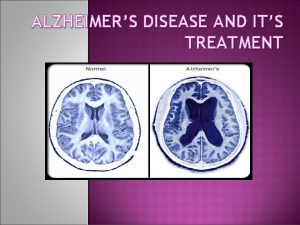
Uni Prot and Alzheimers Disease Linking molecular defects




- Slides: 25

Uni. Prot and Alzheimer’s Disease: Linking molecular defects to disease phenotype Yvonne Lussi help@uniprot. org

Uni. Prot. KB: Universal Protein Knowledgebase a comprehensive, high-quality and freely accessible resource of protein sequence and functional information Expert literature curation on: • Detailed information on protein function, interactions, pathways etc. • Sequences, including isoforms, disease variants and PTMs • Stable identifiers (accessions) European Bioinformatics Institute (EMBL-EBI), Hinxton, Cambridge, UK Protein Information Resource (PIR), Washington DC and Delaware, USA SIB Swiss Institute of Bioinformatics (SIB), Geneva, Switzerland

Uni. Prot. KB: Universal Protein Knowledgebase a comprehensive, high-quality and freely accessible resource of protein sequence and functional information NIH National Institute of Aging (NIA) medical initiative: Capture information of proteins of impact for human health and biomedical research • Proteins involved in Alzheimer’s Disease • Disease variants European Bioinformatics Institute (EMBL-EBI), Hinxton, Cambridge, UK Protein Information Resource (PIR), Washington DC and Delaware, USA SIB Swiss Institute of Bioinformatics (SIB), Geneva, Switzerland

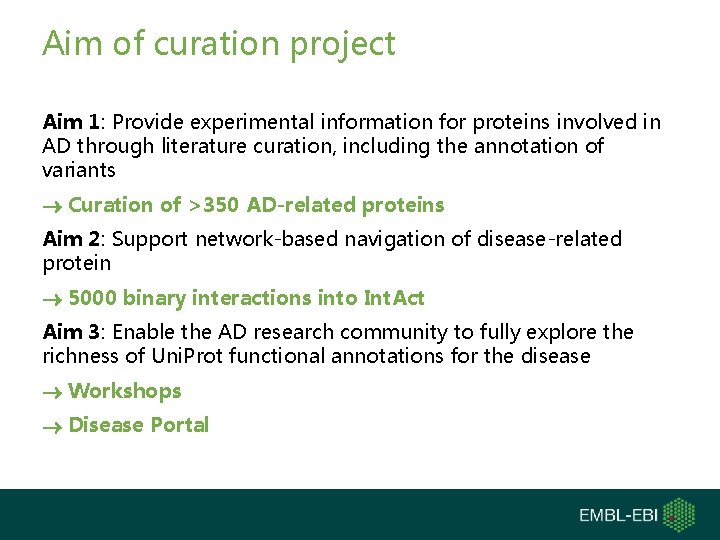
Aim of curation project Aim 1: Provide experimental information for proteins involved in AD through literature curation, including the annotation of variants Curation of >350 AD-related proteins Aim 2: Support network-based navigation of disease-related protein 5000 binary interactions into Int. Act Aim 3: Enable the AD research community to fully explore the richness of Uni. Prot functional annotations for the disease Workshops Disease Portal

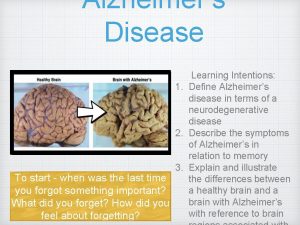
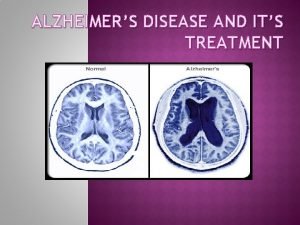
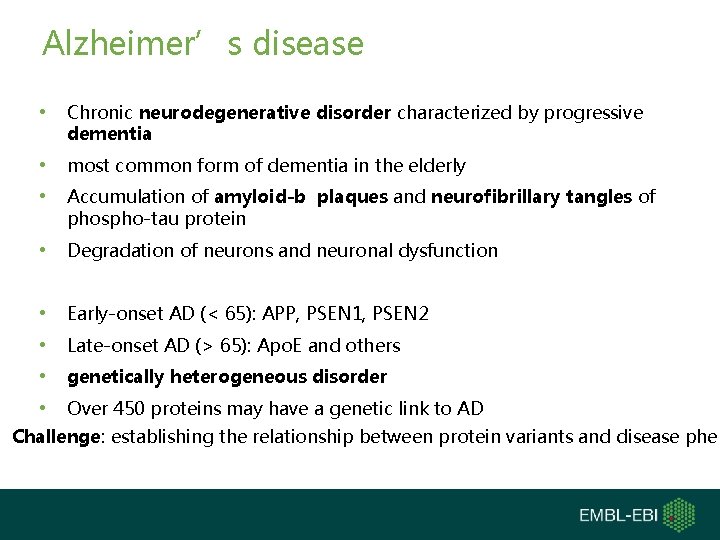
Alzheimer’s disease • Chronic neurodegenerative disorder characterized by progressive dementia • most common form of dementia in the elderly • Accumulation of amyloid-b plaques and neurofibrillary tangles of phospho-tau protein • Degradation of neurons and neuronal dysfunction • • Early-onset AD (< 65): APP, PSEN 1, PSEN 2 Late-onset AD (> 65): Apo. E and others genetically heterogeneous disorder Over 450 proteins may have a genetic link to AD Challenge: establishing the relationship between protein variants and disease phen

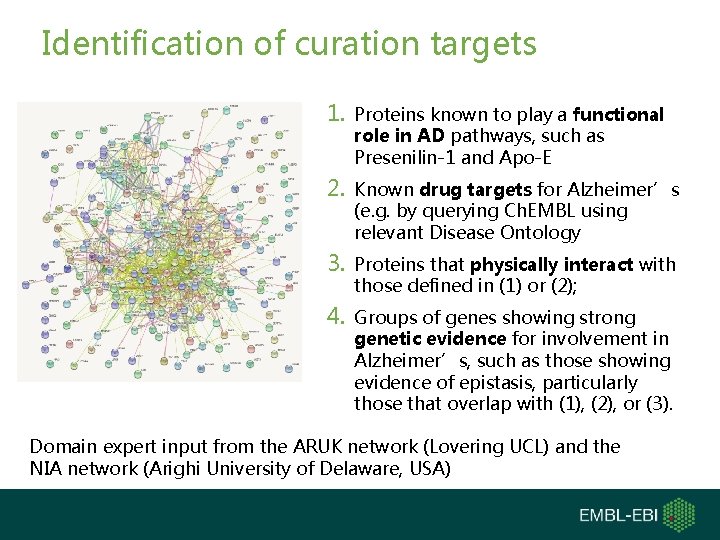
Identification of curation targets 1. Proteins known to play a functional role in AD pathways, such as Presenilin-1 and Apo-E 2. Known drug targets for Alzheimer’s (e. g. by querying Ch. EMBL using relevant Disease Ontology 3. Proteins that physically interact with those defined in (1) or (2); 4. Groups of genes showing strong genetic evidence for involvement in Alzheimer’s, such as those showing evidence of epistasis, particularly those that overlap with (1), (2), or (3). Domain expert input from the ARUK network (Lovering UCL) and the NIA network (Arighi University of Delaware, USA)

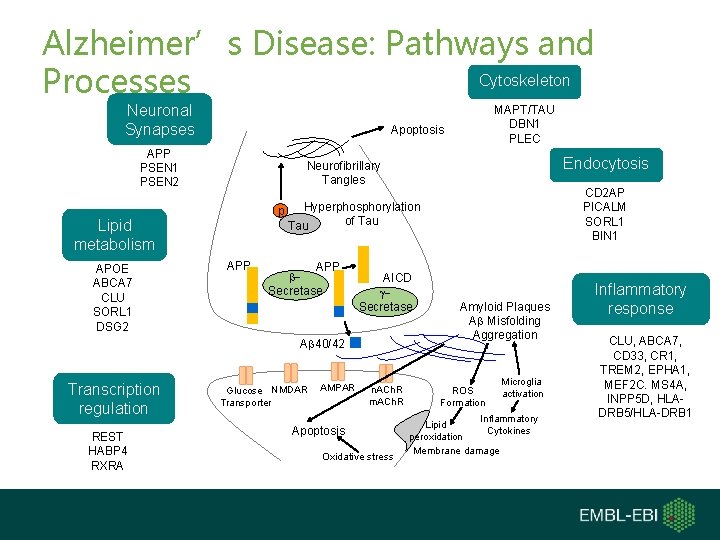
Alzheimer’s Disease: Pathways and Cytoskeleton Processes Neuronal Synapses Apoptosis APP PSEN 1 PSEN 2 REST HABP 4 RXRA CD 2 AP PICALM SORL 1 BIN 1 Hyperphosphorylation p of Tau APP b. Secretase AICD g. Secretase Ab 40/42 Transcription regulation Endocytosis Neurofibrillary Tangles Lipid metabolism APOE ABCA 7 CLU SORL 1 DSG 2 MAPT/TAU DBN 1 PLEC Glucose NMDAR Transporter AMPAR n. ACh. R m. ACh. R Apoptosis Oxidative stress Amyloid Plaques Ab Misfolding Aggregation ROS Formation Microglia activation Inflammatory Lipid Cytokines peroxidation Membrane damage Inflammatory response CLU, ABCA 7, CD 33, CR 1, TREM 2, EPHA 1, MEF 2 C. MS 4 A, INPP 5 D, HLADRB 5/HLA-DRB 1

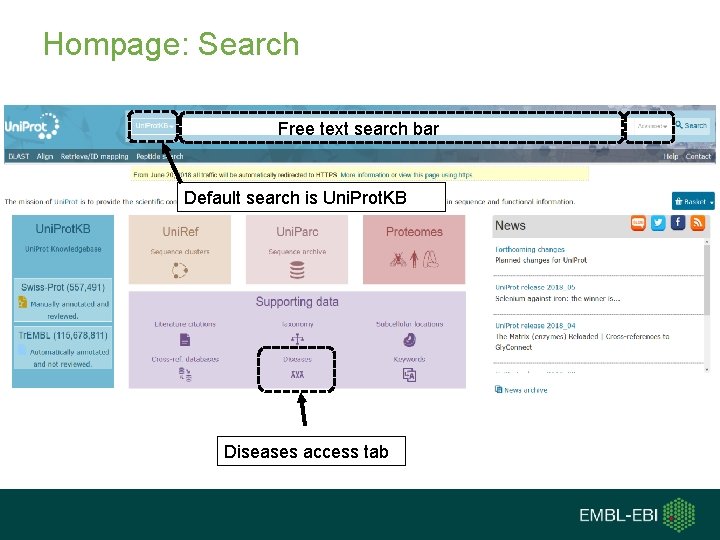
Hompage: Search Free text search bar Default search is Uni. Prot. KB Diseases access tab

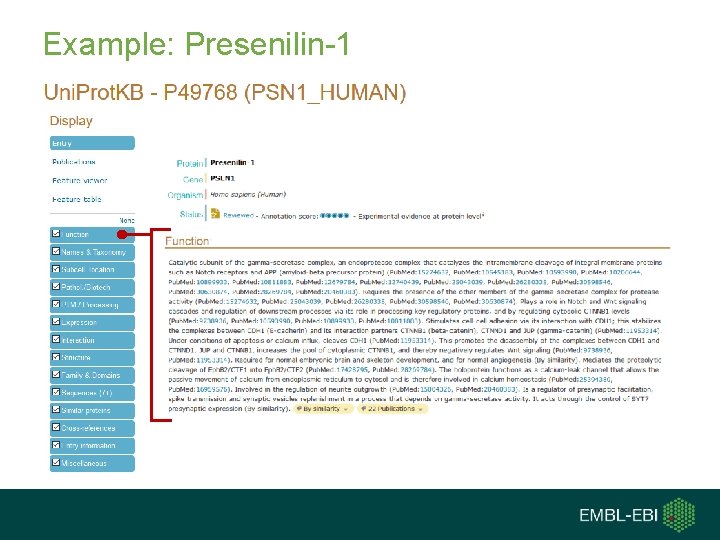
Example: Presenilin-1 Contact us: help@uniprot. org Quick access tabs list entry contents- click to jump to and/or (de)select to toggle content visibility

Example: Presenilin-1

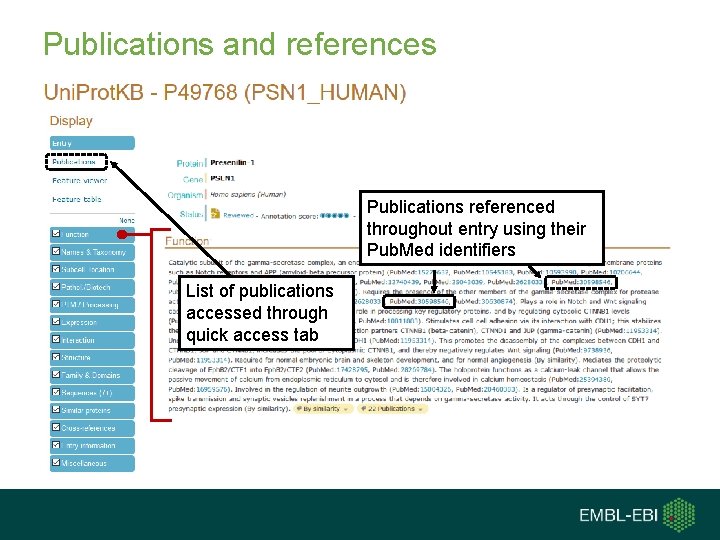
Publications and references Publications referenced throughout entry using their Pub. Med identifiers List of publications accessed through quick access tab

Publications viewer An expanded publications view incorporating filters and access to mapped publications. Hyperlinks to the article in Pub. Med and Europe PMC List of entries that also cite article Indicate the information/data obtained from article and included in entry

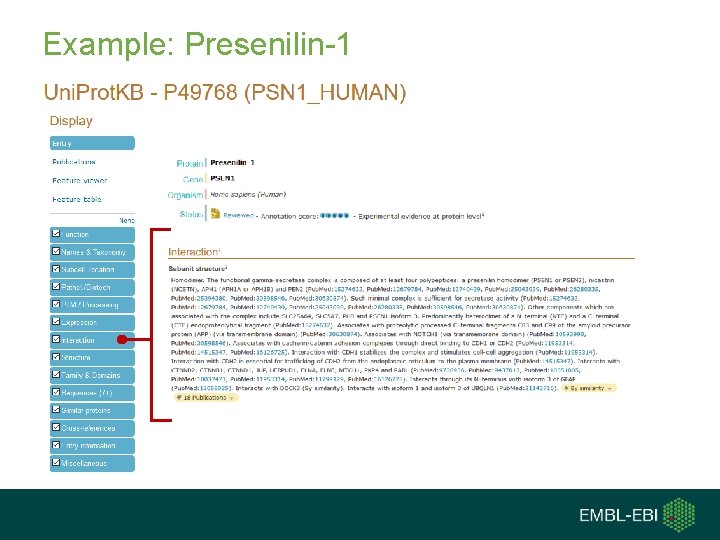
Example: Presenilin-1

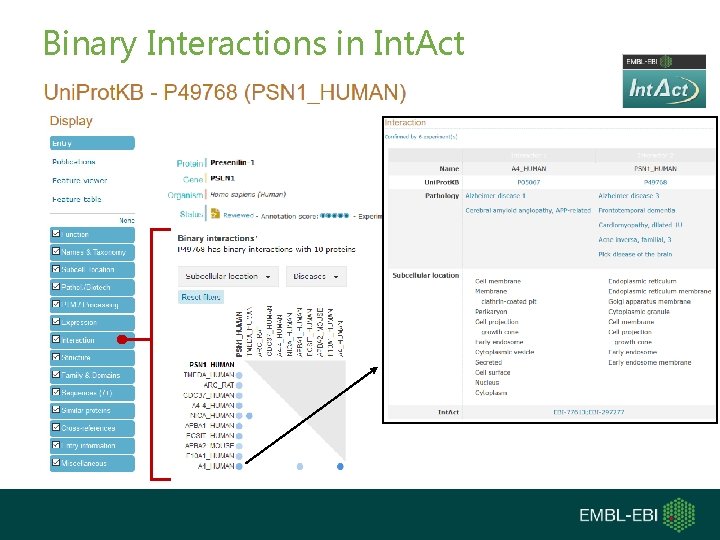
Binary Interactions in Int. Act

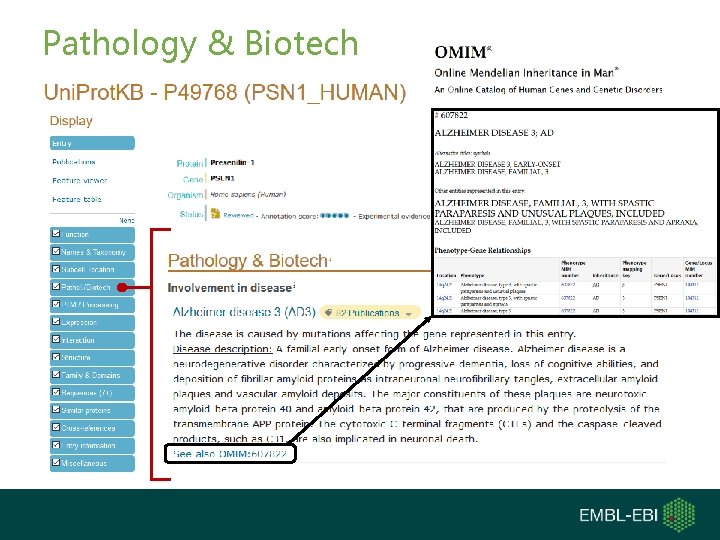
Pathology & Biotech

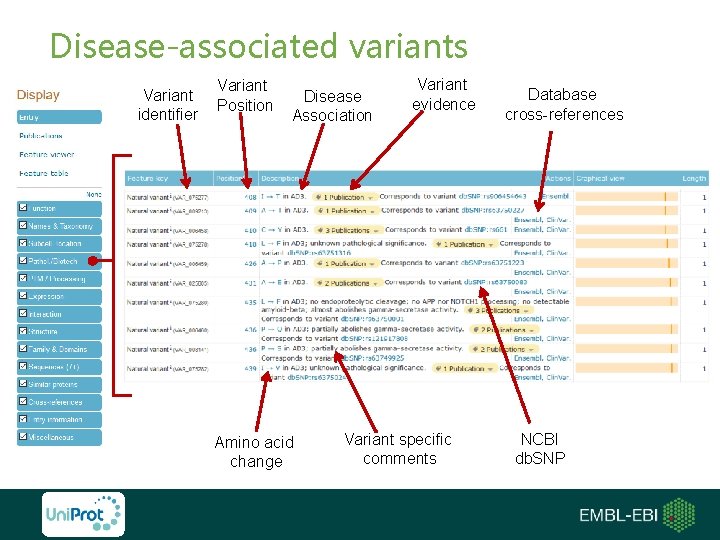
Disease-associated variants Variant identifier Variant Position Disease Association Amino acid change Variant evidence Variant specific comments Database cross-references NCBI db. SNP

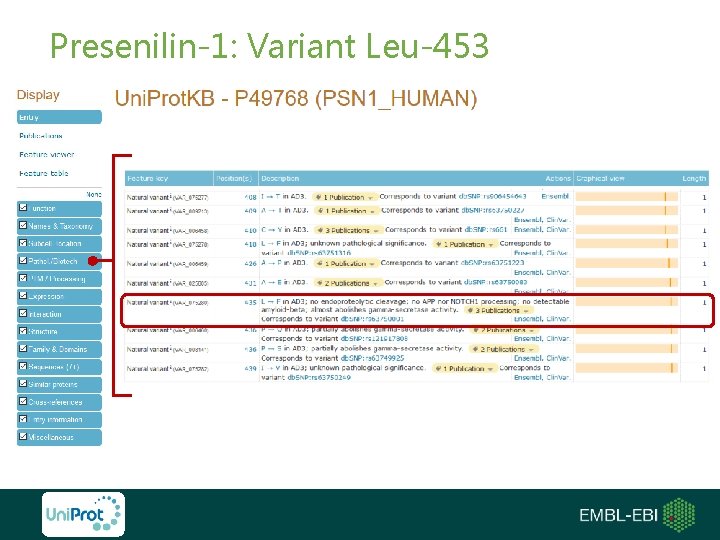
Presenilin-1: Variant Leu-453

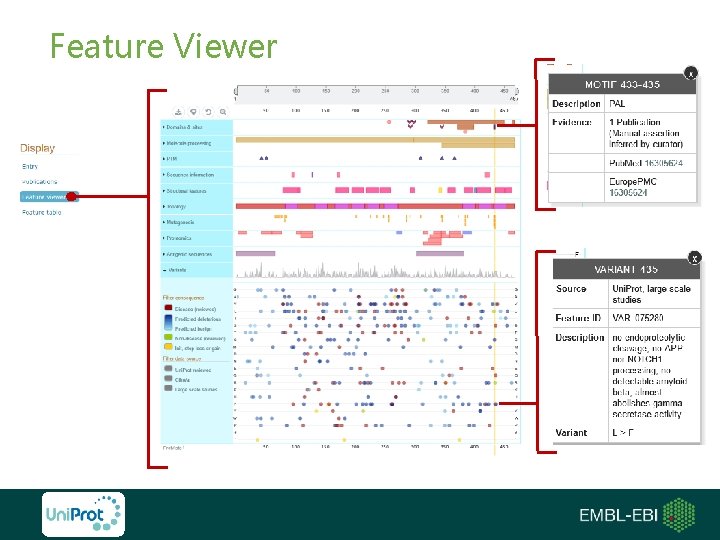
Feature Viewer

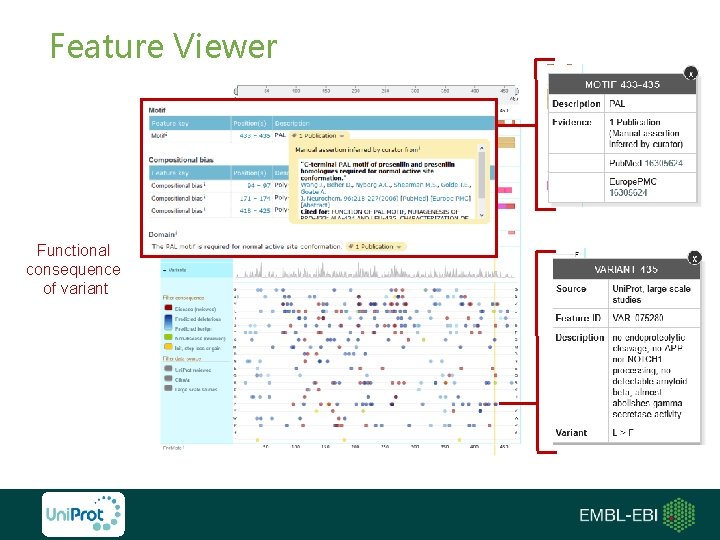
Feature Viewer Functional consequence of variant

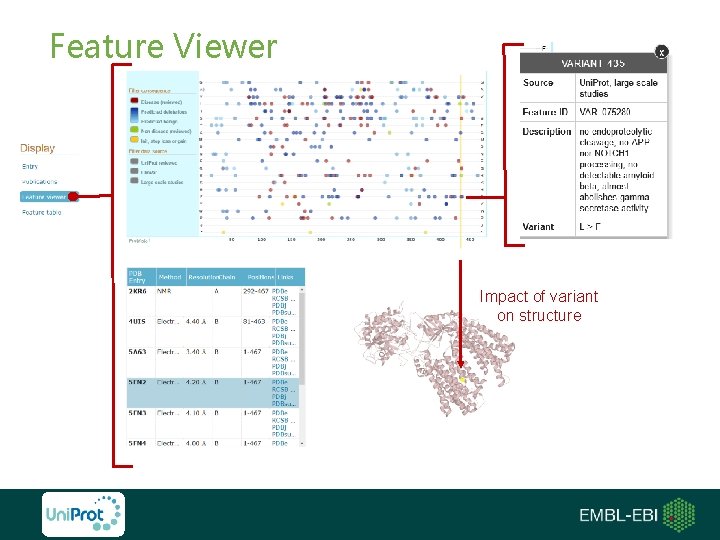
Feature Viewer Impact of variant on structure

Summary • New insights into the molecular function of proteins associated with Alzheimer’s disease and disease mechanisms • Relation between variants and disease and linking molecular defects to disease • Network of proteins and interactions underlying disease development • Overview for researchers in the field of neurodegeneration, clinicians and biomedical researchers

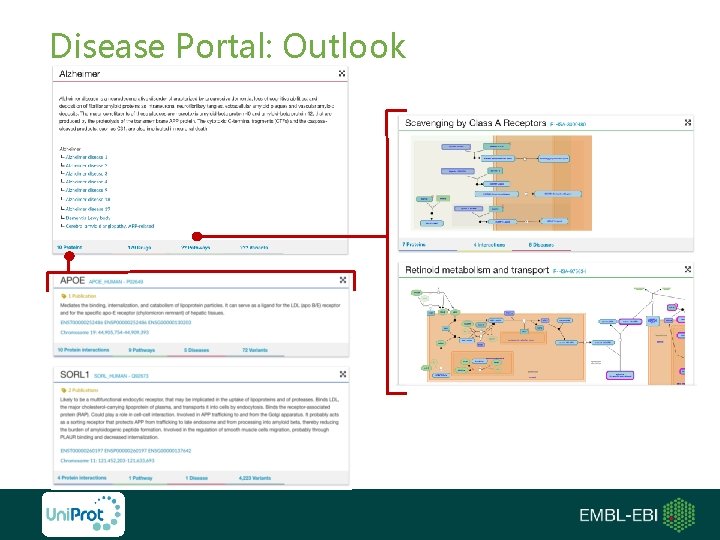
Disease Portal: Outlook

Accessing Uni. Prot Data • Uni. Prot Releases every 4 weeks and is freely available - http: //www. uniprot. org/downloads • Provided in a range of formats - text, XML/RDF, FASTA, GFF, tab-delimited • Web site - Supports simple and complex queries - RESTful API - Documentation and Details at: http: //uniprot. org/help/programmatic_access

Uni. Prot funding This work was supported by the National Eye Institute (NEI), National Human Genome Research Institute (NHGRI), National Heart, Lung, and Blood Institute (NHLBI), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of General Medical Sciences (NIGMS), and National Institute of Mental Health (NIMH) of the National Institutes of Health under Award Number [U 24 HG 007822] (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health). Research reported in this publication was supported by the National Human Genome Research Institute (NHGRI) and the National Institute on Aging (NIA) of the National Institutes of Health under Award Number [3 U 24 HG 00782205 S 1] (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health). National Institutes of Health

Uni. Prot Team PIs: Alex Bateman, Alan Bridge, Cathy Wu Key staff: Cecilia Arighi (Curation), Lionel Breuza (Curation), Elisabeth Coudert (Curation), Hongzhan Huang (Development), Damien Lieberherr (Curation), Michele Magrane (Curation), Maria Martin (Development), Peter Mc. Garvey (Content), Darren Natale (Content), Sandra Orchard (Content), Ivo Pedruzzi (Curation), Sylvain Poux (Curation), Manuela Pruess (Coordination), Shriya Raj (Coordination), Nicole Redaschi (Development) Content / Curation: Lucila Aimo, Ghislaine Argoud-Puy, Andrea Auchincloss, Kristian Axelsen, Emmanuel Boutet, Emily Bowler, Ramona Britto, Hema Bye-A-Jee, Cristina Casals-Casas, Anne Estreicher, Livia Famiglietti, Marc Feuermann, John S. Garavelli, Penelope Garmiri, George Georghiou, Arnaud Gos, Nadine Gruaz, Emma Hatton-Ellis, Ursula Hinz, Chantal Hulo, Nevila Hyka-Nouspikel, Florence Jungo, Kati Laiho, Philippe Lemercier, Yvonne Lussi, Alistair Mac. Dougall, Patrick Masson, Anne Morgat, Sandrine Pilbout, Catherine Rivoire, Karen Ross, Christian Sigrist, Elena Speretta, Shyamala Sundaram, Nidhi Tyagi, C. R. Vinayaka, Qinghua Wang, Kate Warner, Lai-Su Yeh, Rossana Zaru Development: Shadab Ahmed, Emanuele Alpi, Leslie Arminski, Parit Bansal, Delphine Baratin, Teresa Batista Neto, Jerven Bolleman, Borisas Bursteinas, Chuming Chen, Yongxing Chen, Beatrice Cuche, Alan Da Silva, Edouard De Castro, Tunca Dogan, Leyla Garcia Castro, Elisabeth Gasteiger, Sebastien Gehant, Leonardo Gonzales, Alexandr Ignatchenko, Giuseppe Insana, Rizwan Ishtiaq, Vishal Joshi, Dushyanth Jyothi, Arnaud Kerhornou, Thierry Lombardot, Jie Luo, Mahdi Mahmoudy, Andrew Nightingale, Joseph Onwubiko, Monica Pozzato, Sangya Pundir, Guoying Qi, Daniel Rice, Rabie Saidi, Edward Turner, Preethi Vasudev, Vladimir Volynkin, Yuqi Wang, Xavier Watkins, Hermann Zellner, Jian Zhang European Bioinformatics Institute (EMBL-EBI), Hinxton, Cambridge, UK Protein Information Resource (PIR), SIB Swiss Institute of Bioinformatics (SIB), Geneva, Switzerland Washington DC and Delaware, USA
 Uni prot
Uni prot Static vs dynamic linking
Static vs dynamic linking Communicable disease and non communicable disease
Communicable disease and non communicable disease Georgia alzheimers planning
Georgia alzheimers planning Fast scale dementia
Fast scale dementia Alzheimers sjukdom
Alzheimers sjukdom Alzheimers nz conference 2020
Alzheimers nz conference 2020 Site:slidetodoc.com
Site:slidetodoc.com Alzheimers society contented dementia
Alzheimers society contented dementia Alzheimer's eye test joke
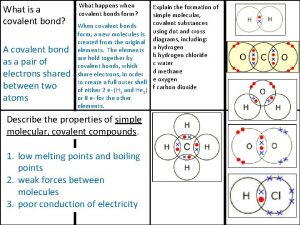
Alzheimer's eye test joke Covalent bond
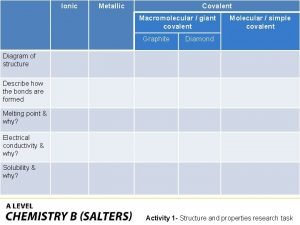
Covalent bond Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Frankel defect
Frankel defect Undercut welding definition
Undercut welding definition Scar defect in casting
Scar defect in casting Diff between schottky and frenkel defects
Diff between schottky and frenkel defects Firmness of the yolk and freedom from yolk defects
Firmness of the yolk and freedom from yolk defects Action verbs and linking verbs
Action verbs and linking verbs Execution defects
Execution defects Twinning tablet defects
Twinning tablet defects What are the aims of secondary education
What are the aims of secondary education Jolt moulding machine
Jolt moulding machine Chain sling defects
Chain sling defects Armany classification
Armany classification What is process sigma
What is process sigma