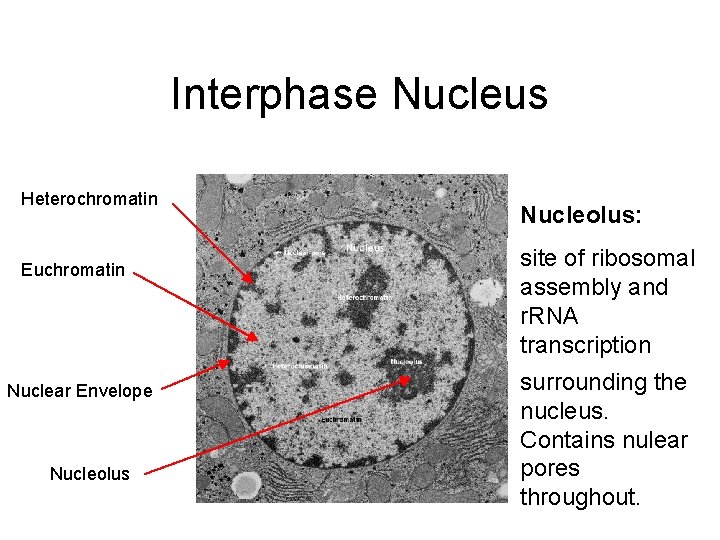
Unit 3 Interphase Nucleus Interphase Nucleus Heterochromatin Euchromatin





- Slides: 17

Unit 3: Interphase Nucleus

Interphase Nucleus Heterochromatin Euchromatin Nuclear Envelope Nucleolus: Heterochromatin: Nuclear Euchromatin: site of ribosomal dark, condensed Envelope: lighter, assembly and DNA that is Contains a transcriptionally r. RNA transcriptionally double active DNA transcription inactive during membrane interphase. surrounding the nucleus. Contains nulear pores throughout.

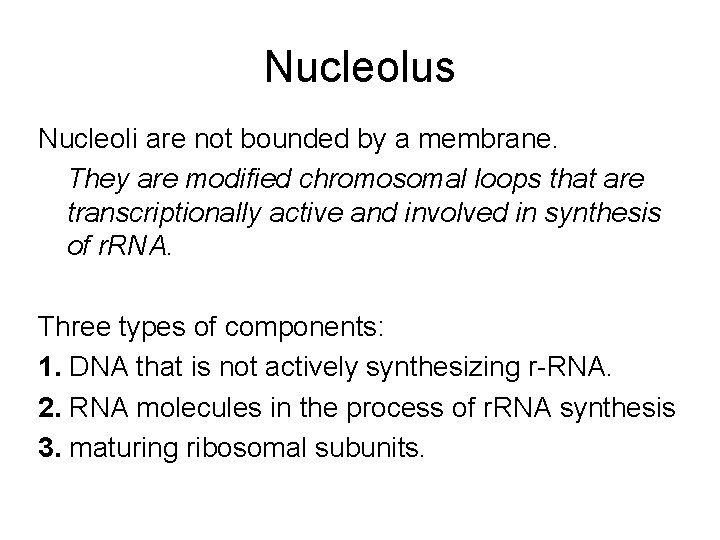
Nucleolus Nucleoli are not bounded by a membrane. They are modified chromosomal loops that are transcriptionally active and involved in synthesis of r. RNA. Three types of components: 1. DNA that is not actively synthesizing r-RNA. 2. RNA molecules in the process of r. RNA synthesis 3. maturing ribosomal subunits.

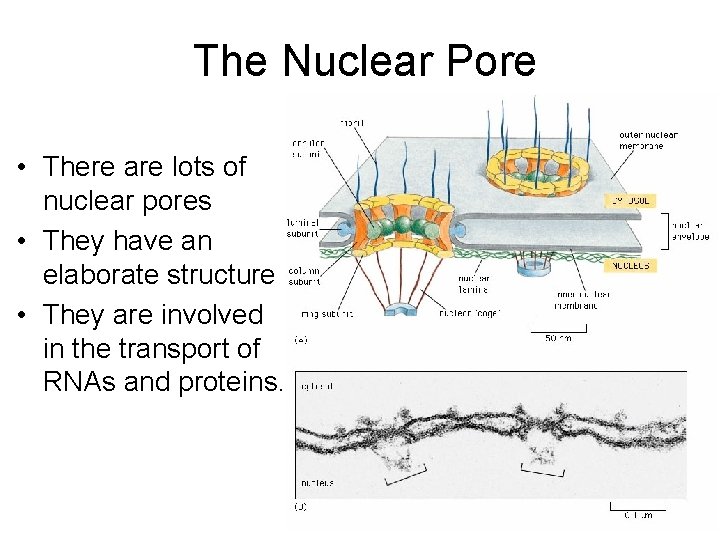
The Nuclear Pore • There are lots of nuclear pores • They have an elaborate structure • They are involved in the transport of RNAs and proteins.

What goes In/Out In: Histone molecules Polymerases and other enzymes required for replication and regulation of replication Ribosomal proteins and other proteins (including sn. RNPs) that complex with newly formed transcripts. Out: Ribosomal subunits m-RNA Protein complexes. Only fully processed and spliced transcripts are allowed out of the nucleus.

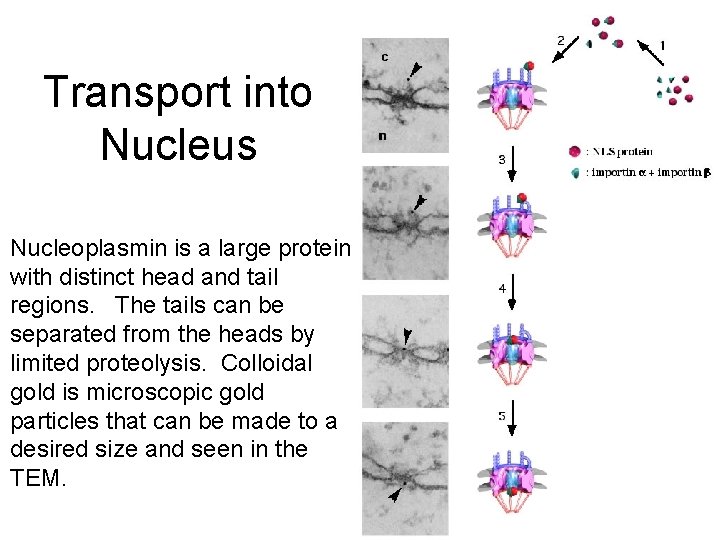
Transport into Nucleus Nucleoplasmin is a large protein with distinct head and tail regions. The tails can be separated from the heads by limited proteolysis. Colloidal gold is microscopic gold particles that can be made to a desired size and seen in the TEM.

Steps of Import: 1. NLS protein combines with nuclear import receptor 2. The complex binds to a fibrils attached to the cytosol surface of the nucelar pore complex. 3. The fibril then bends 4. The protein is transferred to the central component of the nuclear pore complex. 5. The central component of the nuclear pore complex undergoes a conformational change that results in the protein being transferred to the nuclear side of the pore complex.

CHROMATIN and CHROMOSOMES Experimental studies

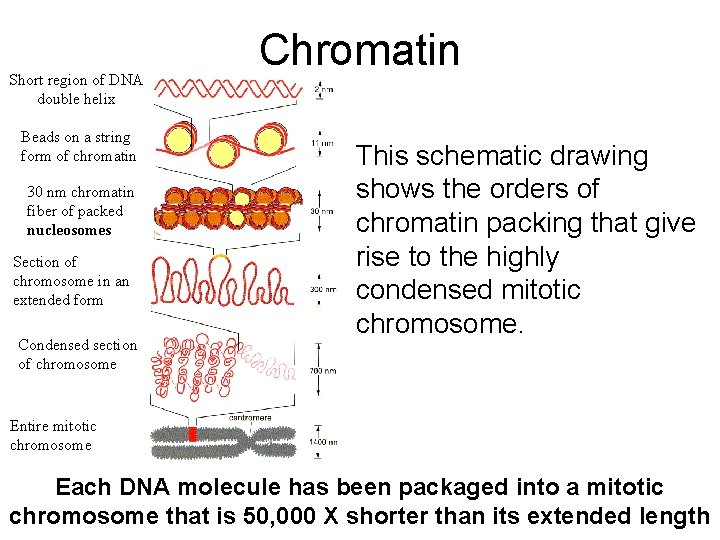
Short region of DNA double helix Beads on a string form of chromatin 30 nm chromatin fiber of packed nucleosomes Section of chromosome in an extended form Condensed section of chromosome Chromatin This schematic drawing shows the orders of chromatin packing that give rise to the highly condensed mitotic chromosome. Entire mitotic chromosome Each DNA molecule has been packaged into a mitotic chromosome that is 50, 000 X shorter than its extended length

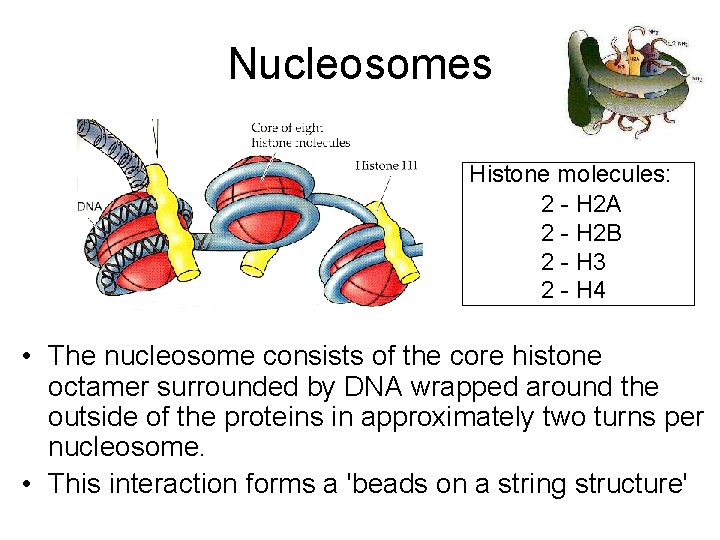
Nucleosomes Histone molecules: 2 - H 2 A 2 - H 2 B 2 - H 3 2 - H 4 • The nucleosome consists of the core histone octamer surrounded by DNA wrapped around the outside of the proteins in approximately two turns per nucleosome. • This interaction forms a 'beads on a string structure'

What are Nucleases? • Enzymes that cleave nucleic acids (RNA or DNA). • Nucleases can be very specific, cutting the DNA or RNA between specific base pairs (targeting a specific nucleotide sequence). • A mixture of different nucleases can be used to create random cuts.

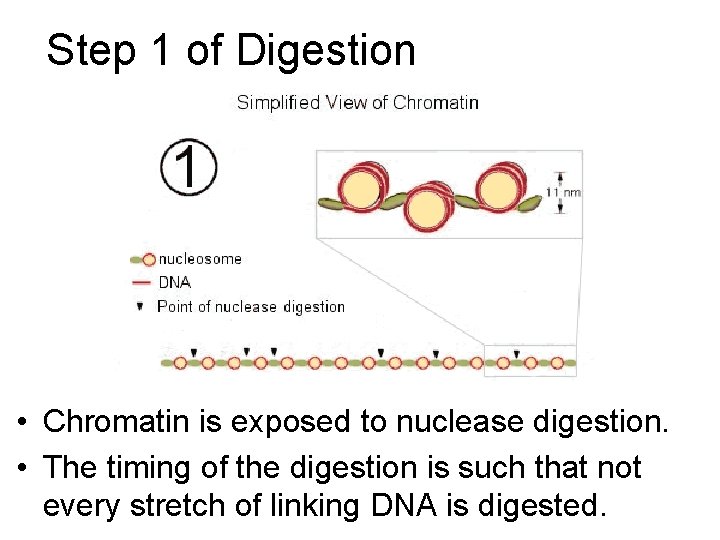
Step 1 of Digestion • Chromatin is exposed to nuclease digestion. • The timing of the digestion is such that not every stretch of linking DNA is digested.

Step 2 • The incomplete digestion of chromatin results in fragments of varying length, but in multiples of the length of one DNA/histone “bead on a string”. • For example, if 100 bp was involved in one “bead on a string”, this digestion will produce fragments of roughly 100, 200, 300 bp etc.

Step 3 • Protein is removed to leave sections of naked DNA of varying length.

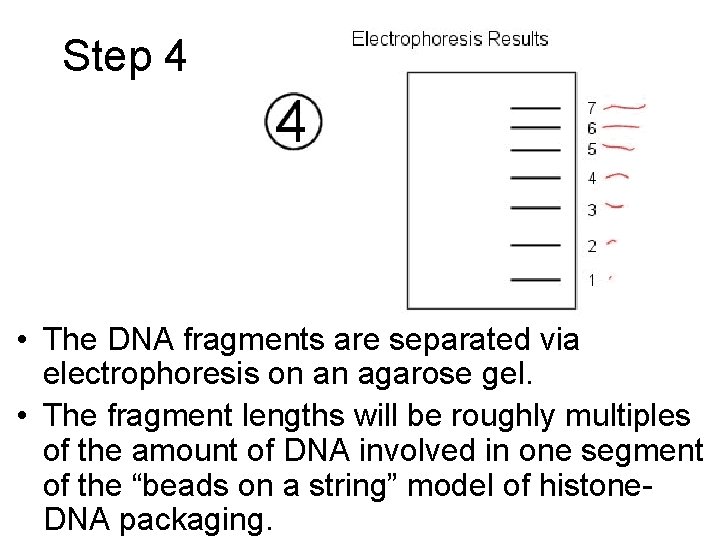
Step 4 • The DNA fragments are separated via electrophoresis on an agarose gel. • The fragment lengths will be roughly multiples of the amount of DNA involved in one segment of the “beads on a string” model of histone. DNA packaging.

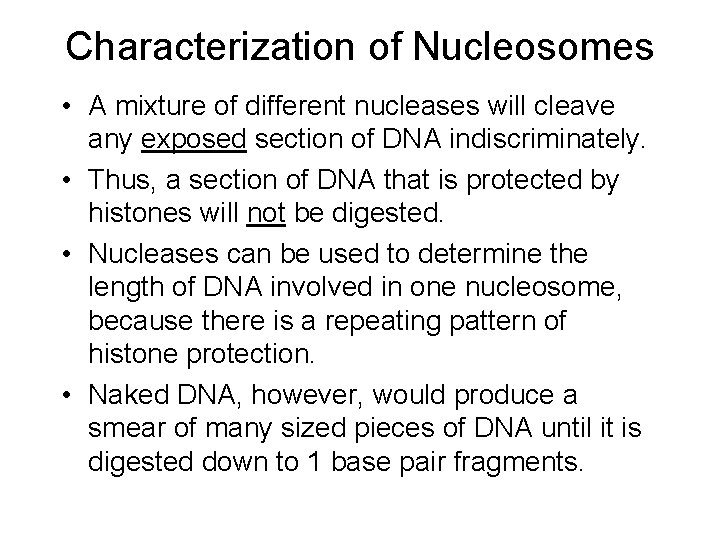
Characterization of Nucleosomes • A mixture of different nucleases will cleave any exposed section of DNA indiscriminately. • Thus, a section of DNA that is protected by histones will not be digested. • Nucleases can be used to determine the length of DNA involved in one nucleosome, because there is a repeating pattern of histone protection. • Naked DNA, however, would produce a smear of many sized pieces of DNA until it is digested down to 1 base pair fragments.

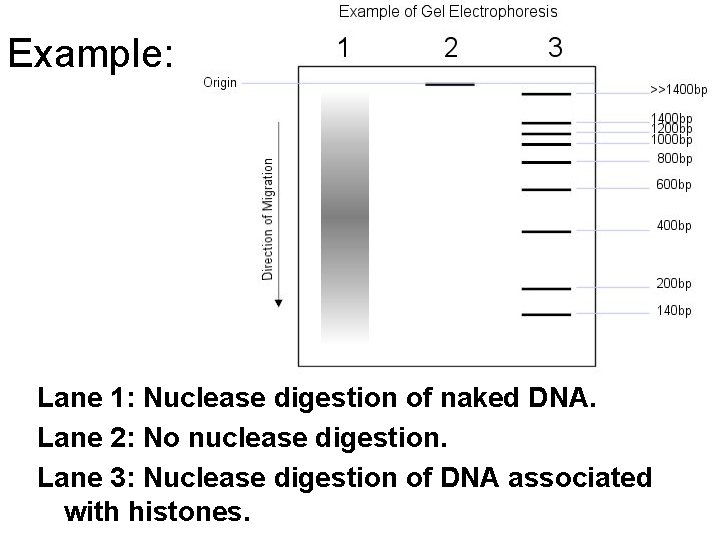
Example: Lane 1: Nuclease digestion of naked DNA. Lane 2: No nuclease digestion. Lane 3: Nuclease digestion of DNA associated with histones.