A Proteomics Toolkit Uni Prot Inter Pro and

















- Slides: 150

A Proteomics Toolkit: Uni. Prot, Inter. Pro and Int. Act Databases at the EBI

Hinxton, U. K.

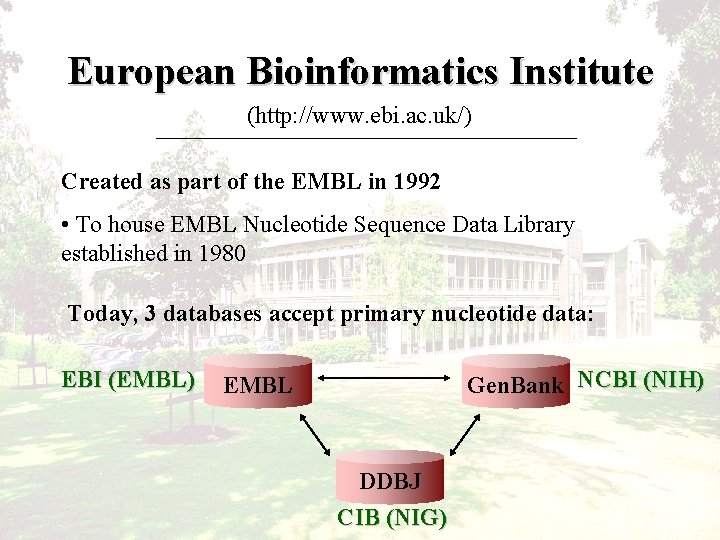
European Bioinformatics Institute (http: //www. ebi. ac. uk/) Created as part of the EMBL in 1992 • To house EMBL Nucleotide Sequence Data Library established in 1980 Today, 3 databases accept primary nucleotide data: EBI (EMBL) Gen. Bank NCBI (NIH) EMBL DDBJ CIB (NIG)

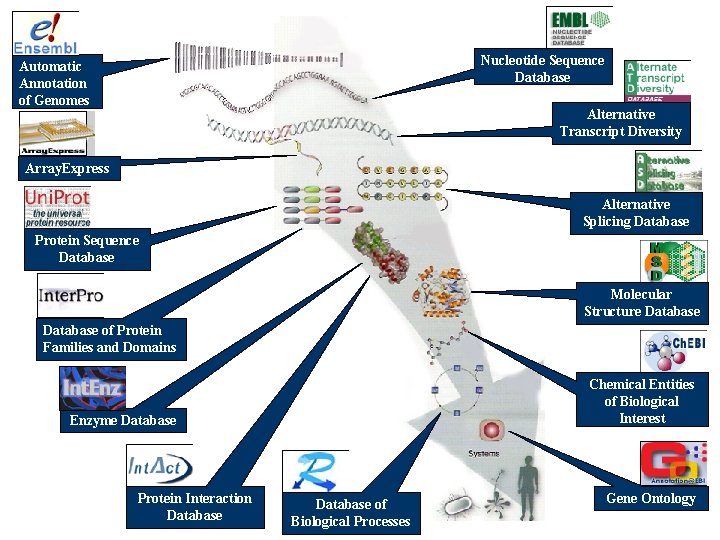
European Bioinformatics Institute (http: //www. ebi. ac. uk/) EMBL-EBI maintains the world’s most comprehensive range of molecular databases

Nucleotide Sequence Database Automatic Annotation of Genomes Alternative Transcript Diversity Array. Express Alternative Splicing Database Protein Sequence Database Molecular Structure Database of Protein Families and Domains Chemical Entities of Biological Interest Enzyme Database Protein Interaction Database of Biological Processes Gene Ontology

http: //www. ebi. ac. uk/services/

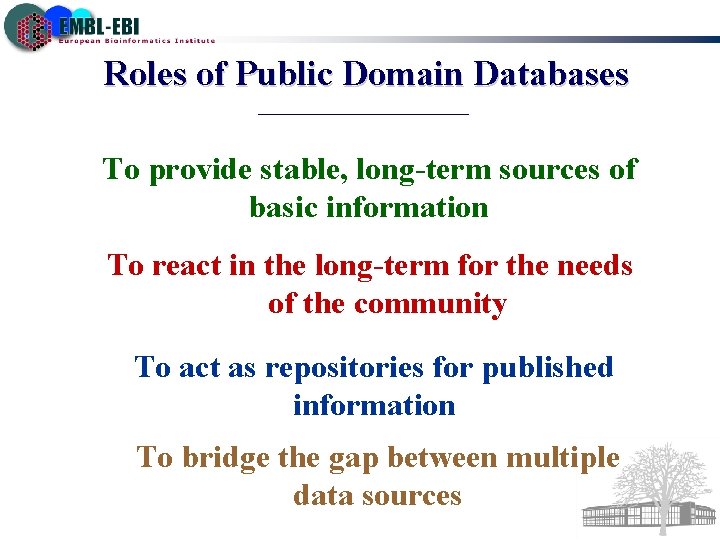
Roles of Public Domain Databases To provide stable, long-term sources of basic information To react in the long-term for the needs of the community To act as repositories for published information To bridge the gap between multiple data sources

Protein Databases Uni. Prot Database of Protein Sequences Inter. Pro Database of Protein Families and Domains Int. Act Database of Protein Interactions

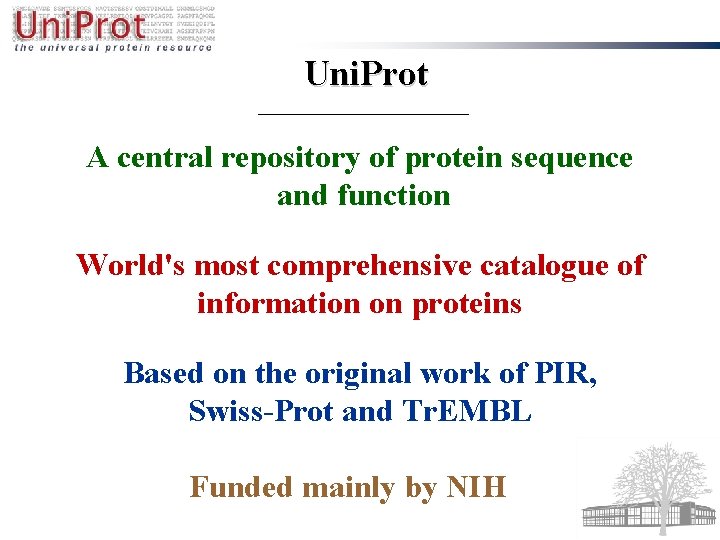
Uni. Prot A central repository of protein sequence and function World's most comprehensive catalogue of information on proteins Based on the original work of PIR, Swiss-Prot and Tr. EMBL Funded mainly by NIH

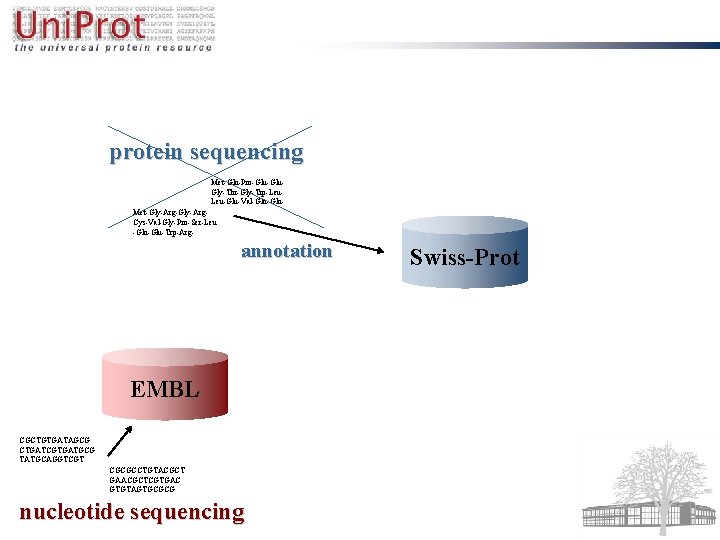
protein sequencing Met-Gln-Pro-Glu. Gly-Thr-Gly-Trp-Leu. Leu-Glu-Val-Gln. Met-Gly-Arg. Cys-Val-Gly-Pro-Ser-Leu -Gln-Glu-Trp-Arg- annotation EMBL CGCTGTGATAGCG CTGATCGTGATGCG TATGCAGGTCGT CGCGCCTGTACGCT GAACGCTCGTGAC GTGTAGTGCGCG nucleotide sequencing Swiss-Prot

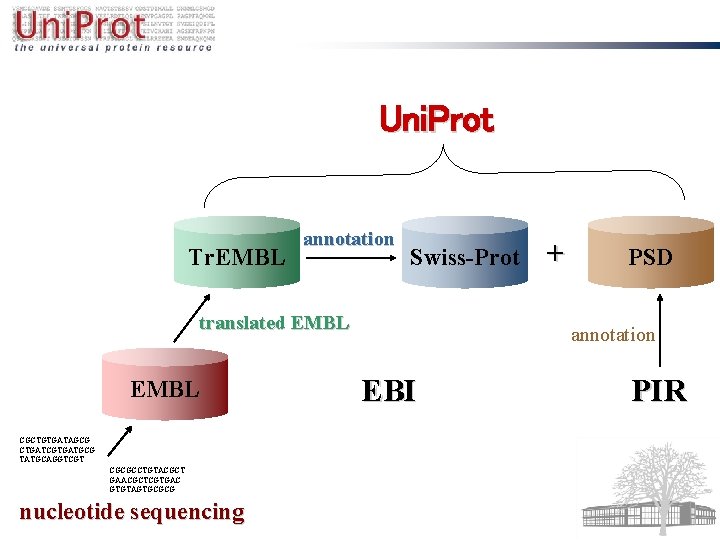
Uni. Prot Tr. EMBL annotation Swiss-Prot translated EMBL CGCTGTGATAGCG CTGATCGTGATGCG TATGCAGGTCGT CGCGCCTGTACGCT GAACGCTCGTGAC GTGTAGTGCGCG nucleotide sequencing + PSD annotation EBI PIR

Uni. Prot Consortium

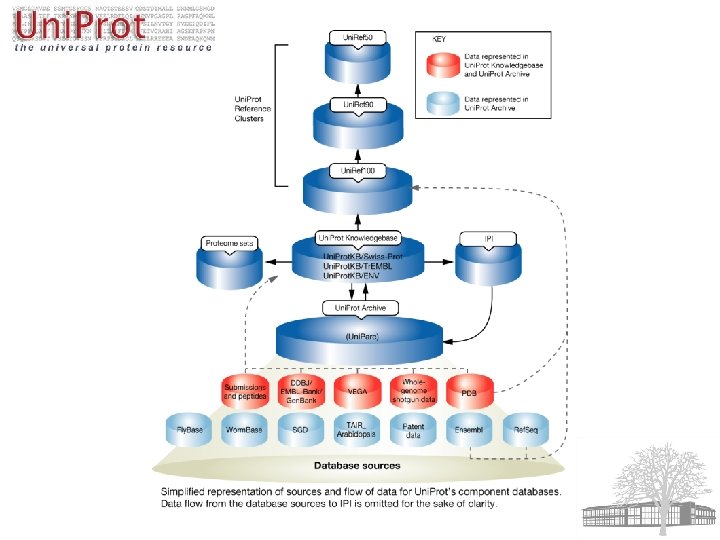
Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) Ø Uni. Prot Reference Clusters (Uni. Ref) Ø Uni. Prot Archive (Uni. Parc)

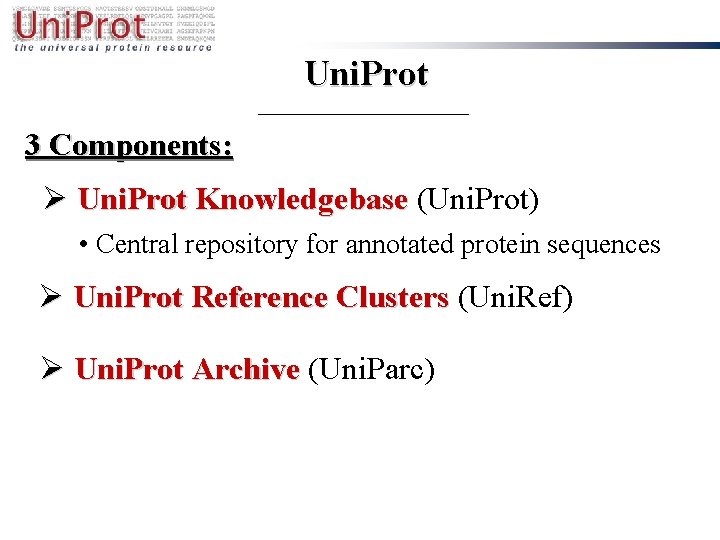
Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) • Central repository for annotated protein sequences Ø Uni. Prot Reference Clusters (Uni. Ref) Ø Uni. Prot Archive (Uni. Parc)

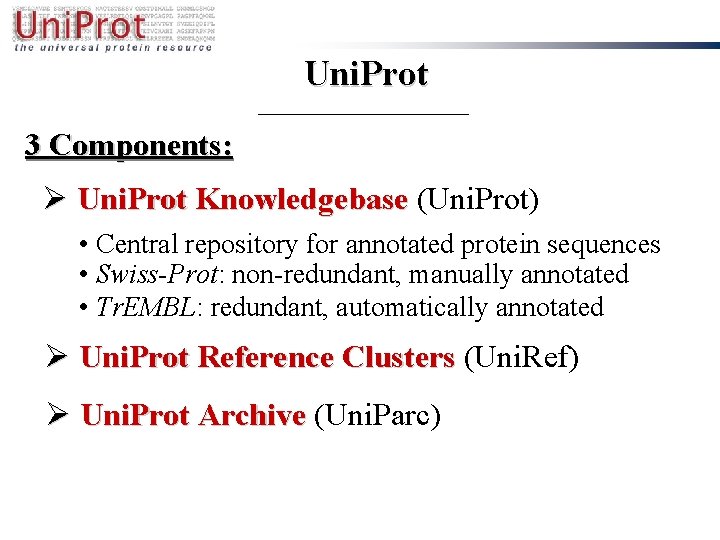
Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) • Central repository for annotated protein sequences • Swiss-Prot: non-redundant, manually annotated • Tr. EMBL: redundant, automatically annotated Ø Uni. Prot Reference Clusters (Uni. Ref) Ø Uni. Prot Archive (Uni. Parc)

Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) • Central repository for annotated protein sequences • Swiss-Prot: non-redundant, manually annotated • Tr. EMBL: redundant, automatically annotated Ø Uni. Prot Reference Clusters (Uni. Ref) • Combines related sequences for speed searching Ø Uni. Prot Archive (Uni. Parc)

Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) • Central repository for annotated protein sequences • Swiss-Prot: non-redundant, manually annotated • Tr. EMBL: redundant, automatically annotated Ø Uni. Prot Reference Clusters (Uni. Ref) • Combines related sequences for speed searching • Uni. Ref 100, Uni. Ref 90, Uni. Ref 50 Ø Uni. Prot Archive (Uni. Parc)

Uni. Prot 3 Components: Ø Uni. Prot Knowledgebase (Uni. Prot) • Central repository for annotated protein sequences • Swiss-Prot: non-redundant, manually annotated • Tr. EMBL: redundant, automatically annotated Ø Uni. Prot Reference Clusters (Uni. Ref) • Combines related sequences for speed searching • Uni. Ref 100, Uni. Ref 90, Uni. Ref 50 Ø Uni. Prot Archive (Uni. Parc) • Comprehensive repository for history of sequences


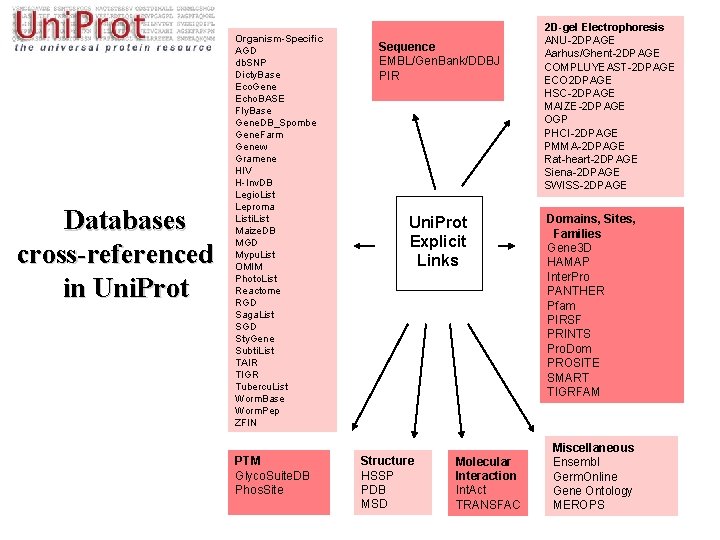
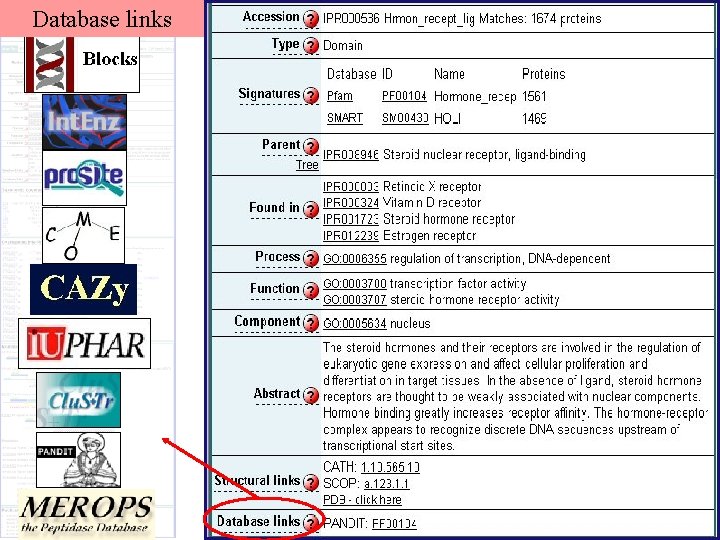
Databases cross-referenced in Uni. Prot Organism-Specific AGD db. SNP Dicty. Base Eco. Gene Echo. BASE Fly. Base Gene. DB_Spombe Gene. Farm Genew Gramene HIV H-Inv. DB Legio. List Leproma Listi. List Maize. DB MGD Mypu. List OMIM Photo. List Reactome RGD Saga. List SGD Sty. Gene Subti. List TAIR TIGR Tubercu. List Worm. Base Worm. Pep ZFIN PTM Glyco. Suite. DB Phos. Site Sequence EMBL/Gen. Bank/DDBJ PIR Uni. Prot Explicit Links Structure HSSP PDB MSD Molecular Interaction Int. Act TRANSFAC 2 D-gel Electrophoresis ANU-2 DPAGE Aarhus/Ghent-2 DPAGE COMPLUYEAST-2 DPAGE ECO 2 DPAGE HSC-2 DPAGE MAIZE-2 DPAGE OGP PHCI-2 DPAGE PMMA-2 DPAGE Rat-heart-2 DPAGE Siena-2 DPAGE SWISS-2 DPAGE Domains, Sites, Families Gene 3 D HAMAP Inter. Pro PANTHER Pfam PIRSF PRINTS Pro. Dom PROSITE SMART TIGRFAM Miscellaneous Ensembl Germ. Online Gene Ontology MEROPS

http: //www. ebi. ac. uk/services/

Searching Uni. Prot Search tools include: • Text Search • Power Search • Blast, Fasta and MPsrch • Links to extra search services (including SRS) http: //www. ebi. uniprot. org/index. shtml

http: //www. ebi. uniprot. org/index. shtml • Text-based searching • Logical operators ‘&’ (and), ‘|’ (or) • (Wildcards and numerical operators not allowed) • Text Search – keyword queries • Power Search – can search for specific entry lines • Warehouse Search – link query to other databases

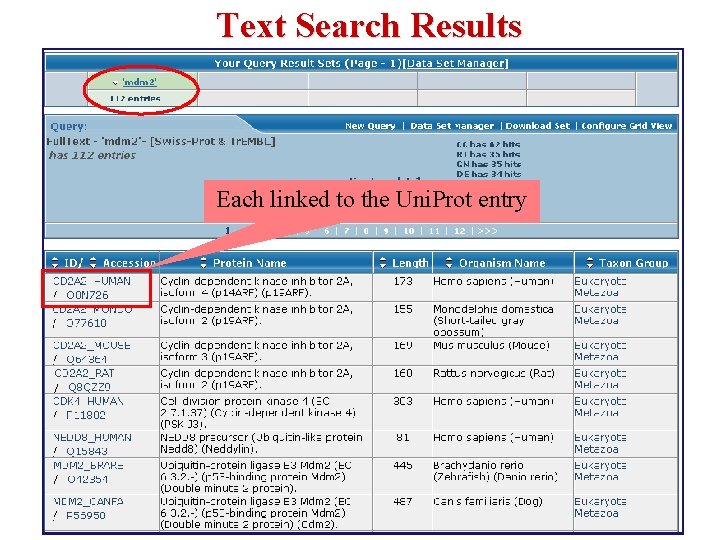
Text Search Results Each linked to the Uni. Prot entry

• Sequence-based searching • BLAST, Fasta, MPsrch

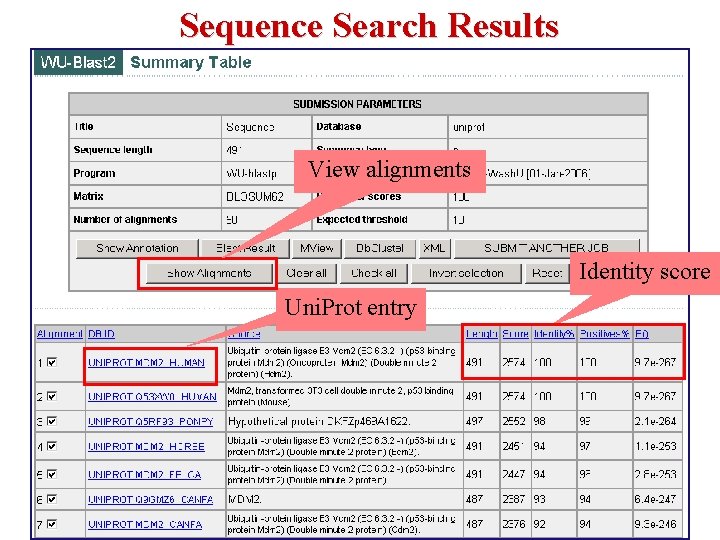
Sequence Search Results View alignments Identity score Uni. Prot entry

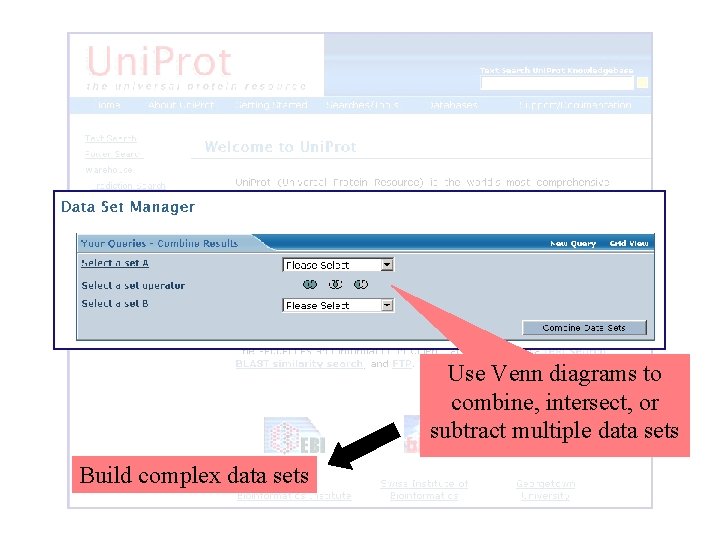
Manipulate multiple data sets

Use Venn diagrams to combine, intersect, or subtract multiple data sets Build complex data sets

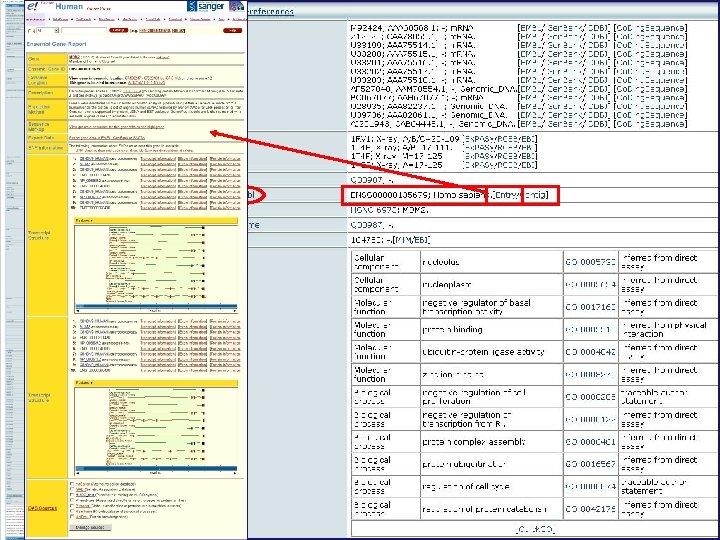
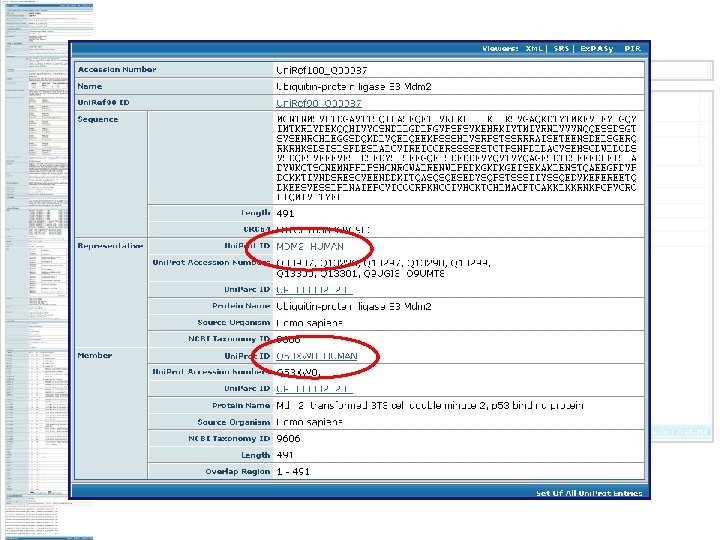
Uni. Prot/Swiss-Prot entry for human ubiquitin-protein ligase E 3 mdm 2

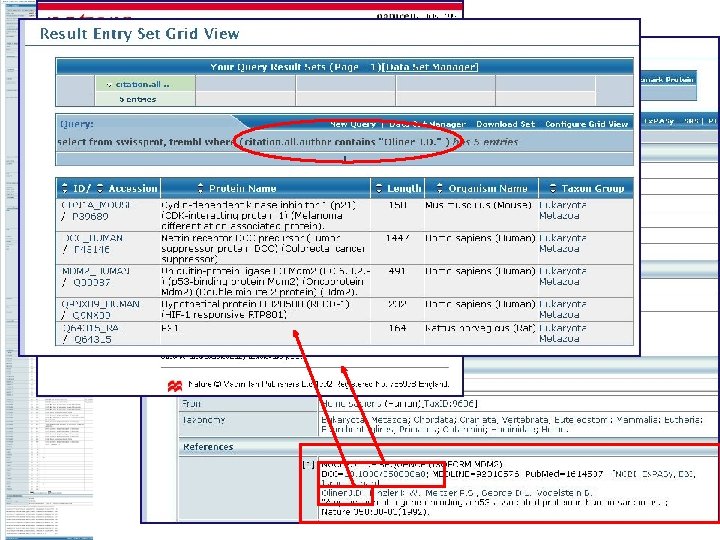
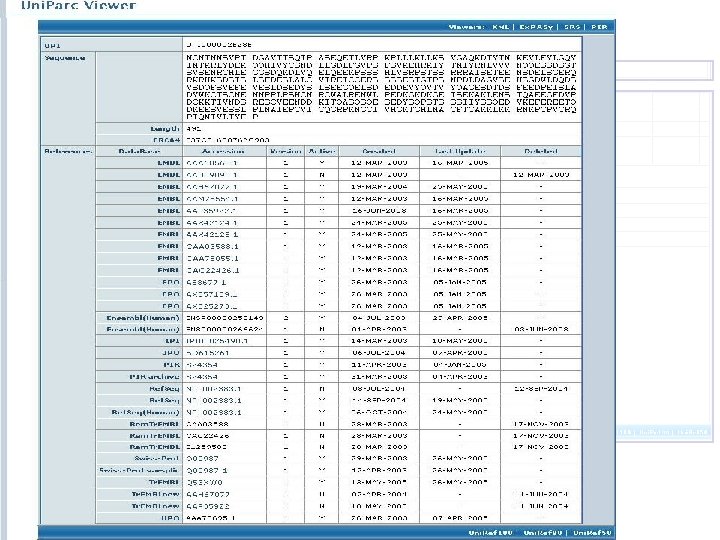
Merged entries: • Remove redundancy • Can still be searched Some literature search engines pull synonyms from Uni. Prot for more complete searching







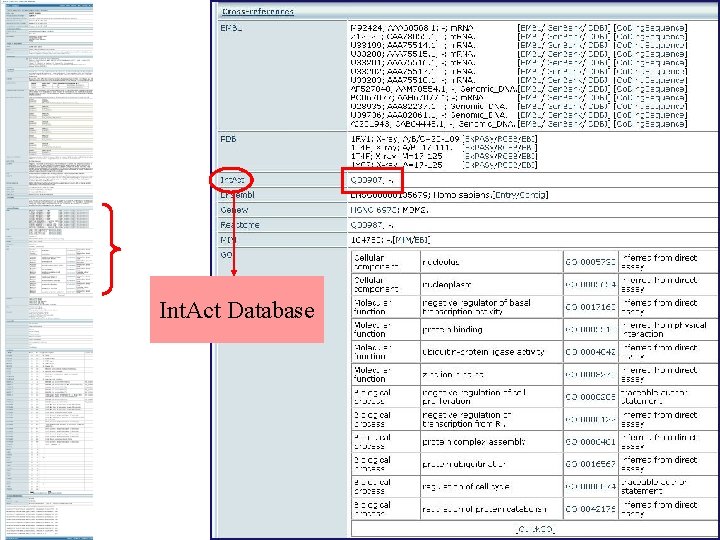
Int. Act Database


Summary of nucleotide data upon which entry is originally based Structural data associated with entry protein

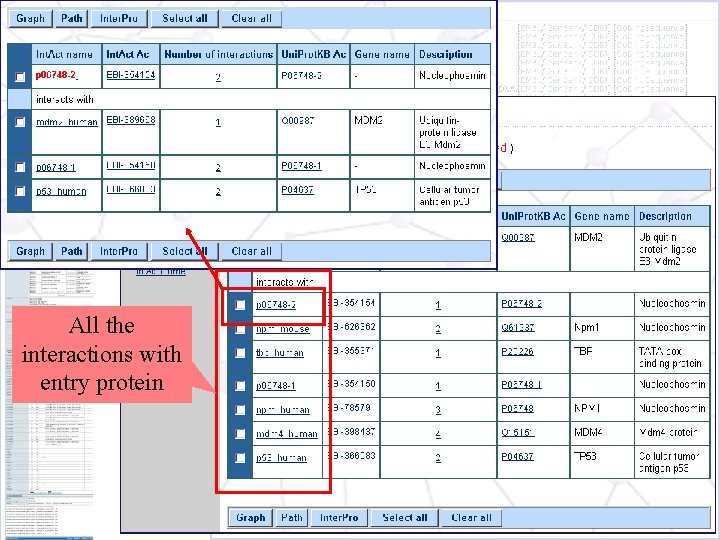
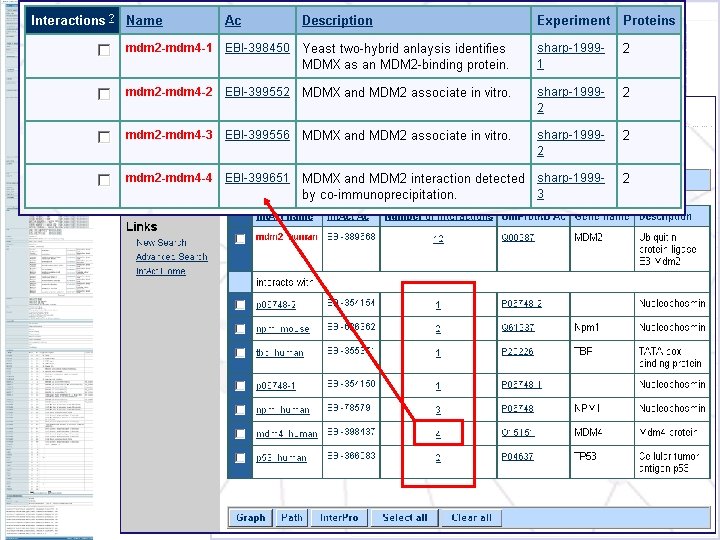
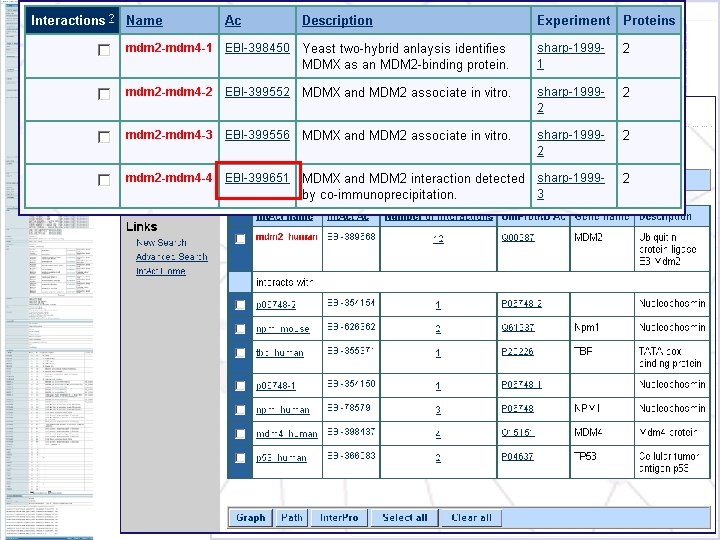
Int. Act Database

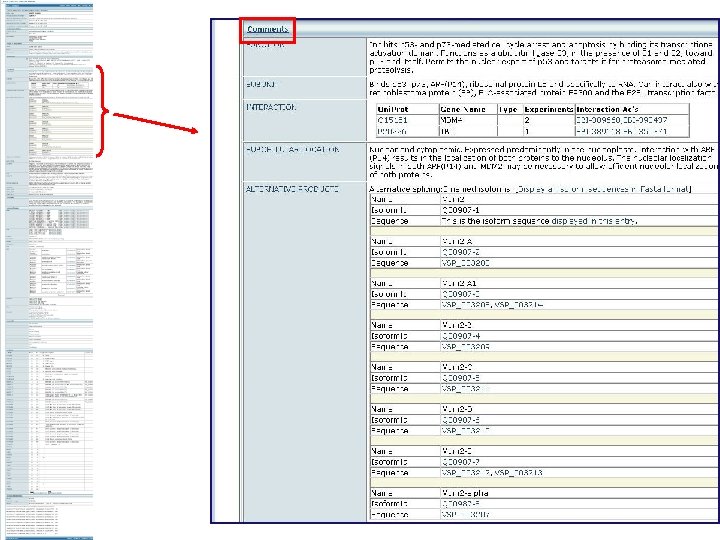
Int. Act Database All the interactions with entry protein

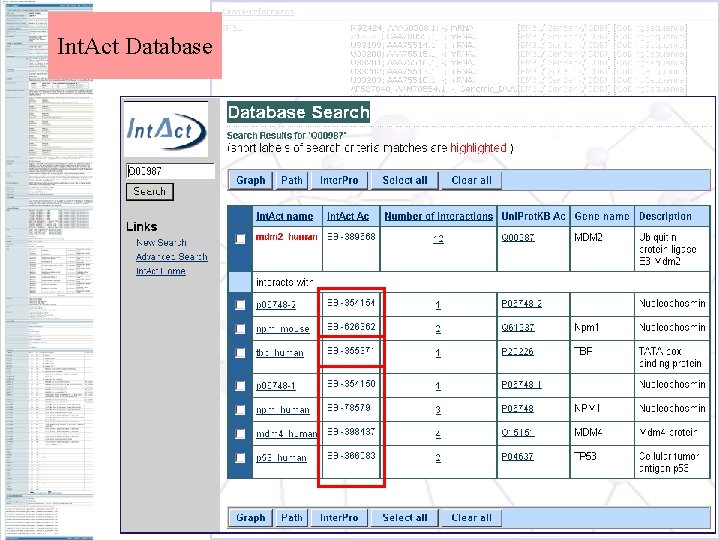
Int. Act Database


Int. Act Database

Int. Act Database

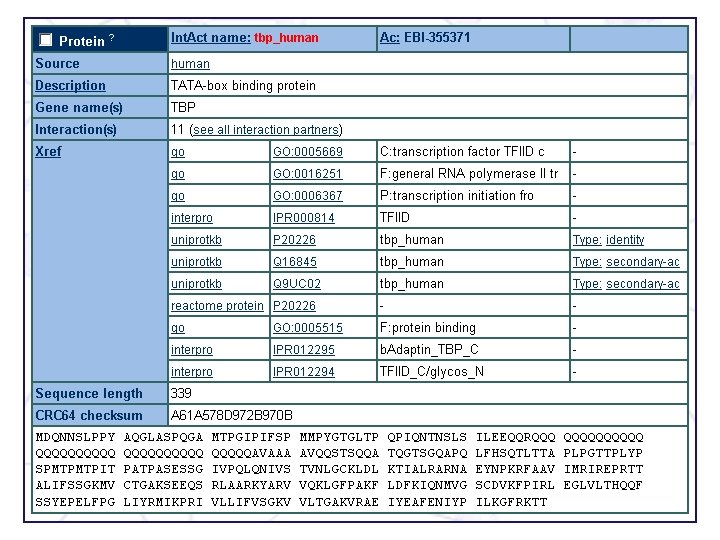
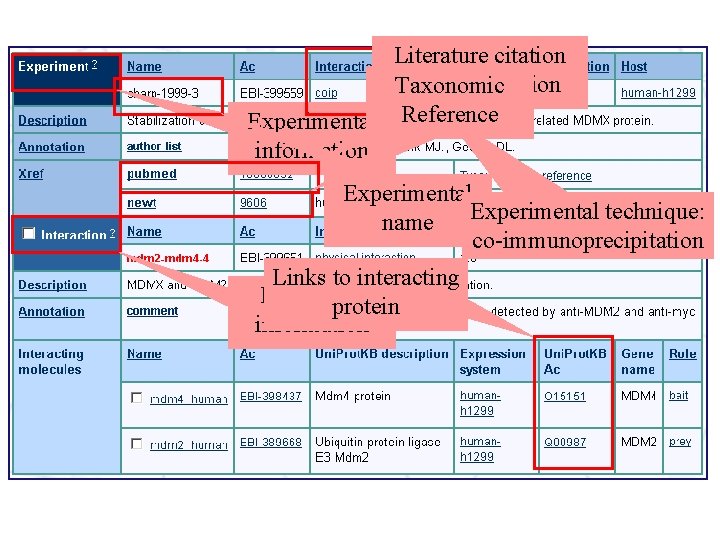
Literature citation used for curation Taxonomic Experimental Reference information Experimental technique: name co-immunoprecipitation Links to interacting Interaction protein information

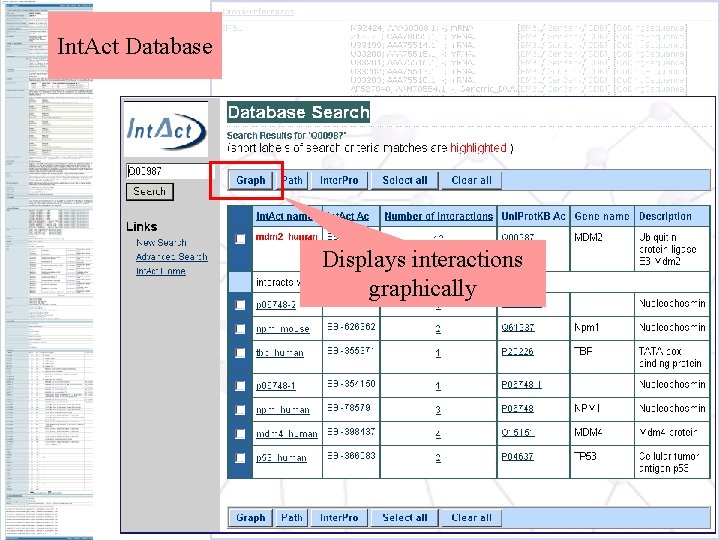
Int. Act Database Displays interactions graphically

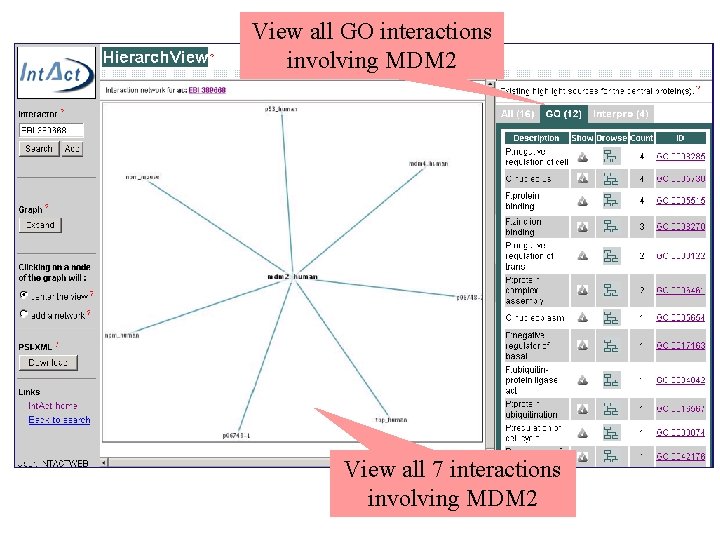
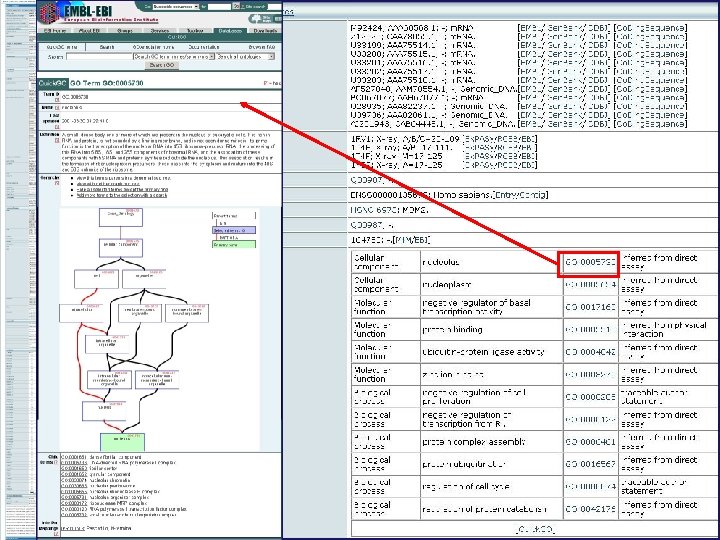
View all GO interactions involving MDM 2 View all 7 interactions involving MDM 2

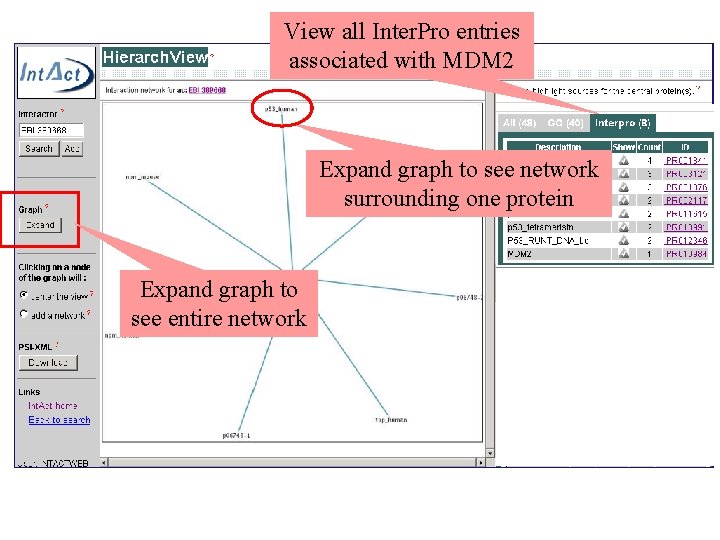
View all Inter. Pro entries associated with MDM 2 Expand graph to see network surrounding one protein Expand graph to see entire network

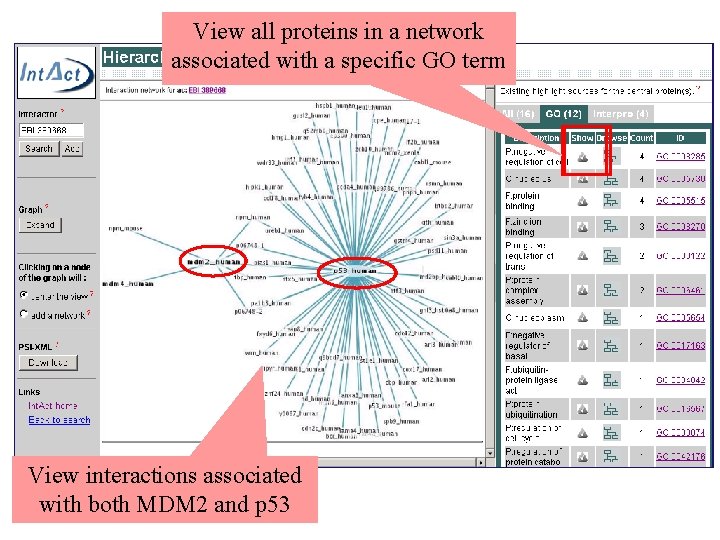
View all proteins in a network associated with a specific GO term View interactions associated with both MDM 2 and p 53

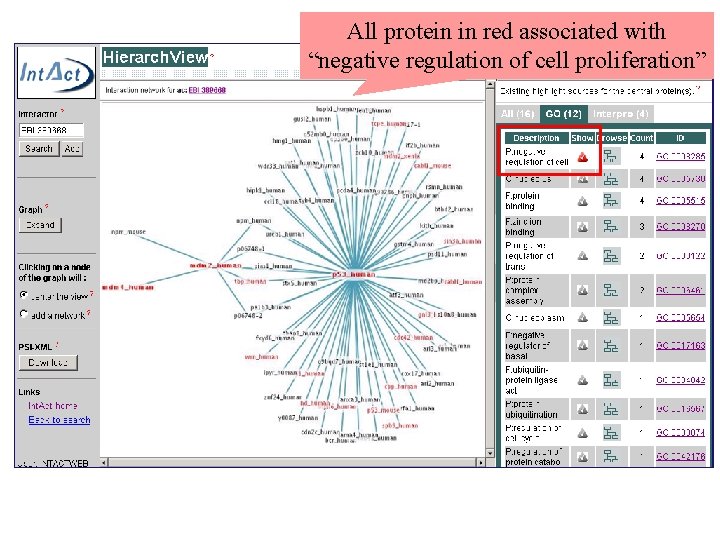
All protein in red associated with “negative regulation of cell proliferation”


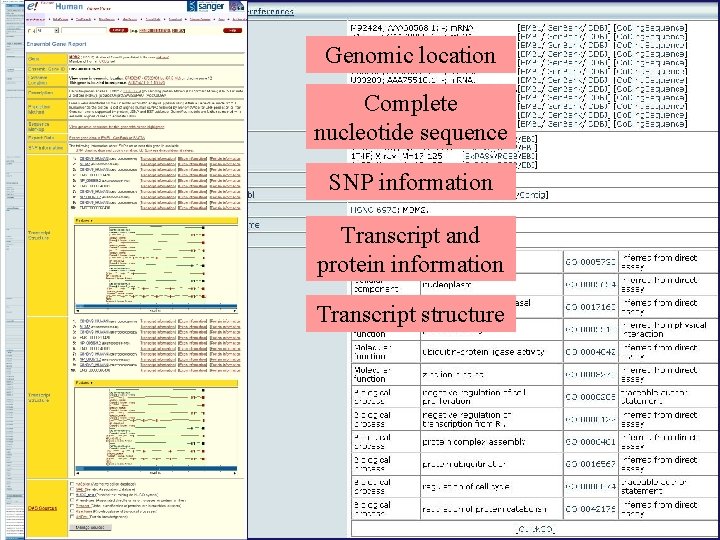
Genomic location Complete nucleotide sequence SNP information Transcript and protein information Transcript structure


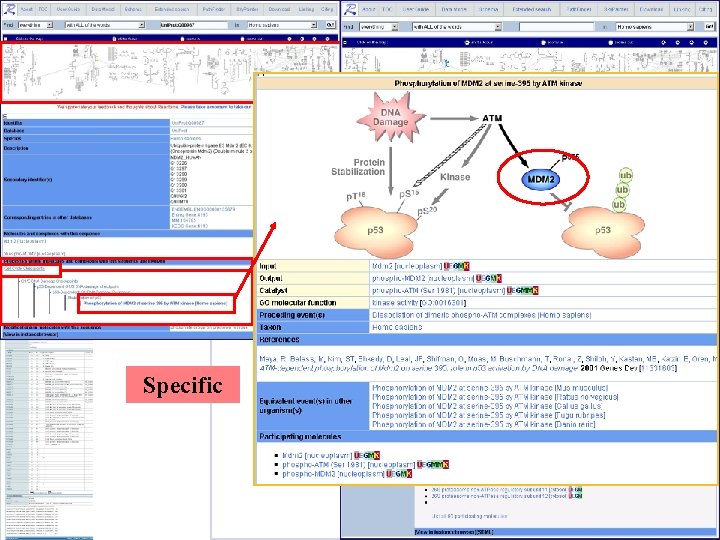
Interactive map. Can zoom in/out, and move around Specific General Summary and links to information about processes involving this molecule (here cell-cycle checkpoints)

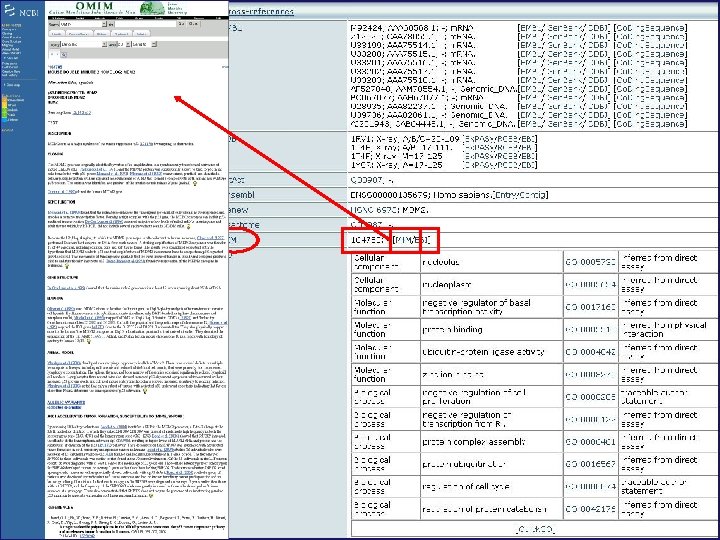
Mendelian Inheritance in Man

Cellular component Molecular function Biological process

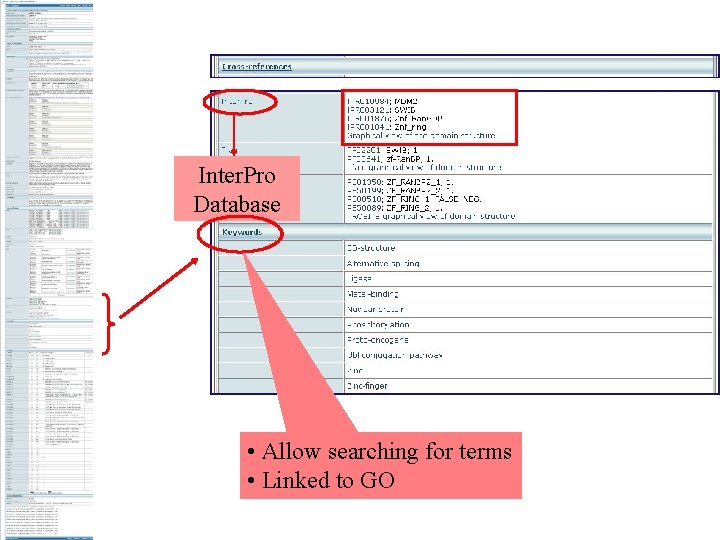
Inter. Pro Database • Allow searching for terms • Linked to GO

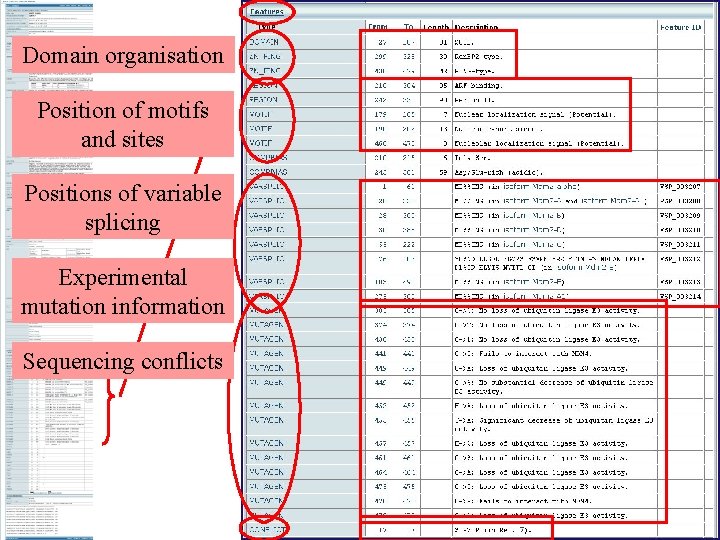
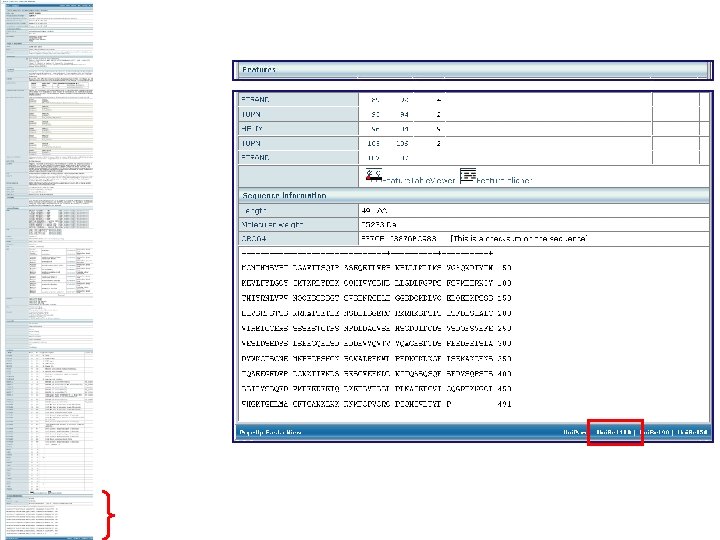
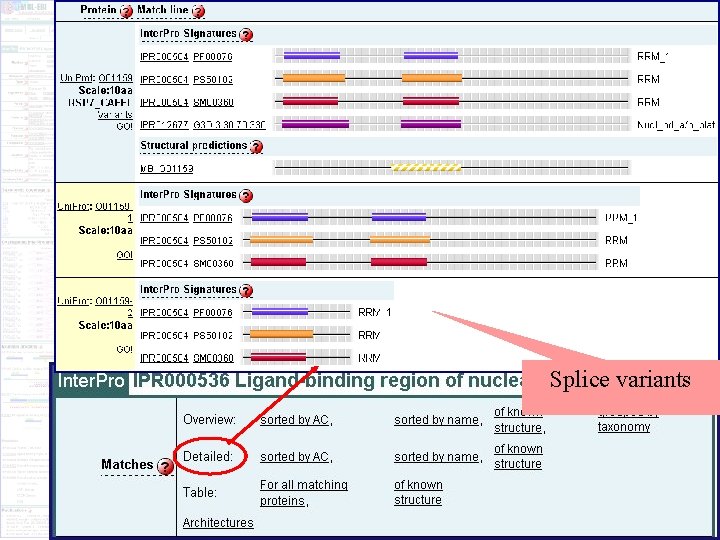
Domain organisation Position of motifs and sites Positions of variable splicing Experimental mutation information Sequencing conflicts

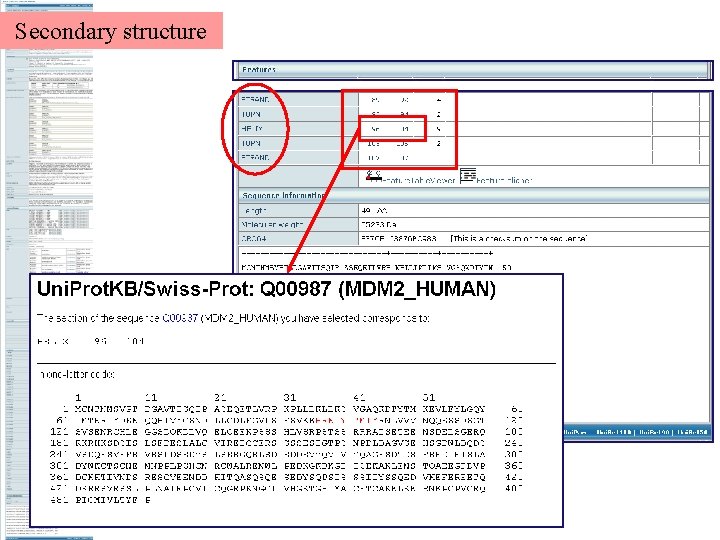
Secondary structure

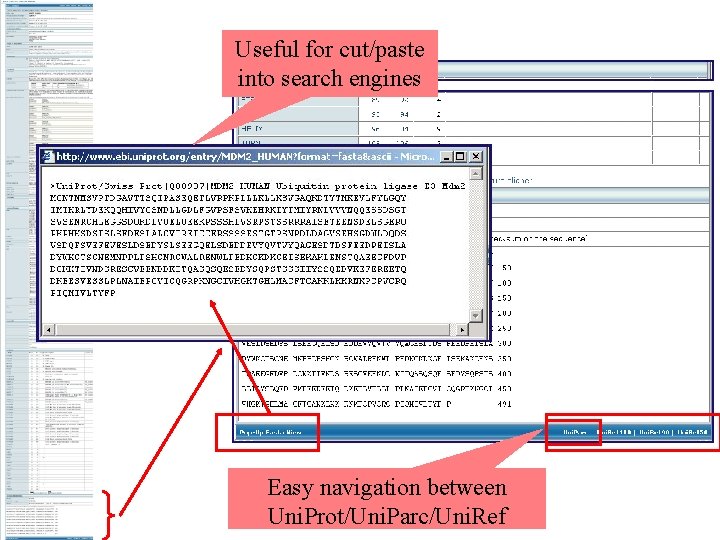
Useful for cut/paste into search engines Easy navigation between Uni. Prot/Uni. Parc/Uni. Ref




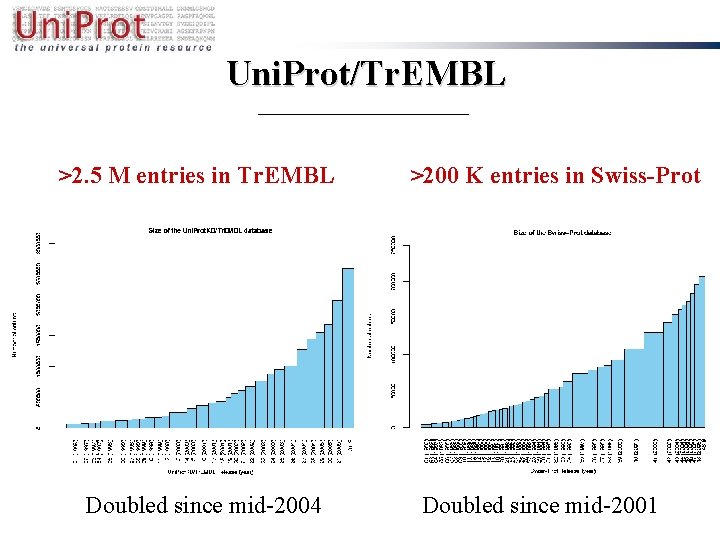
Uni. Prot/Tr. EMBL >2. 5 M entries in Tr. EMBL Doubled since mid-2004 >200 K entries in Swiss-Prot Doubled since mid-2001

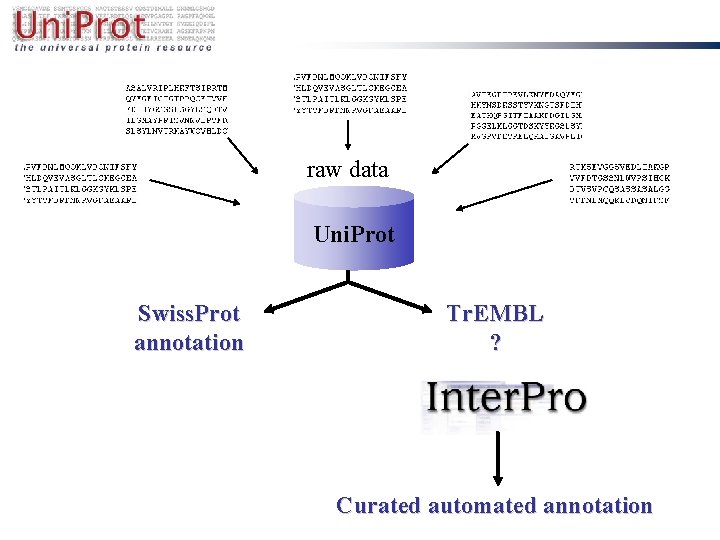
raw data Uni. Prot Swiss. Prot annotation Tr. EMBL ? Curated automated annotation

Uni. Prot/Tr. EMBL Redundancy Automatically maintained • Automatic clean-up of nucleotide data • Inter. Pro run and cross-references updated every 2 weeks • Automatic annotation Ø Recognises common annotation in related Swiss-Prot entries Ø Identifies all members of family using Inter. Pro

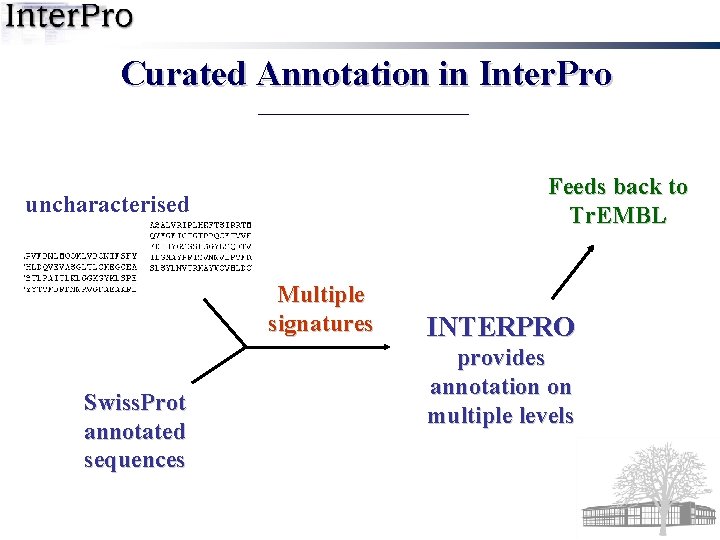
Curated Annotation in Inter. Pro Feeds back to Tr. EMBL uncharacterised Multiple signatures Swiss. Prot annotated sequences INTERPRO provides annotation on multiple levels

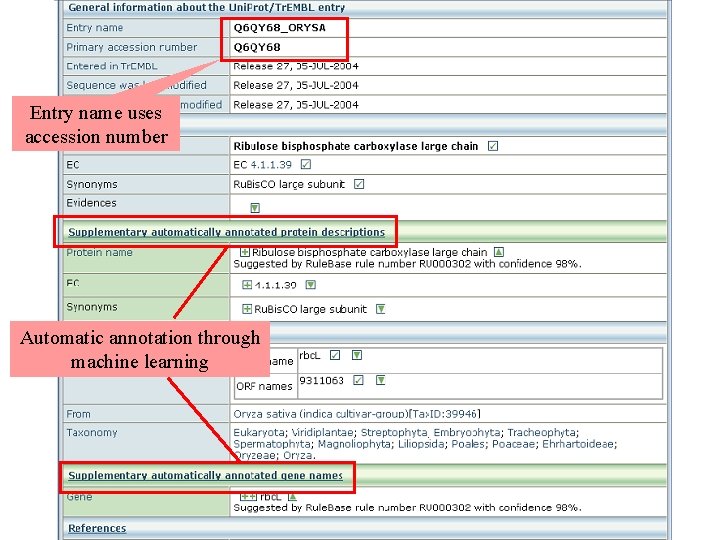
Entry name uses accession number Automatic annotation through machine learning

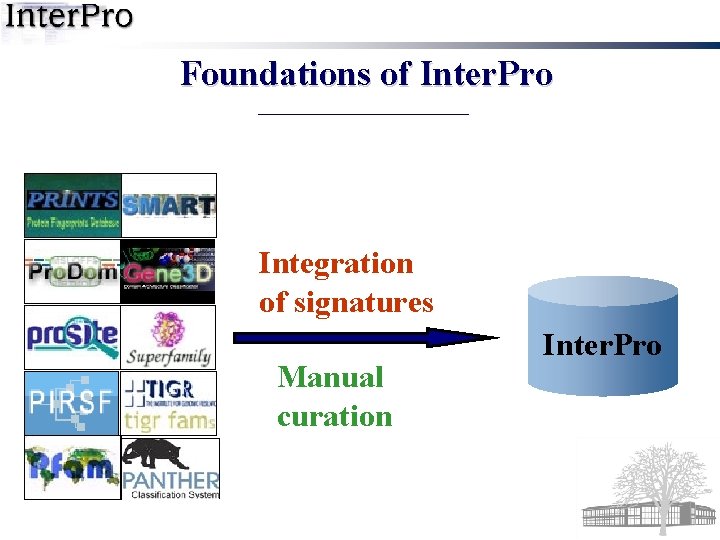
Foundations of Inter. Pro Integration of signatures Manual curation Inter. Pro

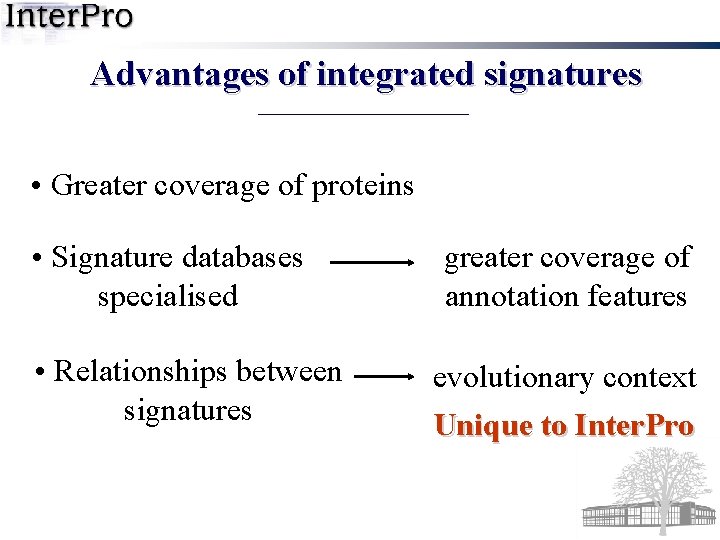
Advantages of integrated signatures • Greater coverage of proteins • Signature databases specialised • Relationships between signatures greater coverage of annotation features evolutionary context Unique to Inter. Pro

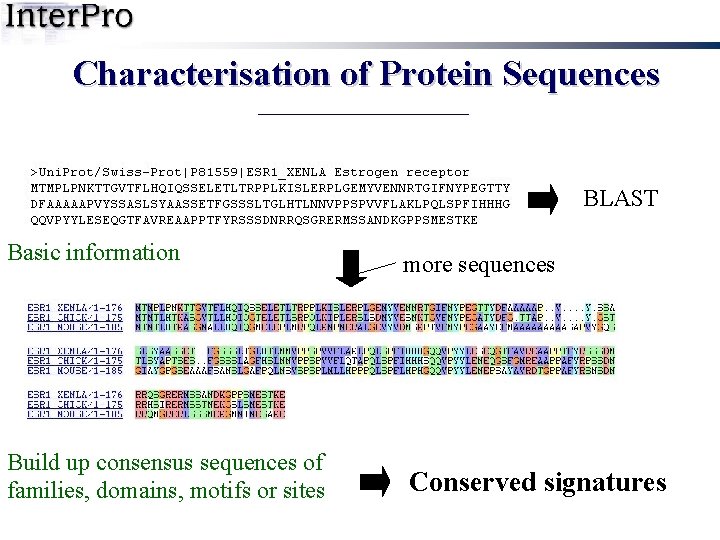
Characterisation of Protein Sequences BLAST Basic information more sequences Build up consensus sequences of families, domains, motifs or sites Conserved signatures

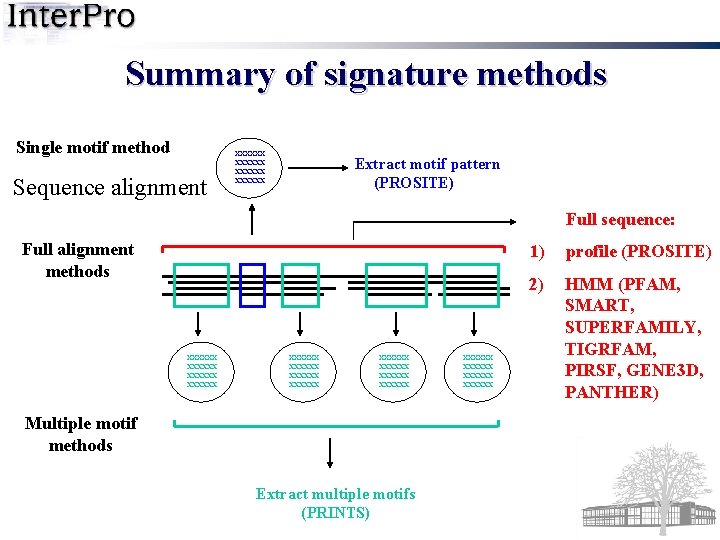
Finding Conserved Signatures • Pattern • Fingerprint Simplest (limited) • Sequence clustering • Profile • HMM More information

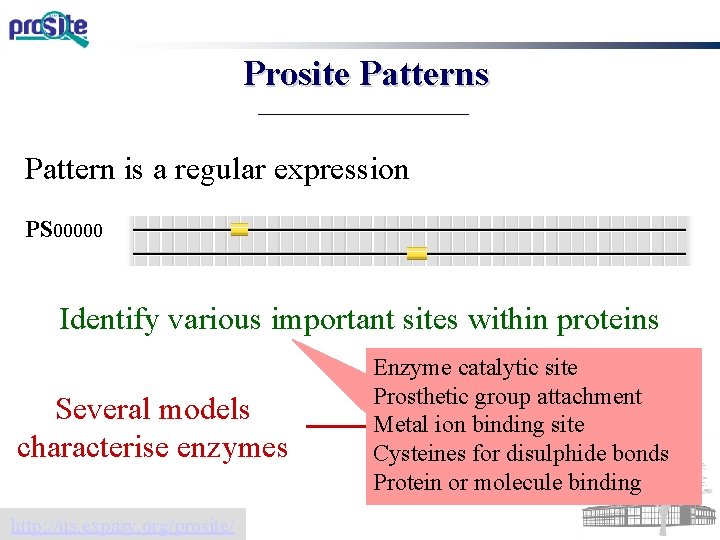
Patterns in sequence regular expressions Often used to define important sites within proteins PROSITE best-known pattern database

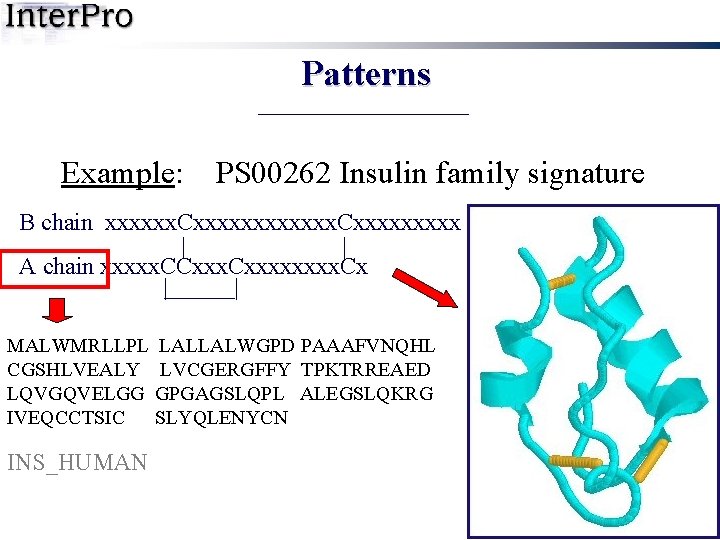
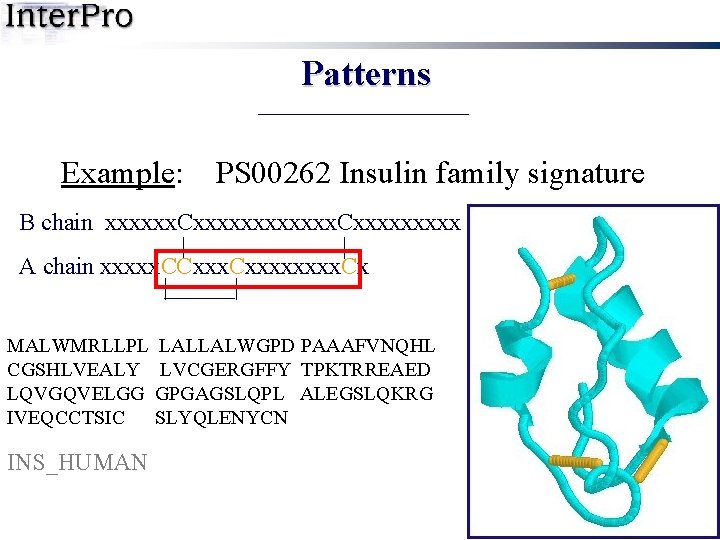
Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQCCTSIC SLYQLENYCN INS_HUMAN

Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQCCTSIC SLYQLENYCN INS_HUMAN

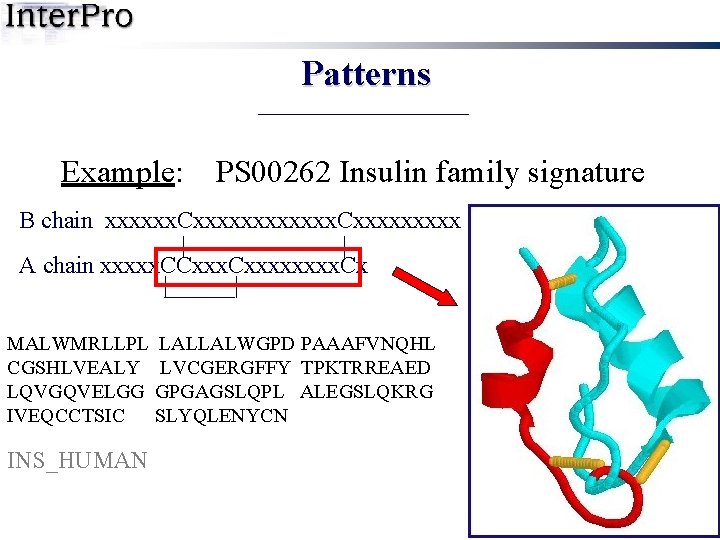
Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQCCTSIC SLYQLENYCN INS_HUMAN

Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQCCTSIC SLYQLENYCN INS_HUMAN

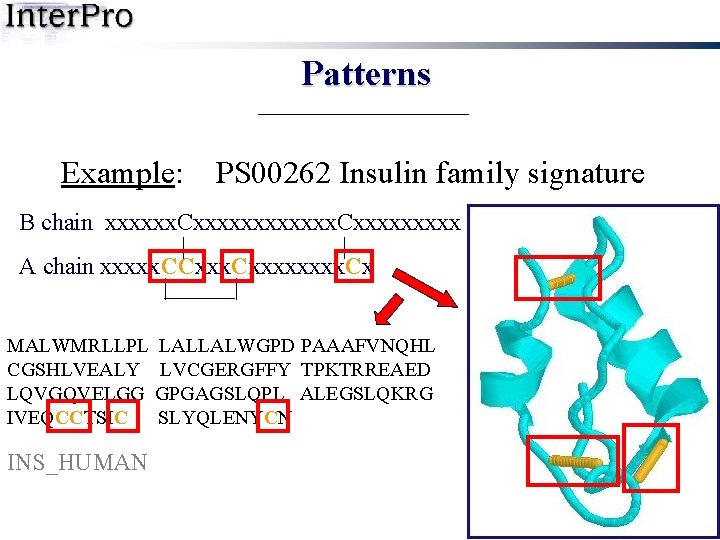
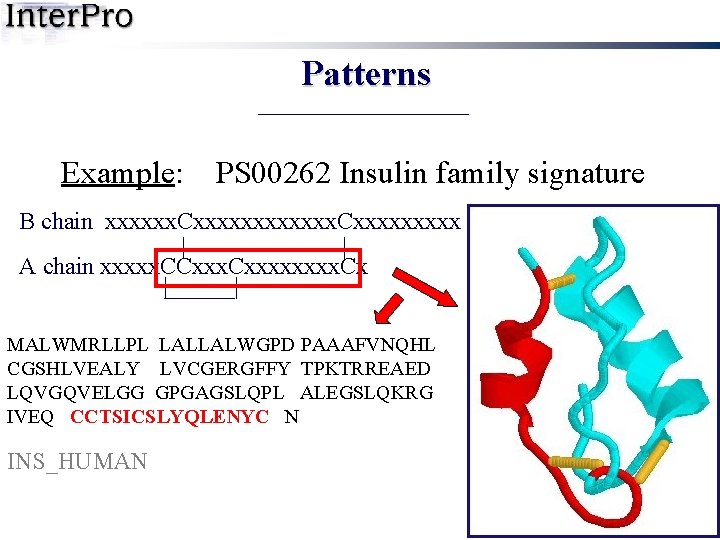
Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQ CCTSICSLYQLENYC N INS_HUMAN

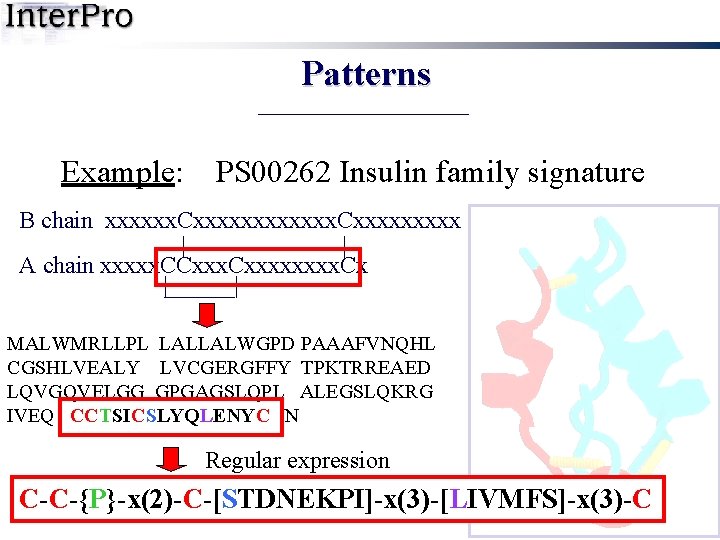
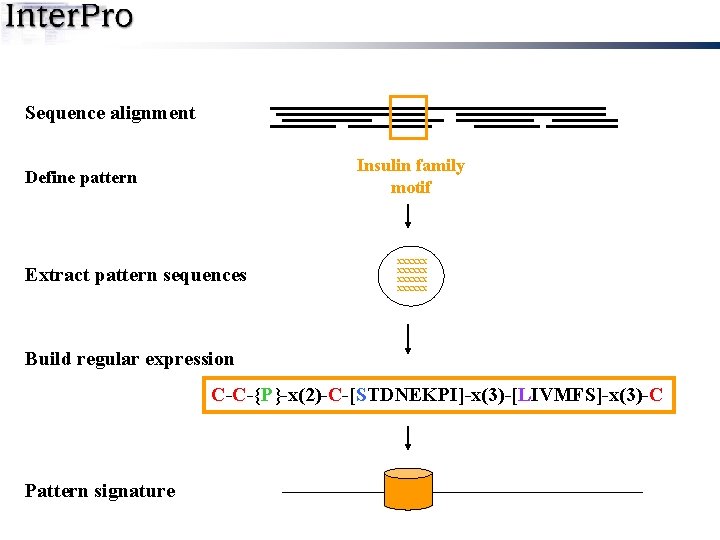
Patterns Example: PS 00262 Insulin family signature B chain xxxxxx. Cxxxxxxxxx | | A chain xxxxx. CCxxxx. Cx | | MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQ CCTSICSLYQLENYC N Regular expression C-C-{P}-x(2)-C-[STDNEKPI]-x(3)-[LIVMFS]-x(3)-C

Sequence alignment Insulin family motif Define pattern Extract pattern sequences xxxxxx Build regular expression C-C-{P}-x(2)-C-[STDNEKPI]-x(3)-[LIVMFS]-x(3)-C Pattern signature

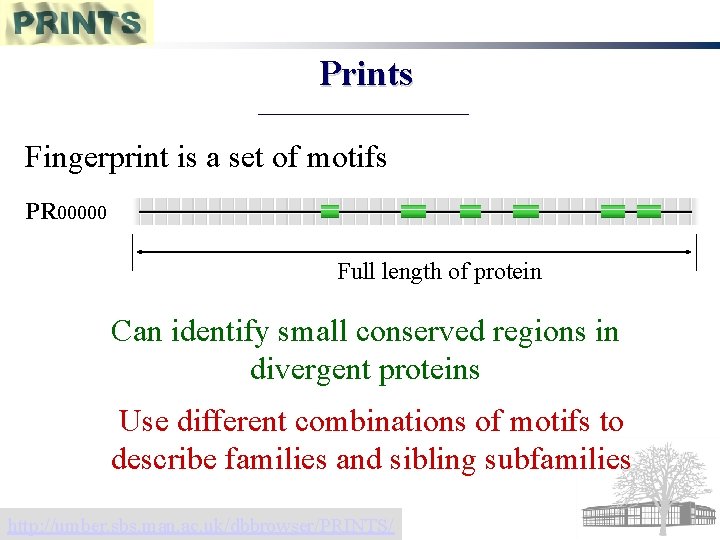
Fingerprints Several discrete motifs characterise family Highly specific matches to small regions of proteins PRINTS best-known fingerprint database

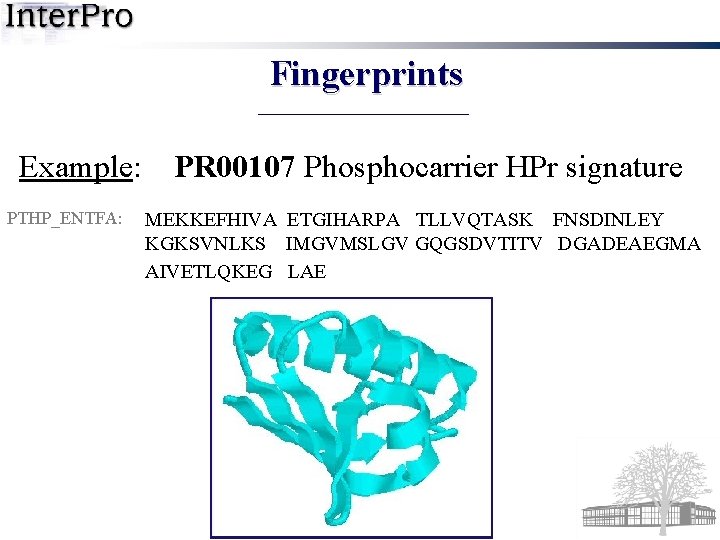
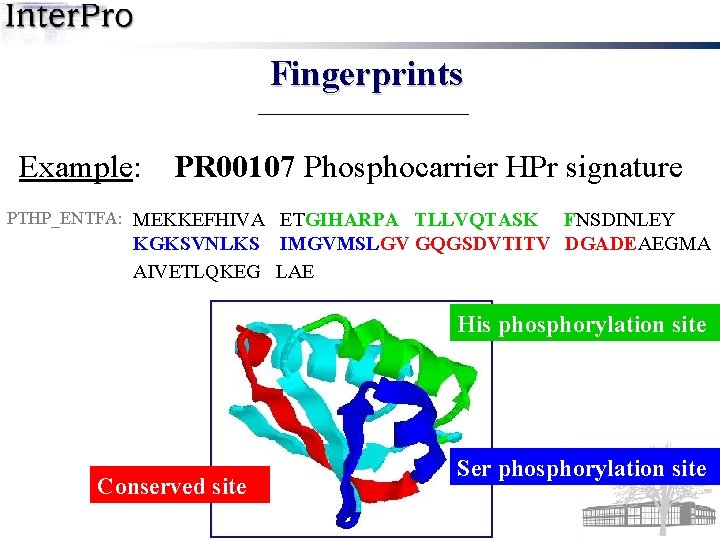
Fingerprints Example: PTHP_ENTFA: PR 00107 Phosphocarrier HPr signature MEKKEFHIVA ETGIHARPA TLLVQTASK FNSDINLEY KGKSVNLKS IMGVMSLGV GQGSDVTITV DGADEAEGMA AIVETLQKEG LAE

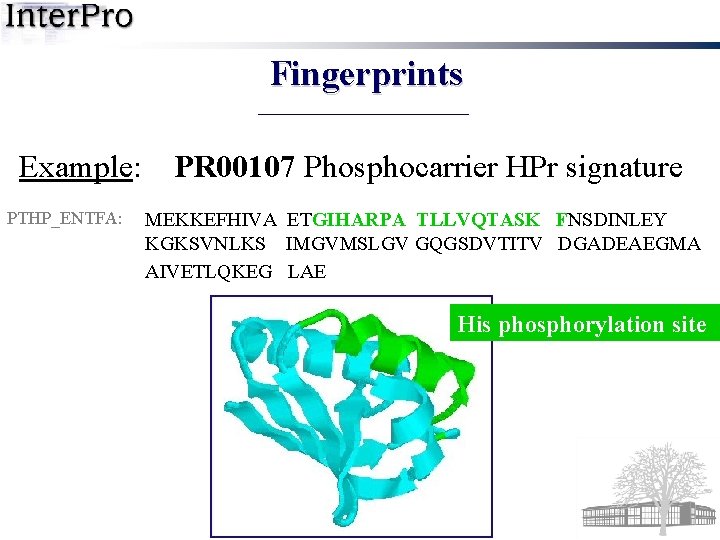
Fingerprints Example: PTHP_ENTFA: PR 00107 Phosphocarrier HPr signature MEKKEFHIVA ETGIHARPA TLLVQTASK FNSDINLEY KGKSVNLKS IMGVMSLGV GQGSDVTITV DGADEAEGMA AIVETLQKEG LAE His phosphorylation site

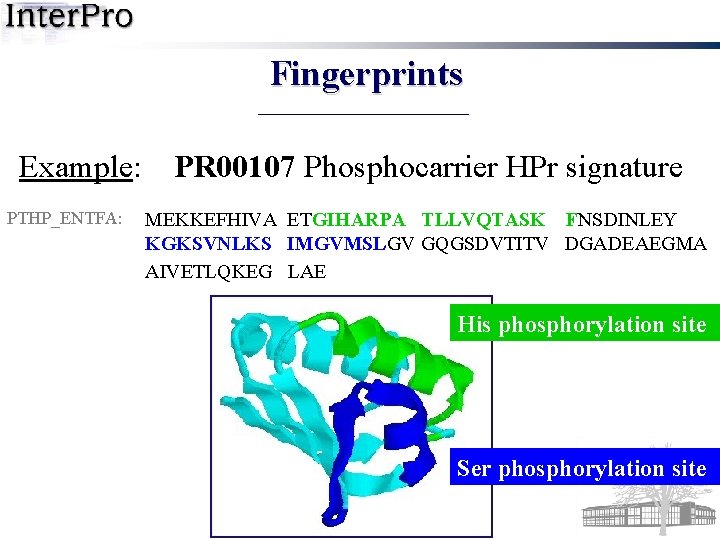
Fingerprints Example: PTHP_ENTFA: PR 00107 Phosphocarrier HPr signature MEKKEFHIVA ETGIHARPA TLLVQTASK FNSDINLEY KGKSVNLKS IMGVMSLGV GQGSDVTITV DGADEAEGMA AIVETLQKEG LAE His phosphorylation site Ser phosphorylation site

Fingerprints Example: PR 00107 Phosphocarrier HPr signature PTHP_ENTFA: MEKKEFHIVA ETGIHARPA TLLVQTASK FNSDINLEY KGKSVNLKS IMGVMSLGV GQGSDVTITV DGADEAEGMA AIVETLQKEG LAE His phosphorylation site Conserved site Ser phosphorylation site

Fingerprints Example: PR 00107 Phosphocarrier HPr signature PTHP_ENTFA: MEKKEFHIVA ETGIHARPA TLLVQTASK FNSDINLEY KGKSVNLKS IMGVMSLGV GQGSDVTITV DGADEAEGMA AIVETLQKEG LAE PR 00107 a fingerprint with three motifs 1) GIHARPATLLVQTASKF 2) KGKSVNLKSIMGVMSL 3) LGVGQGSDVTITVDGADE

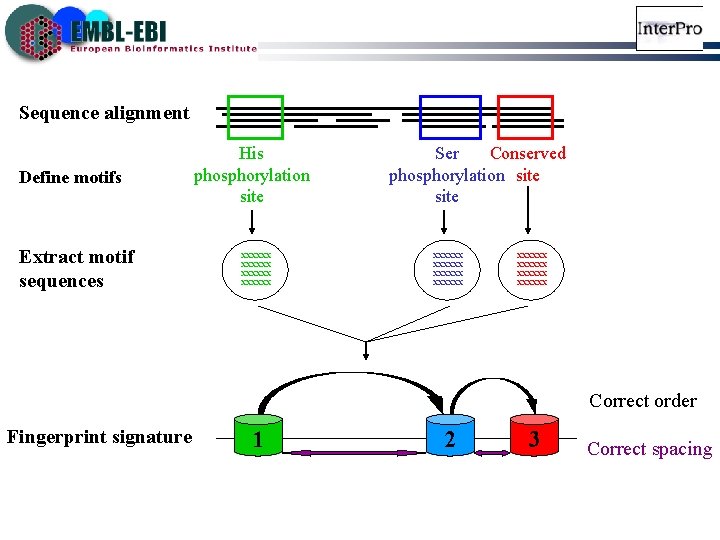
Sequence alignment Define motifs Extract motif sequences His phosphorylation site xxxxxx Ser Conserved phosphorylation site xxxxxx xxxxxx Correct order Fingerprint signature 1 2 3 Correct spacing

Sequence Clustering Automatic clustering of homologous domains Used by Pro. Dom database

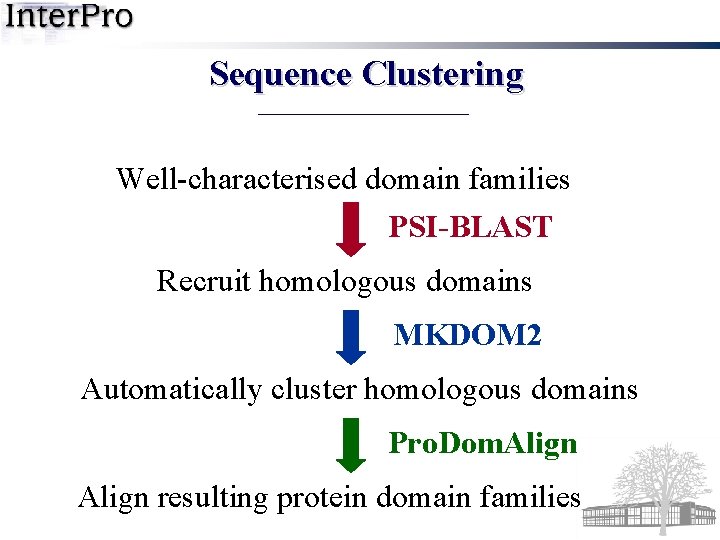
Sequence Clustering Well-characterised domain families PSI-BLAST Recruit homologous domains MKDOM 2 Automatically cluster homologous domains Pro. Dom. Align resulting protein domain families

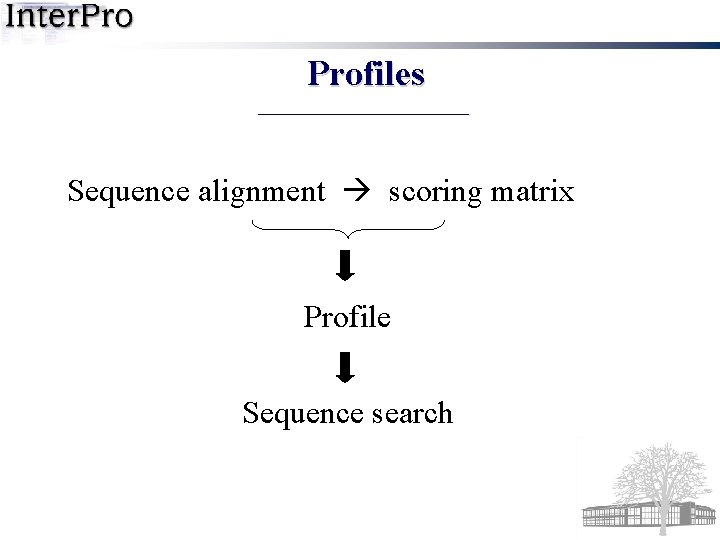
Profiles Sequence alignment scoring matrix Profile Sequence search

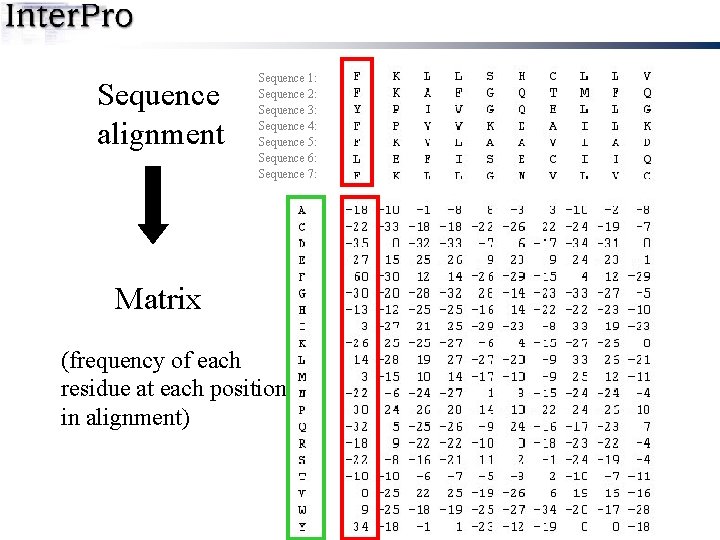
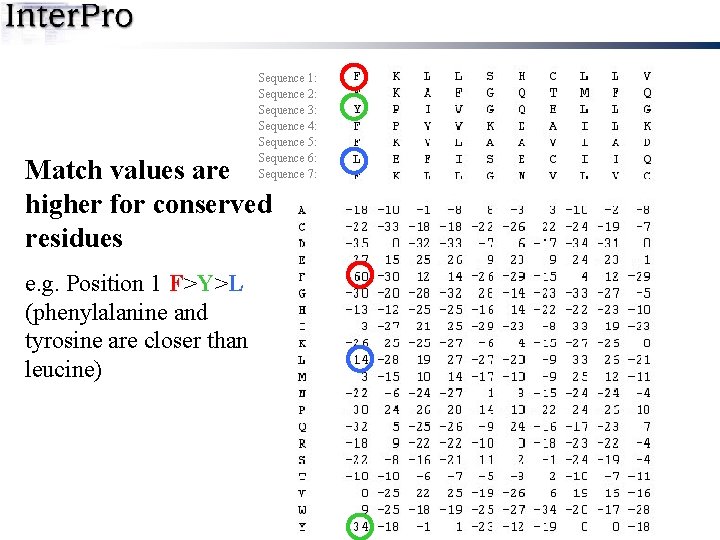
Sequence alignment Sequence 1: Sequence 2: Sequence 3: Sequence 4: Sequence 5: Sequence 6: Sequence 7: Matrix (frequency of each residue at each position in alignment)

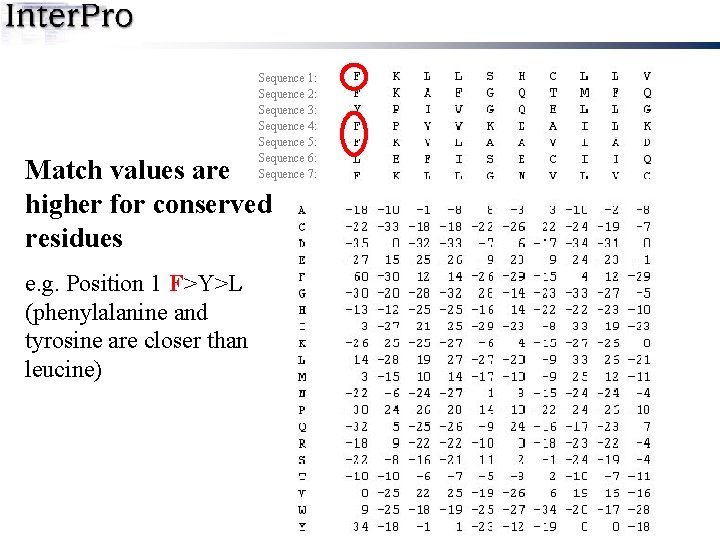
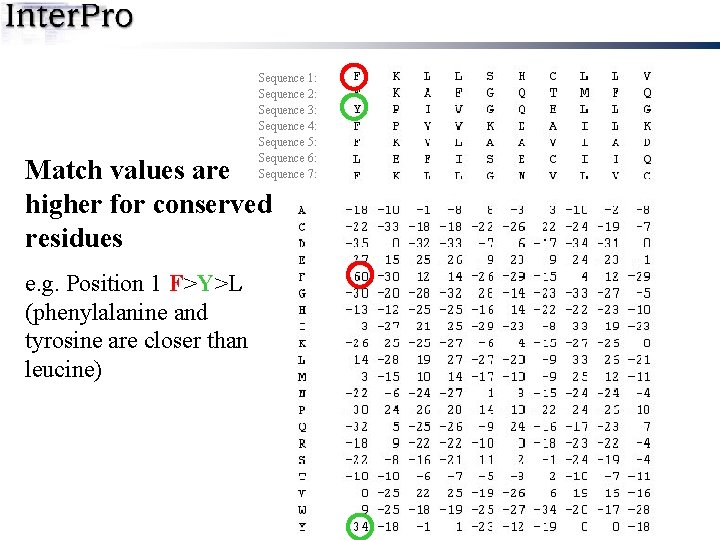
Sequence 1: Sequence 2: Sequence 3: Sequence 4: Sequence 5: Sequence 6: Sequence 7: Match values are higher for conserved residues e. g. Position 1 F>Y>L (phenylalanine and tyrosine are closer than leucine)

Sequence 1: Sequence 2: Sequence 3: Sequence 4: Sequence 5: Sequence 6: Sequence 7: Match values are higher for conserved residues e. g. Position 1 F>Y>L (phenylalanine and tyrosine are closer than leucine)

Sequence 1: Sequence 2: Sequence 3: Sequence 4: Sequence 5: Sequence 6: Sequence 7: Match values are higher for conserved residues e. g. Position 1 F>Y>L (phenylalanine and tyrosine are closer than leucine)

Sequence 1: Sequence 2: Sequence 3: Sequence 4: Sequence 5: Sequence 6: Sequence 7: Match values are higher for conserved residues e. g. Position 1 F>Y>L (phenylalanine and tyrosine are closer than leucine)

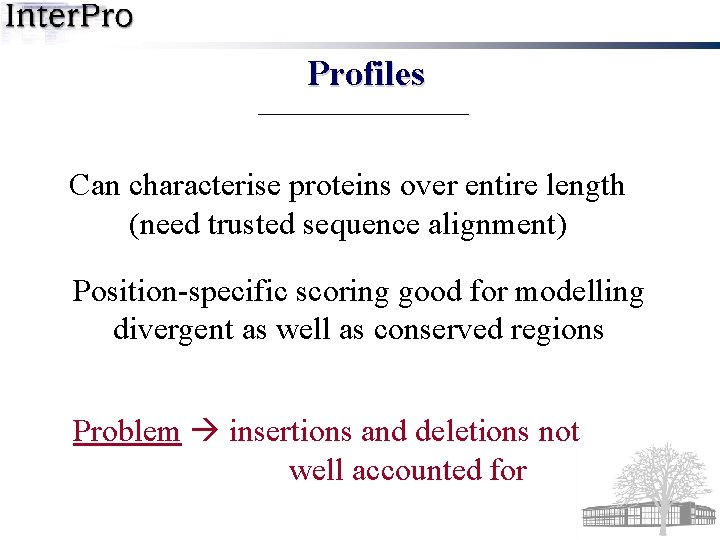
Profiles Can characterise proteins over entire length (need trusted sequence alignment) Position-specific scoring good for modelling divergent as well as conserved regions Problem insertions and deletions not well accounted for

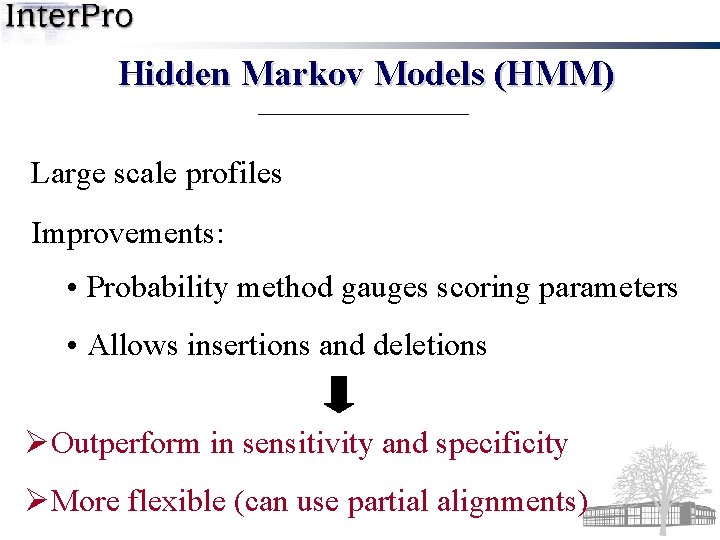
Hidden Markov Models (HMM) Large scale profiles Improvements: • Probability method gauges scoring parameters • Allows insertions and deletions ØOutperform in sensitivity and specificity ØMore flexible (can use partial alignments)

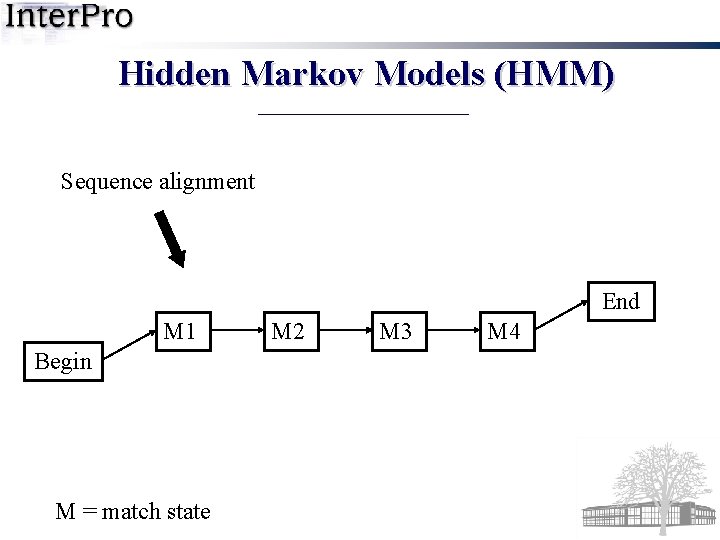
Hidden Markov Models (HMM) Sequence alignment End M 1 Begin M = match state M 2 M 3 M 4

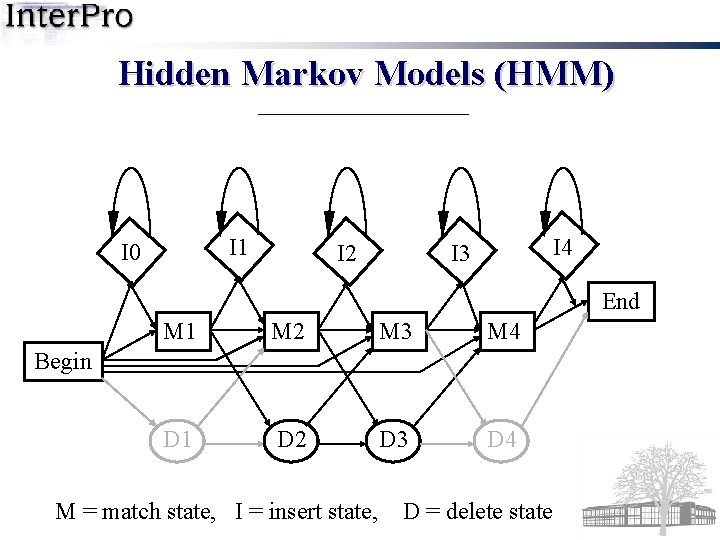
Hidden Markov Models (HMM) I 1 I 0 I 2 I 4 I 3 End M 1 M 2 M 3 M 4 D 1 D 2 D 3 D 4 Begin M = match state, I = insert state, D = delete state

Hidden Markov Models (HMM) HMMER 2 package: HMMbuild HMMcalibrate Database search http: //hmmer. wustl. edu/

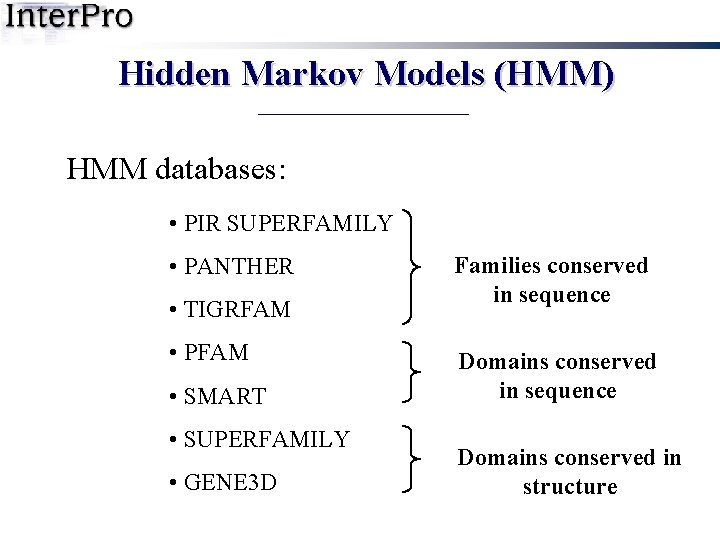
Hidden Markov Models (HMM) HMM databases: • PIR SUPERFAMILY • PANTHER • TIGRFAM • PFAM • SMART • SUPERFAMILY • GENE 3 D Families conserved in sequence Domains conserved in structure

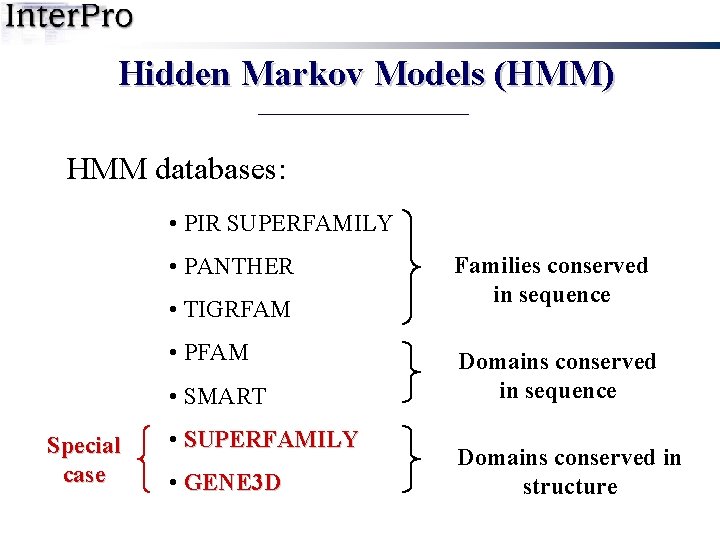
Hidden Markov Models (HMM) HMM databases: • PIR SUPERFAMILY • PANTHER • TIGRFAM • PFAM • SMART Special case • SUPERFAMILY • GENE 3 D Families conserved in sequence Domains conserved in structure

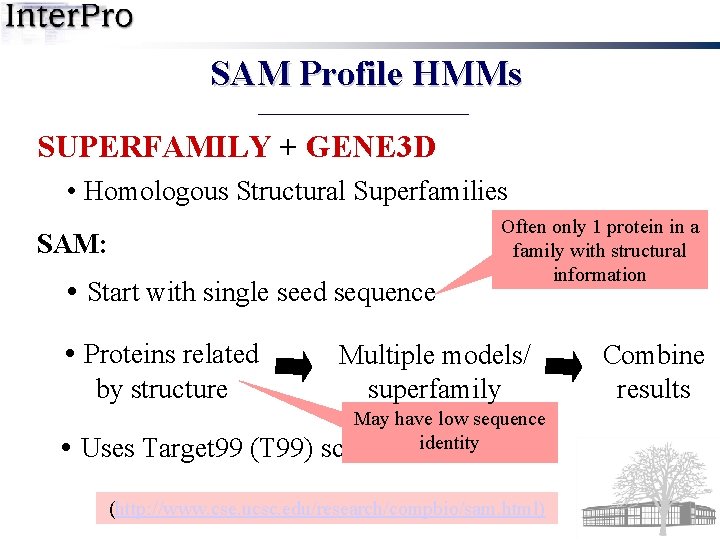
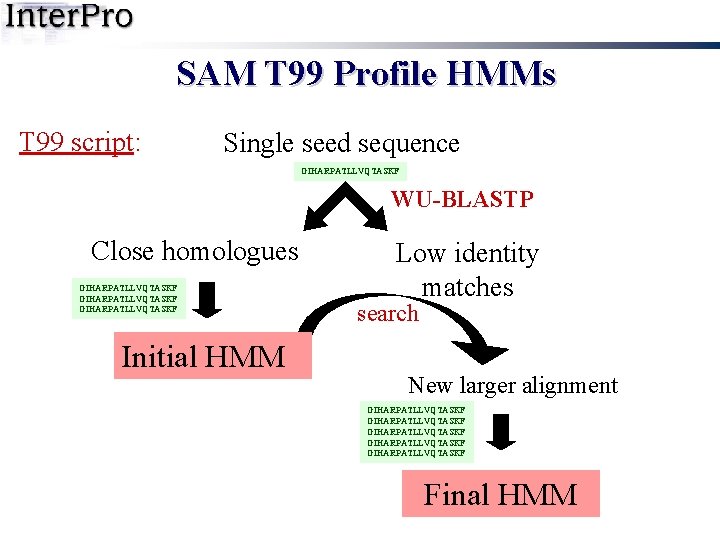
SAM Profile HMMs SUPERFAMILY + GENE 3 D • Homologous Structural Superfamilies SAM: • Start with single seed sequence • Proteins related by structure • Uses Target 99 (T 99) Often only 1 protein in a family with structural information Multiple models/ superfamily May have low sequence script identity (http: //www. cse. ucsc. edu/research/compbio/sam. html) Combine results

SAM T 99 Profile HMMs T 99 script: Single seed sequence GIHARPATLLVQTASKF WU-BLASTP Close homologues GIHARPATLLVQTASKF Initial HMM Low identity matches search New larger alignment GIHARPATLLVQTASKF GIHARPATLLVQTASKF Final HMM

Summary of signature methods Single motif method Sequence alignment xxxxxx Extract motif pattern (PROSITE) Full sequence: Full alignment methods xxxxxx xxxxxx xxxxxx Multiple motif methods Extract multiple motifs (PRINTS) xxxxxx 1) profile (PROSITE) 2) HMM (PFAM, SMART, SUPERFAMILY, TIGRFAM, PIRSF, GENE 3 D, PANTHER)

Protein Signature Databases Patterns Prosite Fingerprints Prints Sequence clustering Pro. Dom Profiles HMM T 99 -SAM HMM Prosite PIR Superfamily Tigrfam Smart Gene 3 D Panther Pfam Superfamily

Prints Fingerprint is a set of motifs PR 00000 Full length of protein Can identify small conserved regions in divergent proteins Use different combinations of motifs to describe families and sibling subfamilies http: //umber. sbs. man. ac. uk/dbbrowser/PRINTS/

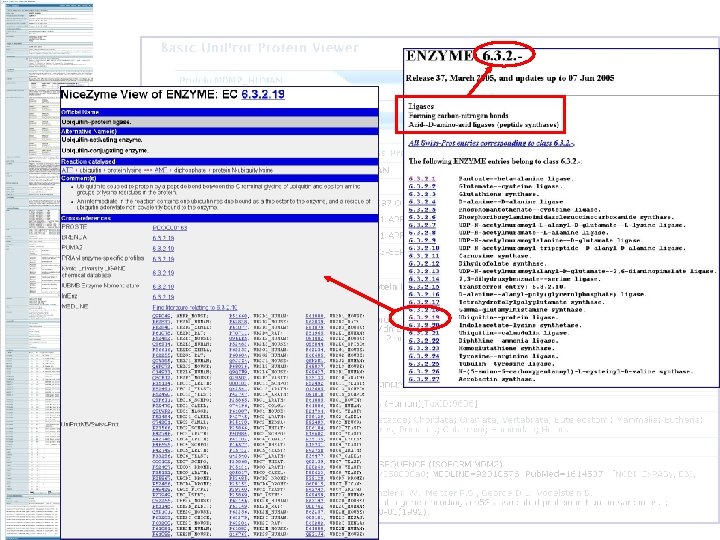
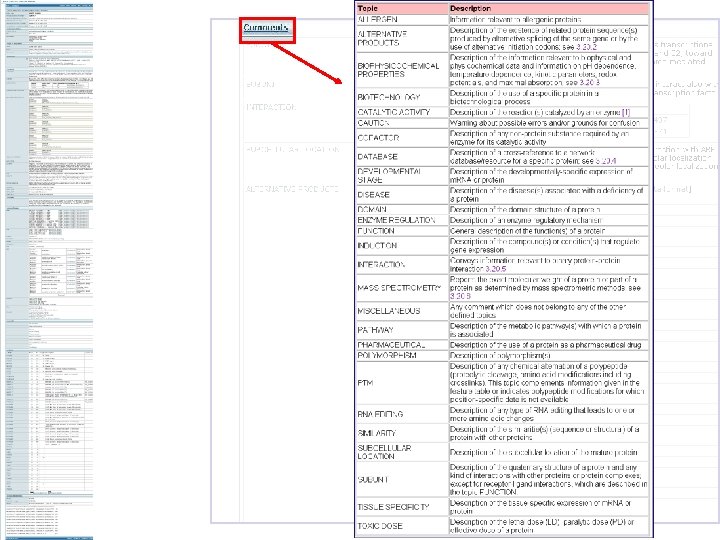
Prosite Patterns Pattern is a regular expression PS 00000 Identify various important sites within proteins Several models characterise enzymes http: //us. expasy. org/prosite/ Enzyme catalytic site Prosthetic group attachment Used by Uni. Prot to Metal ion binding site Cysteines disulphide sites bonds defineforcatalytic Protein or molecule binding

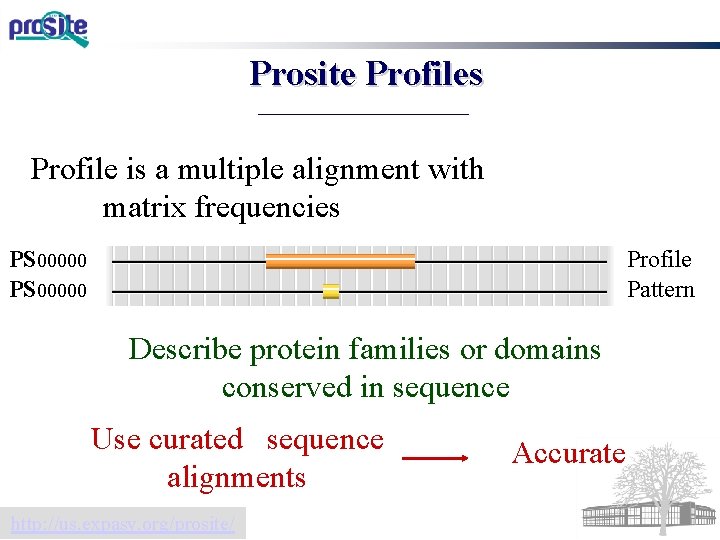
Prosite Profiles Profile is a multiple alignment with matrix frequencies Profile Pattern PS 00000 Describe protein families or domains conserved in sequence Use curated sequence alignments http: //us. expasy. org/prosite/ Accurate

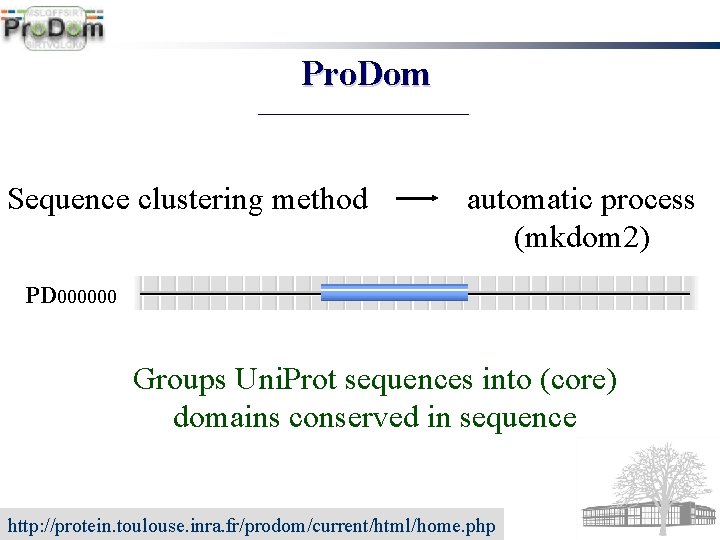
Pro. Dom Sequence clustering method automatic process (mkdom 2) PD 000000 Groups Uni. Prot sequences into (core) domains conserved in sequence http: //protein. toulouse. inra. fr/prodom/current/html/home. php

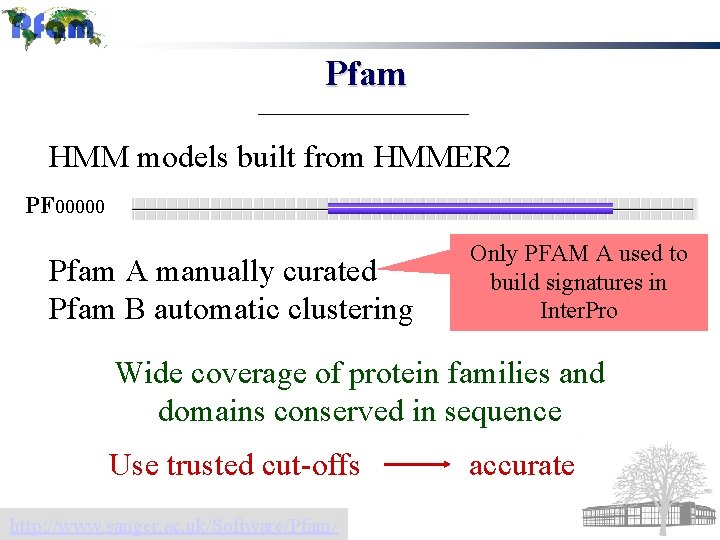
Pfam HMM models built from HMMER 2 PF 00000 Pfam A manually curated Pfam B automatic clustering Only PFAM A used to build signatures in Inter. Pro Wide coverage of protein families and domains conserved in sequence Use trusted cut-offs http: //www. sanger. ac. uk/Software/Pfam/ accurate

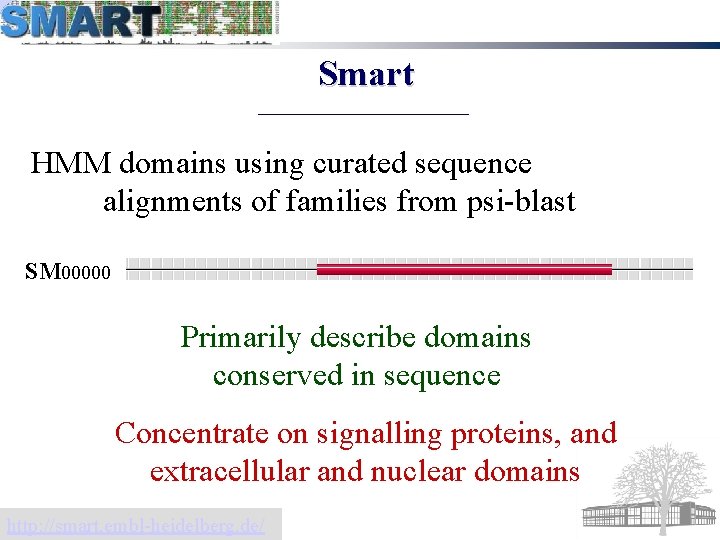
Smart HMM domains using curated sequence alignments of families from psi-blast SM 00000 Primarily describe domains conserved in sequence Concentrate on signalling proteins, and extracellular and nuclear domains http: //smart. embl-heidelberg. de/

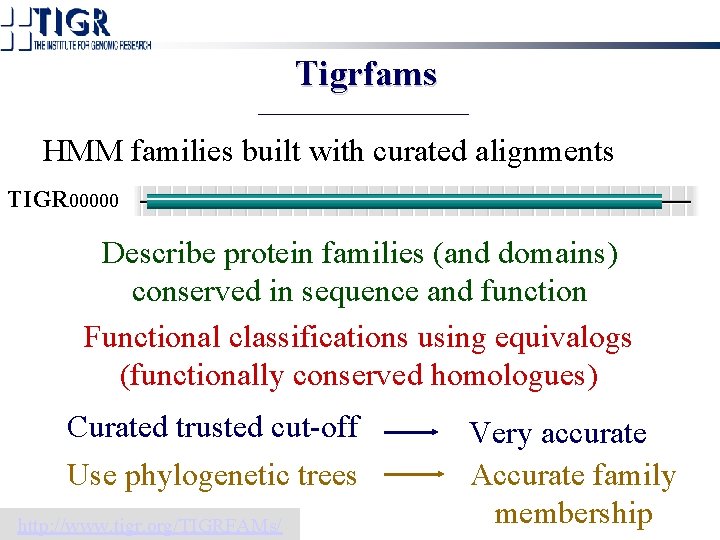
Tigrfams HMM families built with curated alignments TIGR 00000 Describe protein families (and domains) conserved in sequence and function Functional classifications using equivalogs (functionally conserved homologues) Curated trusted cut-off Use phylogenetic trees http: //www. tigr. org/TIGRFAMs/ Very accurate Accurate family membership

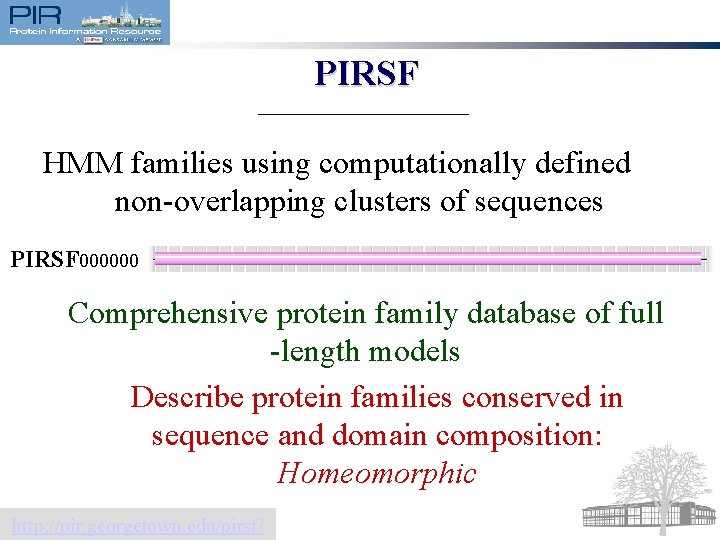
PIRSF HMM families using computationally defined non-overlapping clusters of sequences PIRSF 000000 Comprehensive protein family database of full -length models Describe protein families conserved in sequence and domain composition: Homeomorphic http: //pir. georgetown. edu/pirsf/

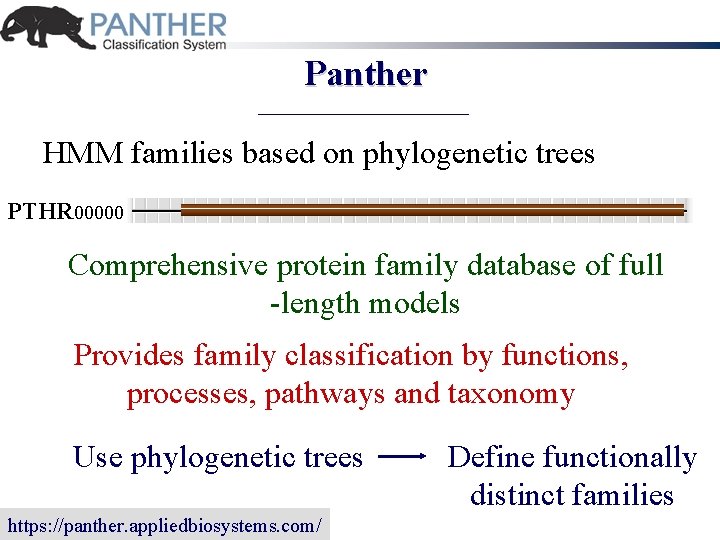
Panther HMM families based on phylogenetic trees PTHR 00000 Comprehensive protein family database of full -length models Provides family classification by functions, processes, pathways and taxonomy Use phylogenetic trees https: //panther. appliedbiosystems. com/ Define functionally distinct families

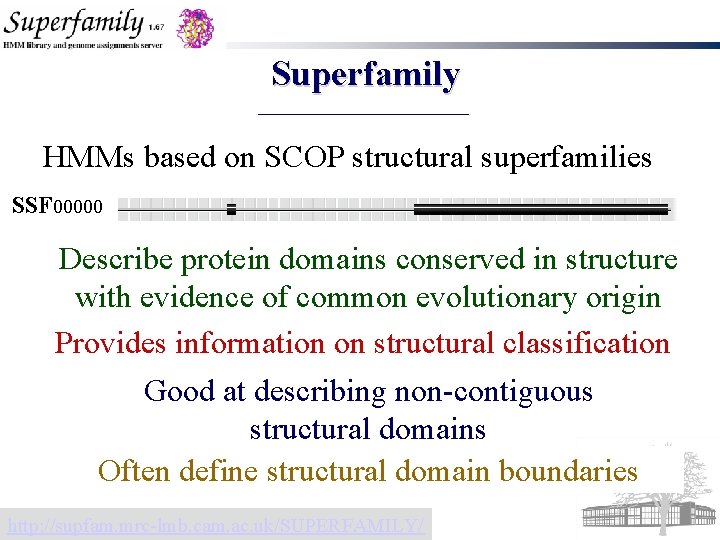
Superfamily HMMs based on SCOP structural superfamilies SSF 00000 Describe protein domains conserved in structure with evidence of common evolutionary origin Provides information on structural classification Good at describing non-contiguous structural domains Often define structural domain boundaries http: //supfam. mrc-lmb. cam. ac. uk/SUPERFAMILY/

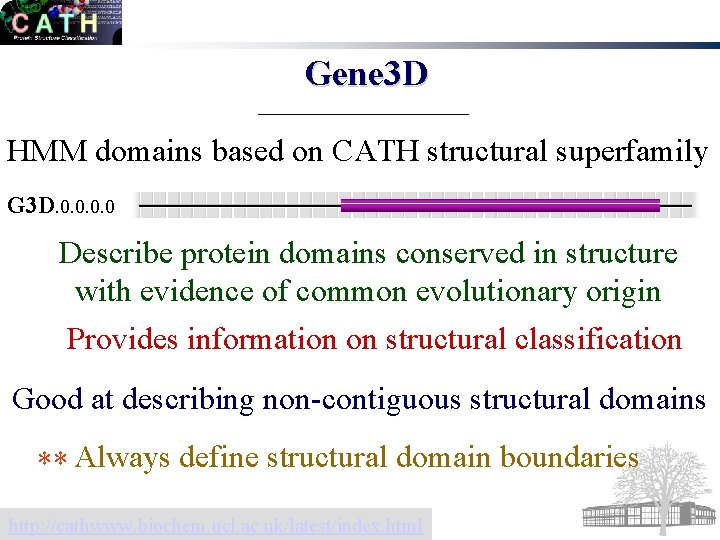
Gene 3 D HMM domains based on CATH structural superfamily G 3 D. 0. 0 Describe protein domains conserved in structure with evidence of common evolutionary origin Provides information on structural classification Good at describing non-contiguous structural domains ** Always define structural domain boundaries http: //cathwww. biochem. ucl. ac. uk/latest/index. html

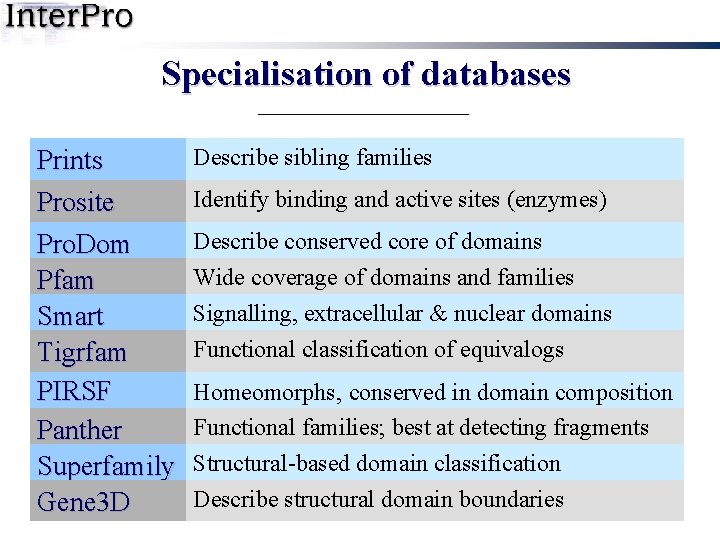
Specialisation of databases Prints Prosite Pro. Dom Pfam Smart Tigrfam PIRSF Panther Superfamily Gene 3 D Describe sibling families Identify binding and active sites (enzymes) Describe conserved core of domains Wide coverage of domains and families Signalling, extracellular & nuclear domains Functional classification of equivalogs Homeomorphs, conserved in domain composition Functional families; best at detecting fragments Structural-based domain classification Describe structural domain boundaries

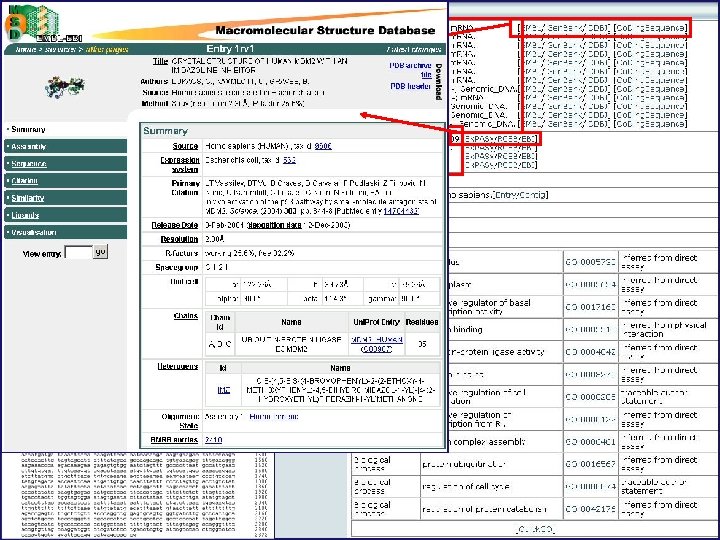
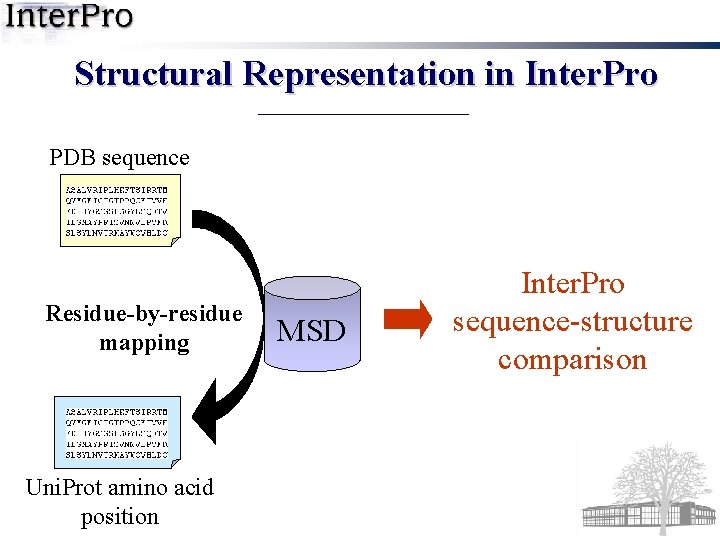
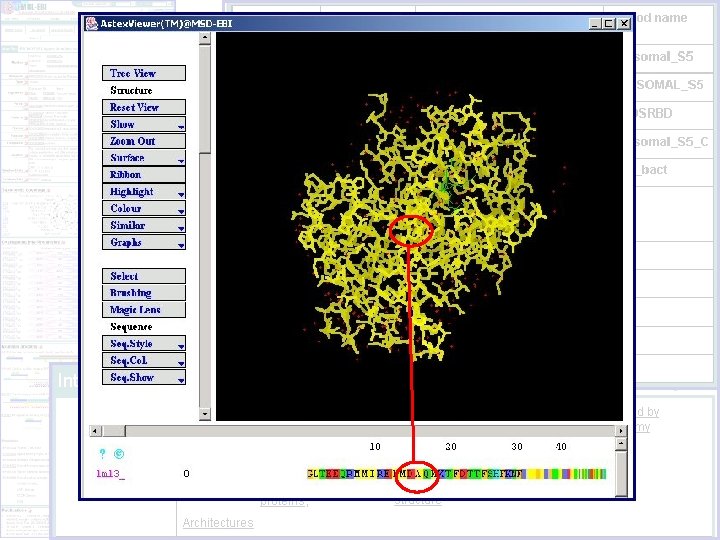
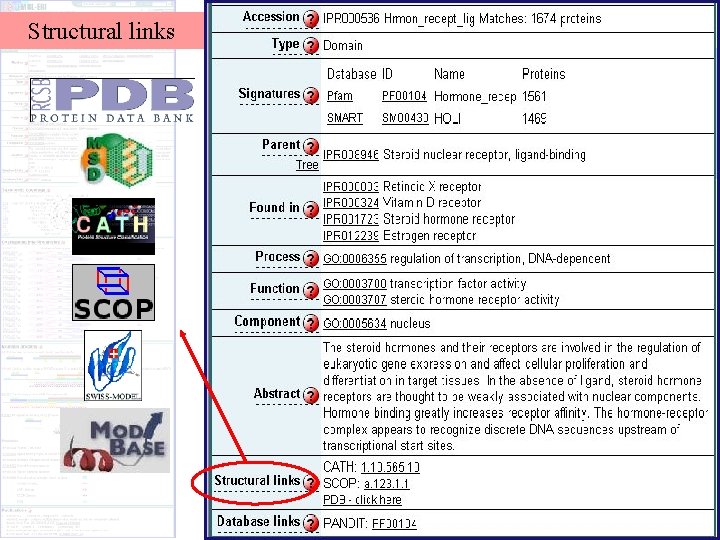
Structural Representation in Inter. Pro PDB sequence Residue-by-residue mapping Uni. Prot amino acid position MSD Inter. Pro sequence-structure comparison

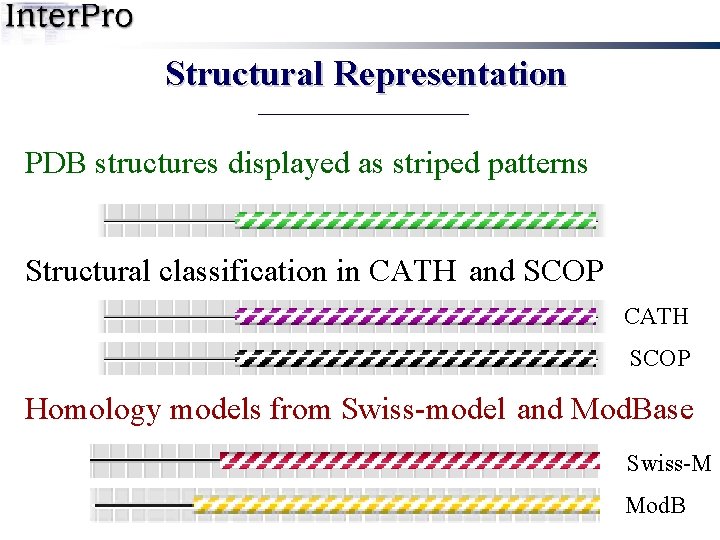
Structural Representation PDB structures displayed as striped patterns Structural classification in CATH and SCOP CATH SCOP Homology models from Swiss-model and Mod. Base Swiss-M Mod. B

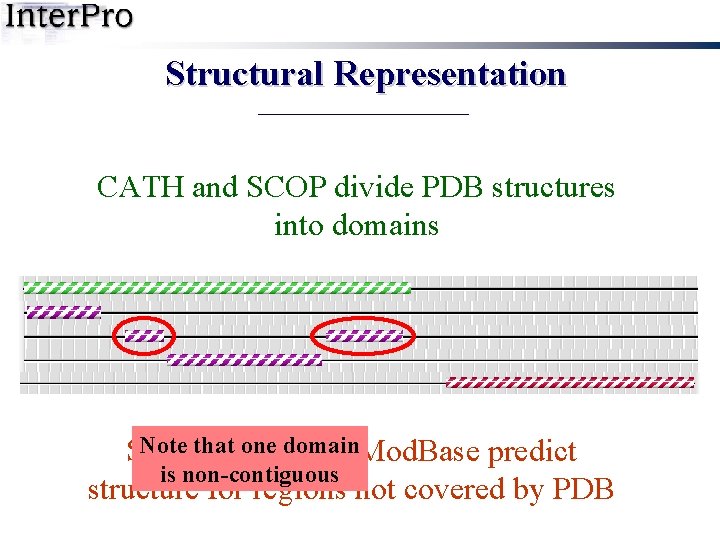
Structural Representation CATH and SCOP divide PDB structures into domains Note that one domain Swiss-Model and Mod. Base predict is non-contiguous structure for regions not covered by PDB

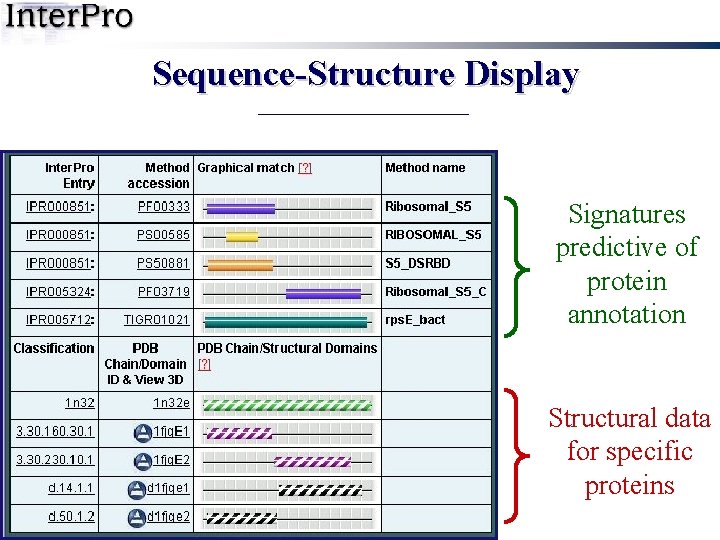
Sequence-Structure Display Signatures predictive of protein annotation Structural data for specific proteins

Searching Inter. Pro Search tools include: • Text Search • Inter. Pro. Scan (sequence search) • SRS (multiple database search) http: //www. ebi. ac. uk/interpro/

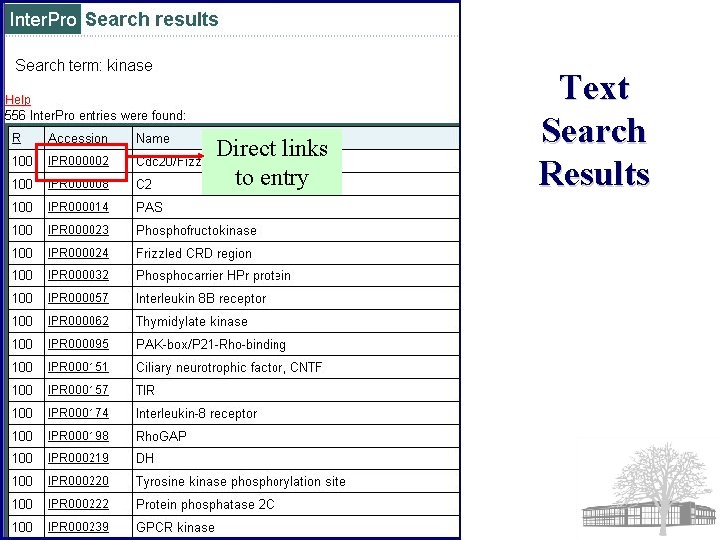
Direct links to entry Text Search Results

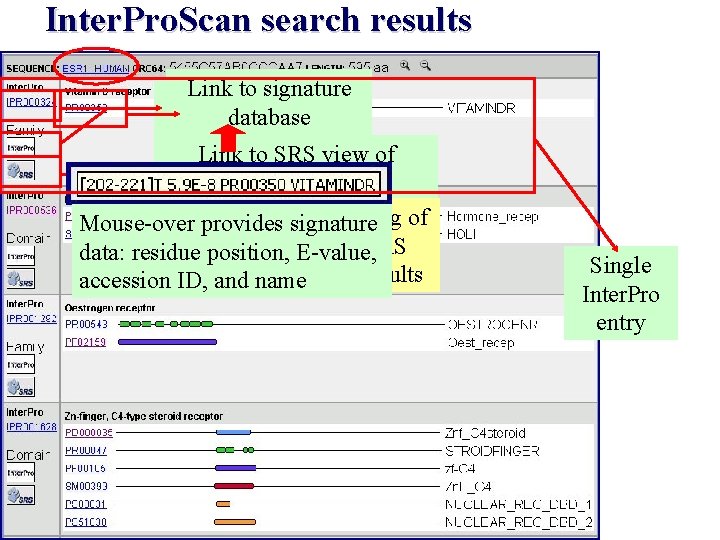
Inter. Pro. Scan search results Linktotosignature database Inter. Pro entry Link to SRS view of Inter. Pro entry Enables direct searching of Mouse-over provides signature other databases in SRS data: residue position, E-value, Inter. Pro. Scan results accessionusing ID, and name Single Inter. Pro entry

Inter. Pro Entry • Groups similar signatures together and provide relationships between signatures • Provides extensive manual annotation • Provides links to other databases • Provides structural information and viewers

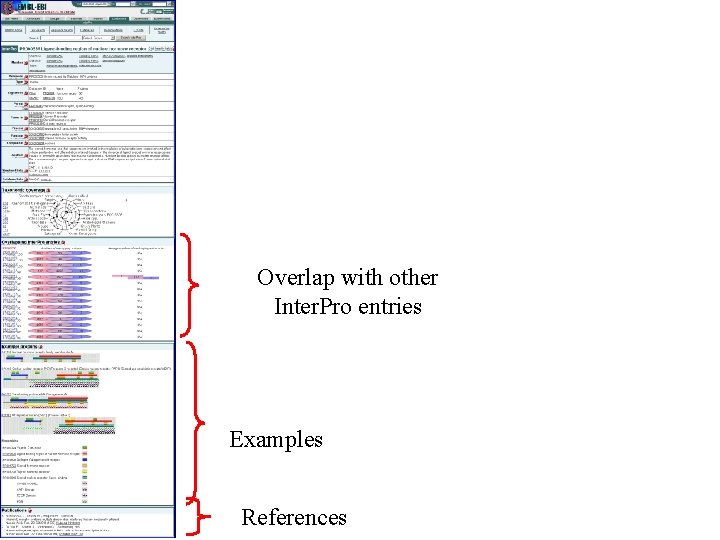
Annotation Fields in Inter. Pro • • • Name and short name Entry type Relationships GO mapping Abstract Structural links Database links Taxonomy Examples Publications

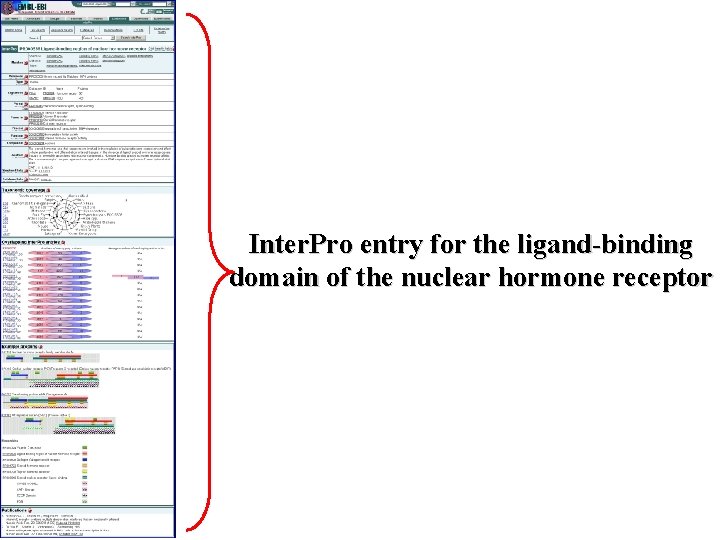
Inter. Pro entry for the ligand-binding domain of the nuclear hormone receptor

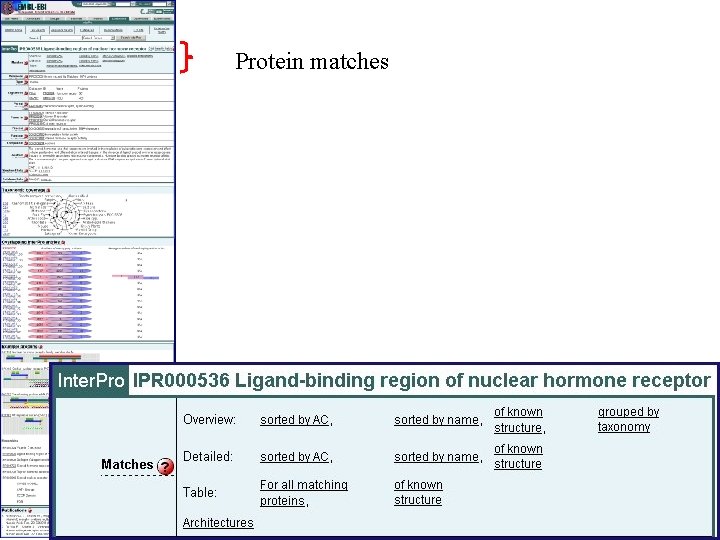
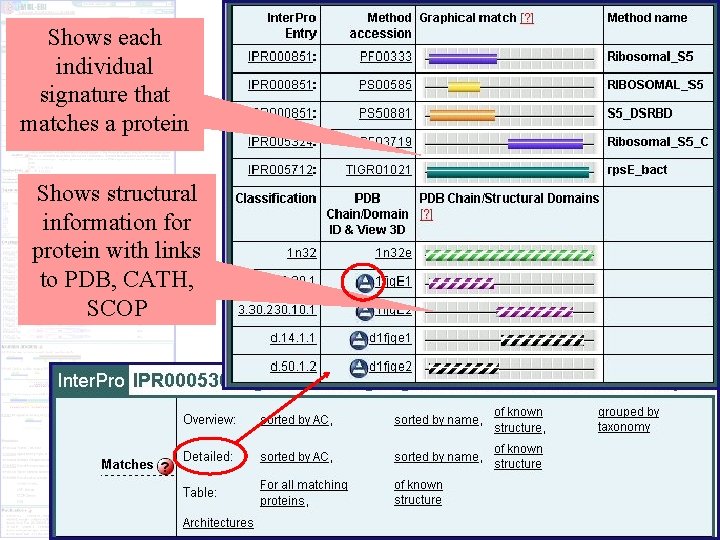
Protein matches

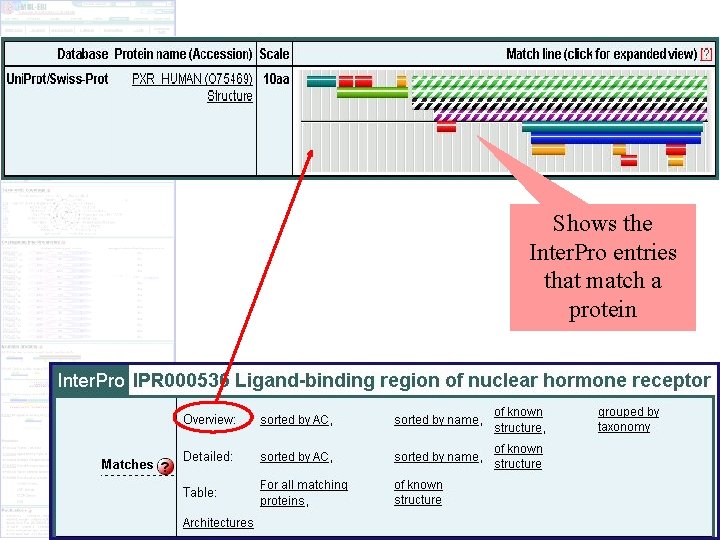
Shows the Inter. Pro entries that match a protein

Shows each individual signature that matches a protein Shows structural information for protein with links to PDB, CATH, SCOP Protein matches

Protein matches

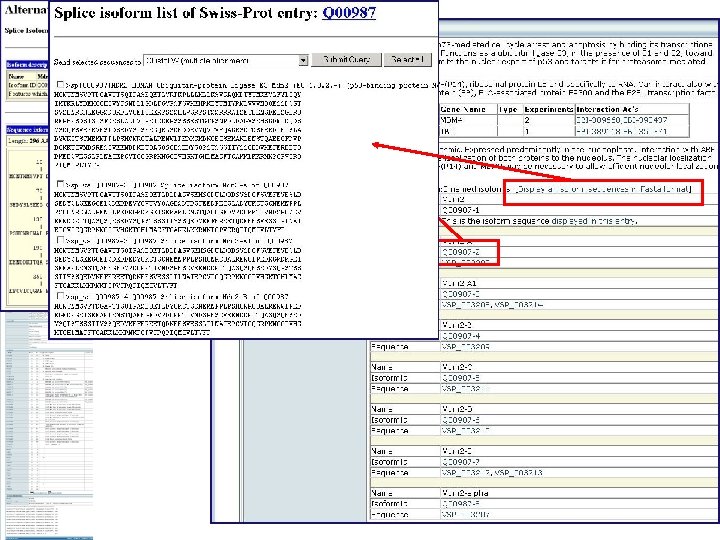
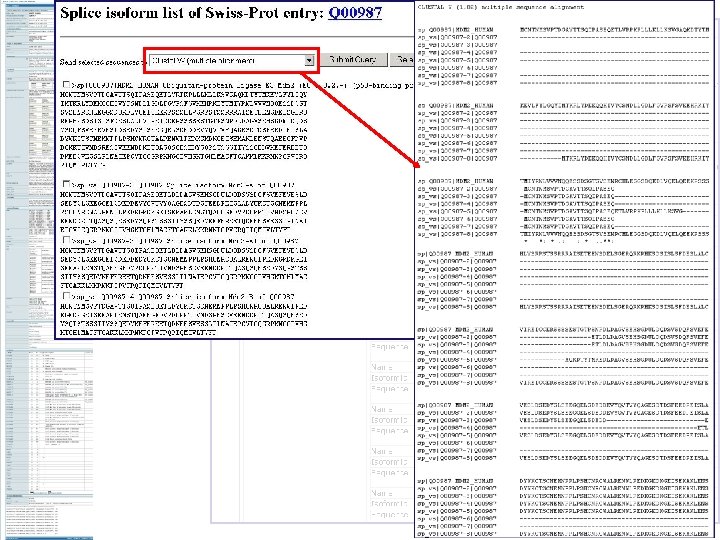
Protein matches Splice variants

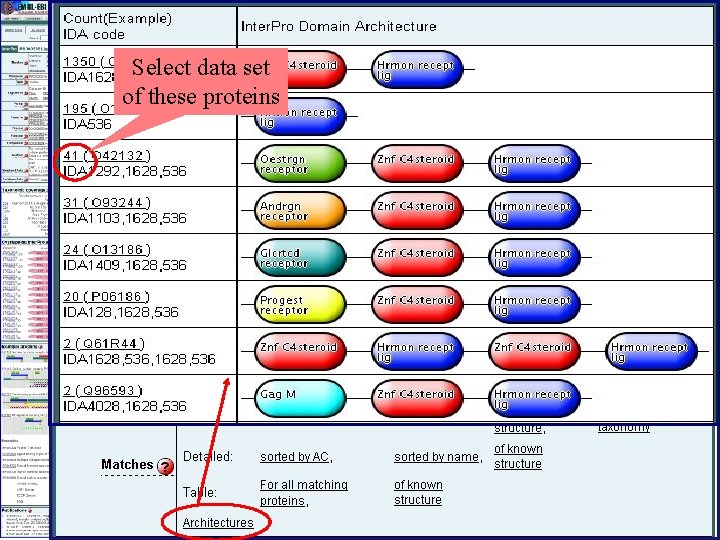
Select data set of these proteins

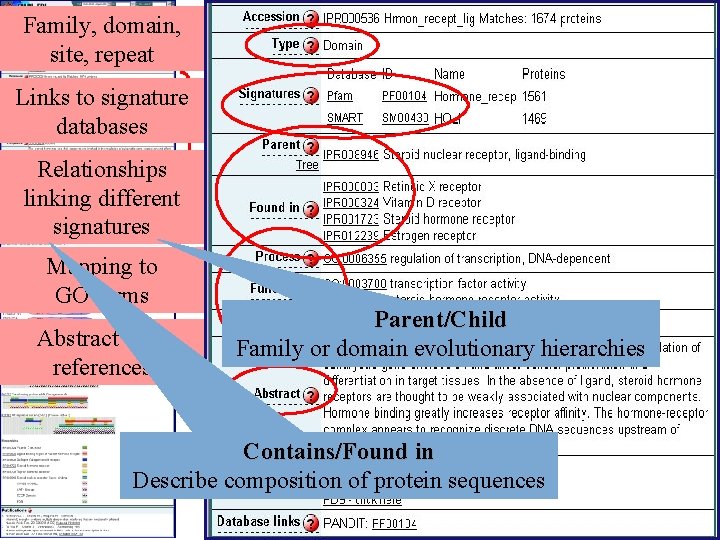
Family, domain, site, repeat Links to signature databases Detailed information Relationships linking different signatures Mapping to GO terms Abstract with references Parent/Child Family or domain evolutionary hierarchies Contains/Found in Describe composition of protein sequences

Structural links

Database links

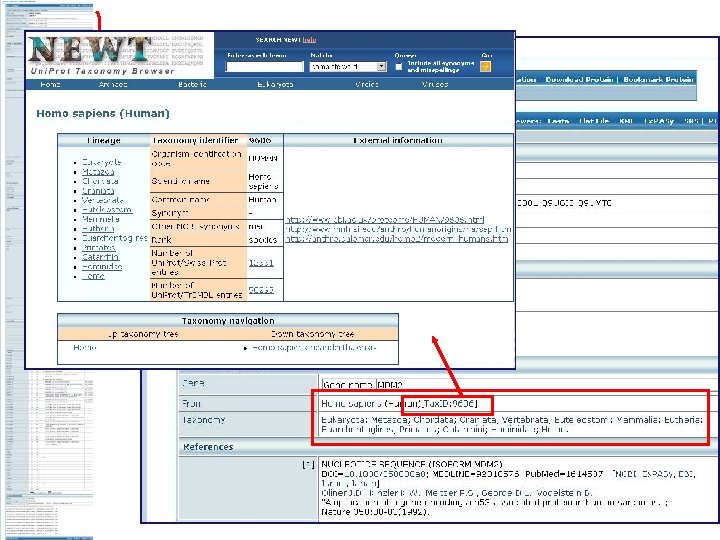
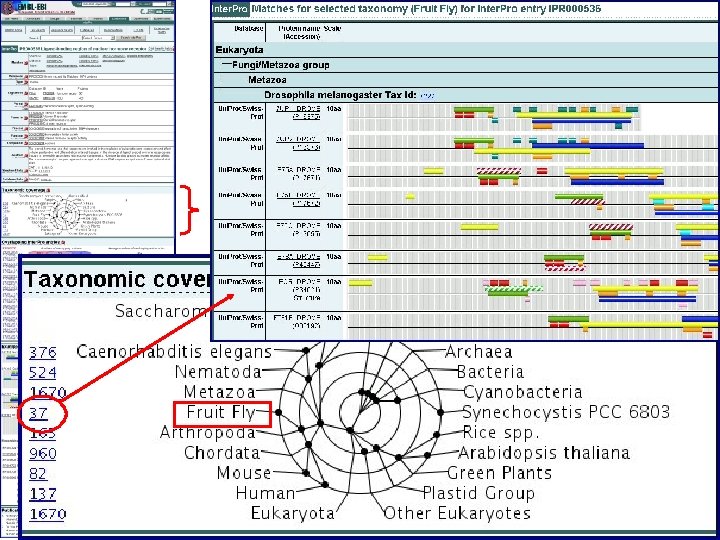
Taxonomy

Overlap with other Inter. Pro entries Examples References

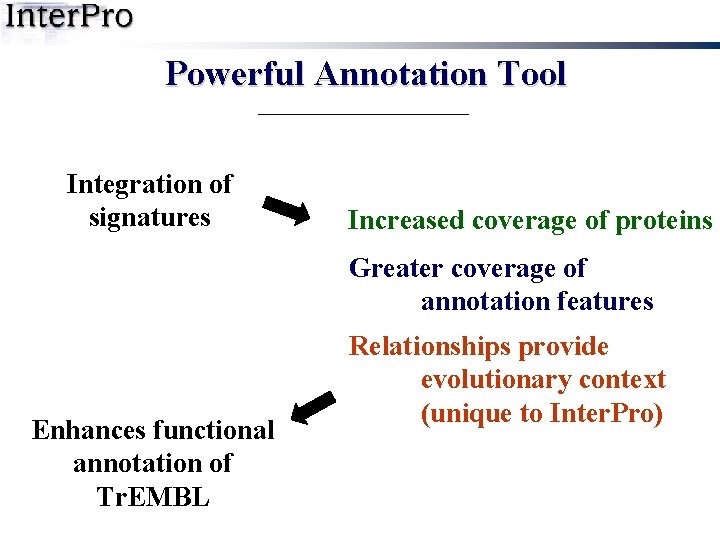
Powerful Annotation Tool Integration of signatures Increased coverage of proteins Greater coverage of annotation features Enhances functional annotation of Tr. EMBL Relationships provide evolutionary context (unique to Inter. Pro)

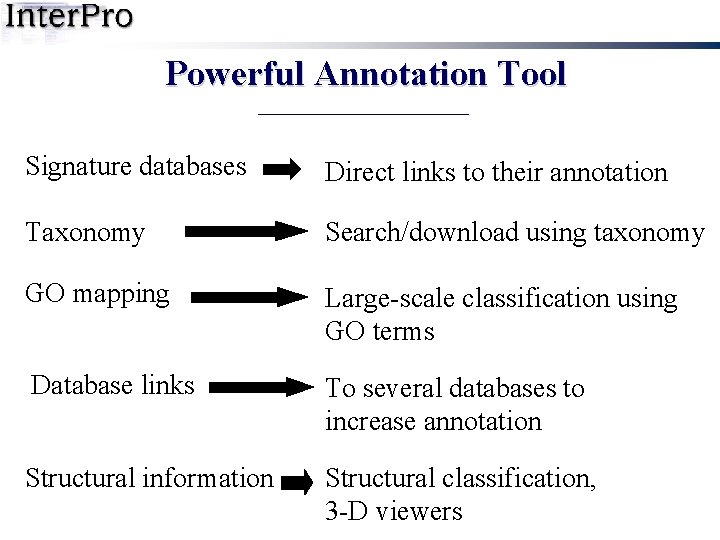
Powerful Annotation Tool Signature databases Direct links to their annotation Taxonomy Search/download using taxonomy GO mapping Large-scale classification using GO terms Database links To several databases to increase annotation Structural information Structural classification, 3 -D viewers

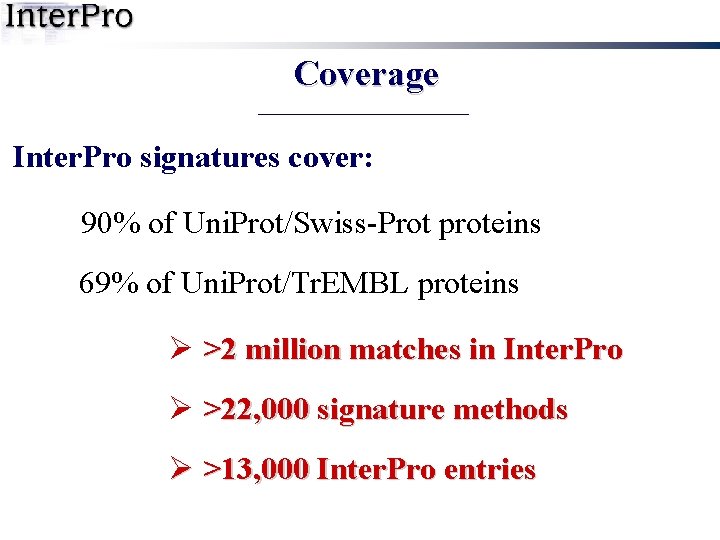
Coverage Inter. Pro signatures cover: 90% of Uni. Prot/Swiss-Prot proteins 69% of Uni. Prot/Tr. EMBL proteins Ø >2 million matches in Inter. Pro Ø >22, 000 signature methods Ø >13, 000 Inter. Pro entries

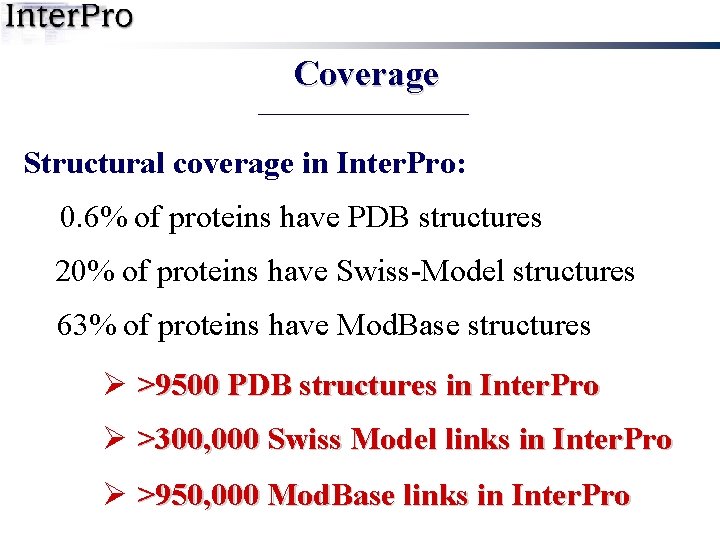
Coverage Structural coverage in Inter. Pro: 0. 6% of proteins have PDB structures 20% of proteins have Swiss-Model structures 63% of proteins have Mod. Base structures Ø >9500 PDB structures in Inter. Pro Ø >300, 000 Swiss Model links in Inter. Pro Ø >950, 000 Mod. Base links in Inter. Pro

Availability and downloads Web access Tool/Databases: http: //www. ebi. ac. uk/services/ Downloads ftp site: ftp: //ftp. ebi. ac. uk/pub/databases/

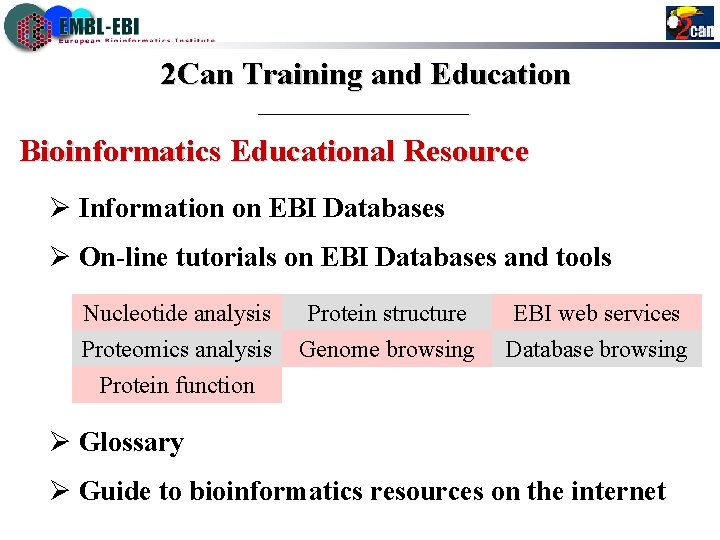
2 Can Training and Education Bioinformatics Educational Resource Ø Information on EBI Databases Ø On-line tutorials on EBI Databases and tools Nucleotide analysis Proteomics analysis Protein function Protein structure Genome browsing EBI web services Database browsing Ø Glossary Ø Guide to bioinformatics resources on the internet

http: //www. ebi. ac. uk/

http: //www. ebi. ac. uk/2 can/

http: //www. ebi. ac. uk/interpro/

Acknowledgements Rolf Apweiler Henning Hermajakob Nicky Mulder Amos Bairoch Int. Act Team Inter. Pro Team Cathy Wu +100 annotators Inter. Pro Consortium