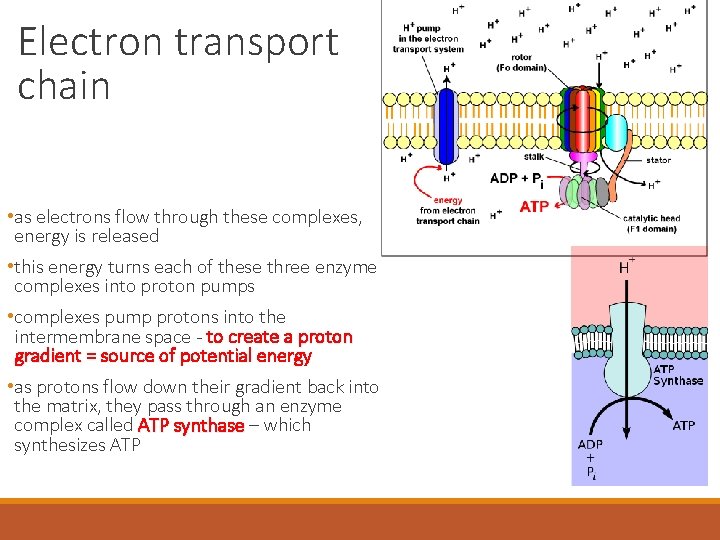
Electron transport chain • as electrons flow through these complexes, energy is released • this energy turns each of these three enzyme complexes into proton pumps • complexes pump protons into the intermembrane space - to create a proton gradient = source of potential energy • as protons flow down their gradient back into the matrix, they pass through an enzyme complex called ATP synthase – which synthesizes ATP

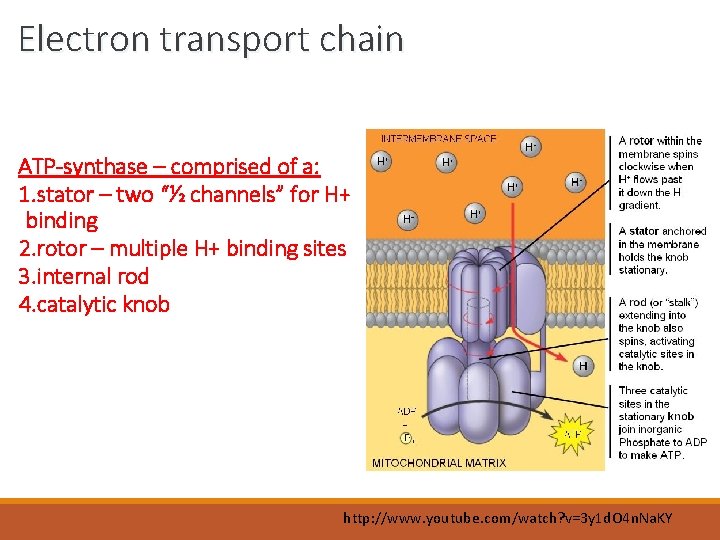
Electron transport chain ATP-synthase – comprised of a: 1. stator – two “½ channels” for H+ binding 2. rotor – multiple H+ binding sites 3. internal rod 4. catalytic knob http: //www. youtube. com/watch? v=3 y 1 d. O 4 n. Na. KY

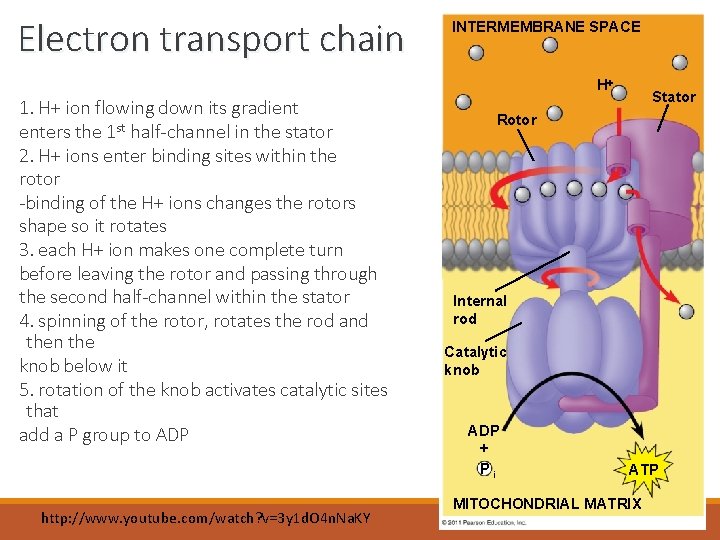
Electron transport chain INTERMEMBRANE SPACE H 1. H+ ion flowing down its gradient enters the 1 st half-channel in the stator 2. H+ ions enter binding sites within the rotor -binding of the H+ ions changes the rotors shape so it rotates 3. each H+ ion makes one complete turn before leaving the rotor and passing through the second half-channel within the stator 4. spinning of the rotor, rotates the rod and then the knob below it 5. rotation of the knob activates catalytic sites that add a P group to ADP http: //www. youtube. com/watch? v=3 y 1 d. O 4 n. Na. KY Stator Rotor Internal rod Catalytic knob ADP + Pi ATP MITOCHONDRIAL MATRIX

ETC animations http: //www. youtube. com/watch? v=xb. J 0 nbzt 5 Kw&feature=relmfu http: //www. youtube. com/watch? v=3 y 1 d. O 4 n. Na. KY http: //biology. about. com/gi/dynamic/offsite. htm? site=http: //www. sp. uconn. edu/%7 Eterry/images/anim/ETS. html

Preference for fats • in response to hormones like adrenalin – fats are hydrolyzed in adipocytes to form free fatty acids and glycerol • the fatty acids are released into the blood where they are taken up easily by most cells and oxidized by the mitochondria • in humans the oxidation of free fatty acids is quantitatively more important than the oxidation of glucose

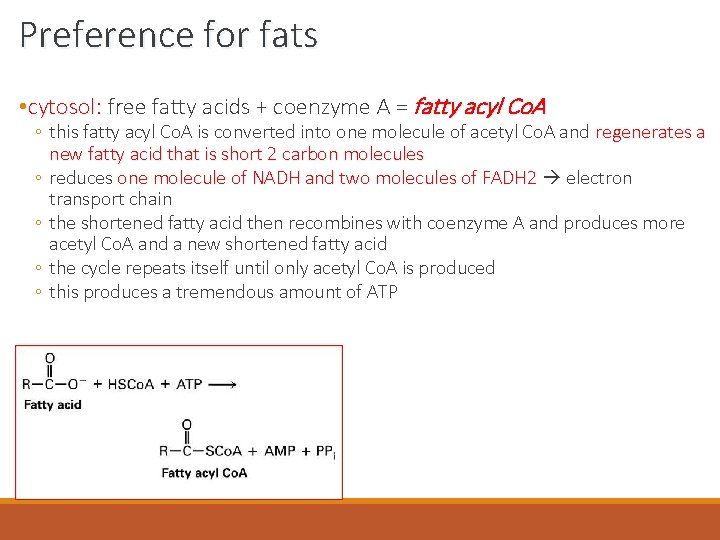
Preference for fats • cytosol: free fatty acids + coenzyme A = fatty acyl Co. A ◦ this fatty acyl Co. A is converted into one molecule of acetyl Co. A and regenerates a new fatty acid that is short 2 carbon molecules ◦ reduces one molecule of NADH and two molecules of FADH 2 electron transport chain ◦ the shortened fatty acid then recombines with coenzyme A and produces more acetyl Co. A and a new shortened fatty acid ◦ the cycle repeats itself until only acetyl Co. A is produced ◦ this produces a tremendous amount of ATP