www uniprot org Uni Prot Nonredundant Reference Cluster
- Slides: 1

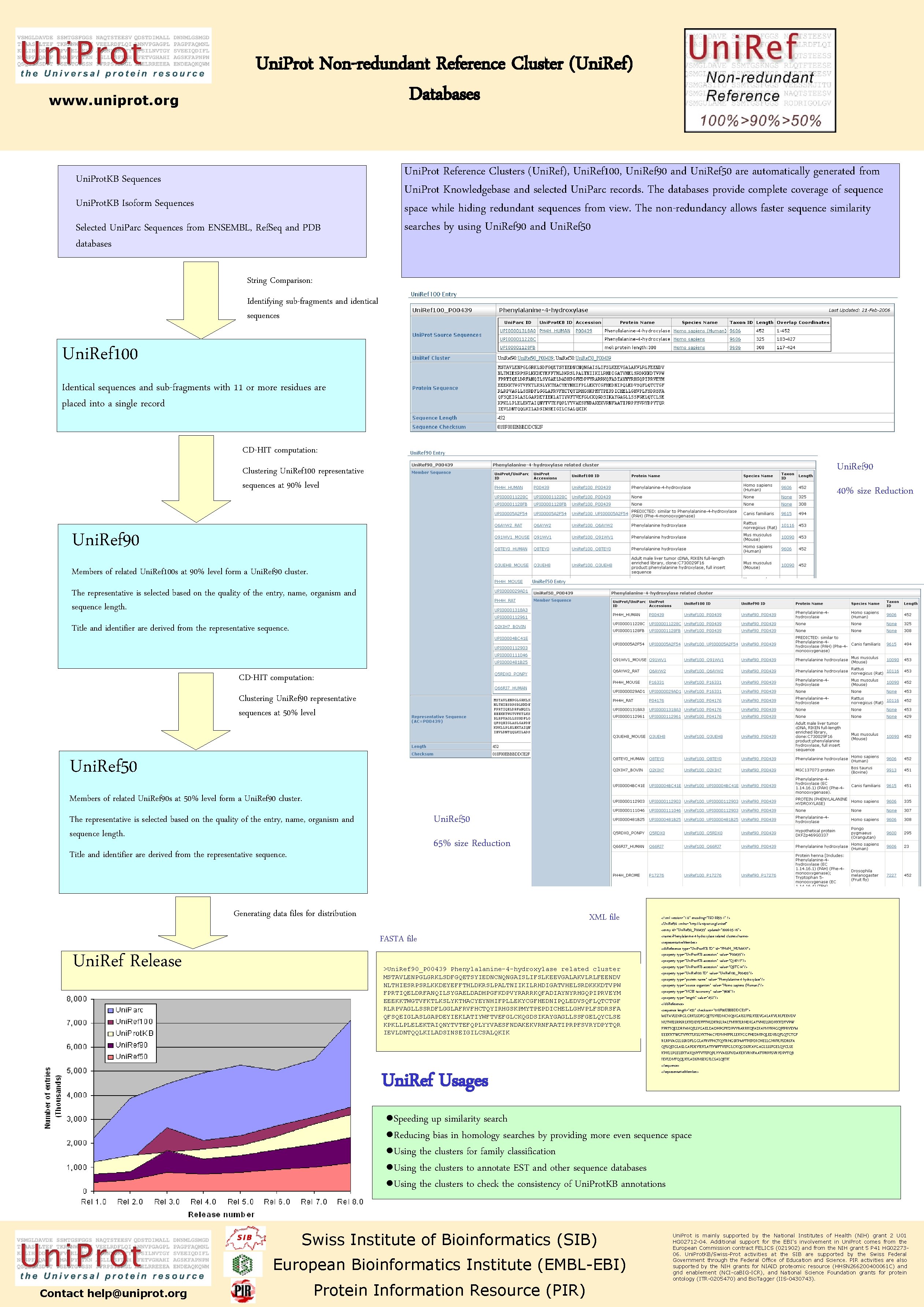
www. uniprot. org Uni. Prot Non-redundant Reference Cluster (Uni. Ref) Databases Uni. Prot. KB Sequences Uni. Prot. KB Isoform Sequences Selected Uni. Parc Sequences from ENSEMBL, Ref. Seq and PDB databases Uni. Prot Reference Clusters (Uni. Ref), Uni. Ref 100, Uni. Ref 90 and Uni. Ref 50 are automatically generated from Uni. Prot Knowledgebase and selected Uni. Parc records. The databases provide complete coverage of sequence space while hiding redundant sequences from view. The non-redundancy allows faster sequence similarity searches by using Uni. Ref 90 and Uni. Ref 50 String Comparison: Identifying sub-fragments and identical sequences Uni. Ref 100 Identical sequences and sub-fragments with 11 or more residues are placed into a single record CD-HIT computation: Uni. Ref 90 Clustering Uni. Ref 100 representative sequences at 90% level 40% size Reduction Uni. Ref 90 Members of related Uni. Ref 100 s at 90% level form a Uni. Ref 90 cluster. The representative is selected based on the quality of the entry, name, organism and sequence length. Title and identifier are derived from the representative sequence. CD-HIT computation: Clustering Uni. Ref 90 representative sequences at 50% level Uni. Ref 50 Members of related Uni. Ref 90 s at 50% level form a Uni. Ref 90 cluster. Uni. Ref 50 The representative is selected based on the quality of the entry, name, organism and sequence length. 65% size Reduction Title and identifier are derived from the representative sequence. Generating data files for distribution XML file FASTA file Uni. Ref Release >Uni. Ref 90_P 00439 Phenylalanine-4 -hydroxylase related cluster MSTAVLENPGLGRKLSDFGQETSYIEDNCNQNGAISLIFSLKEEVGALAKVLRLFEENDV NLTHIESRPSRLKKDEYEFFTHLDKRSLPALTNIIKILRHDIGATVHELSRDKKKDTVPW FPRTIQELDRFANQILSYGAELDADHPGFKDPVYRARRKQFADIAYNYRHGQPIPRVEYM EEEKKTWGTVFKTLKSLYKTHACYEYNHIFPLLEKYCGFHEDNIPQLEDVSQFLQTCTGF RLRPVAGLLSSRDFLGGLAFRVFHCTQYIRHGSKPMYTPEPDICHELLGHVPLFSDRSFA QFSQEIGLASLGAPDEYIEKLATIYWFTVEFGLCKQGDSIKAYGAGLLSSFGELQYCLSE KPKLLPLELEKTAIQNYTVTEFQPLYYVAESFNDAKEKVRNFAATIPRPFSVRYDPYTQR IEVLDNTQQLKILADSINSEIGILCSALQKIK Uni. Ref Usages <? xml version="1. 0" encoding="ISO-8859 -1" ? > <Uni. Ref 90 xmlns="http: //uniprot. org/uniref" <entry id="Uni. Ref 90_P 00439" updated="2006 -05 -16"> <name>Phenylalanine-4 -hydroxylase related cluster</name> <representative. Member> <db. Reference type="Uni. Prot. KB ID" id="PH 4 H_HUMAN"> <property type="Uni. Prot. KB accession" value="P 00439"/> <property type="Uni. Prot. KB accession" value="Q 16717"/> <property type="Uni. Prot. KB accession" value="Q 8 TC 14"/> <property type="Uni. Ref 100 ID" value="Uni. Ref 100_P 00439"/> <property type="protein name" value="Phenylalanine-4 -hydroxylase"/> <property type="source organism" value="Homo sapiens (Human)"/> <property type="NCBI taxonomy" value="9606"/> <property type="length" value="452"/> </db. Reference> <sequence length="452" checksum="018 F 00 EBBBDDCE 2 F"> MSTAVLENPGLGRKLSDFGQETSYIEDNCNQNGAISLIFSLKEEVGALAKVLRLFEENDV NLTHIESRPSRLKKDEYEFFTHLDKRSLPALTNIIKILRHDIGATVHELSRDKKKDTVPW FPRTIQELDRFANQILSYGAELDADHPGFKDPVYRARRKQFADIAYNYRHGQPIPRVEYM EEEKKTWGTVFKTLKSLYKTHACYEYNHIFPLLEKYCGFHEDNIPQLEDVSQFLQTCTGF RLRPVAGLLSSRDFLGGLAFRVFHCTQYIRHGSKPMYTPEPDICHELLGHVPLFSDRSFA QFSQEIGLASLGAPDEYIEKLATIYWFTVEFGLCKQGDSIKAYGAGLLSSFGELQYCLSE KPKLLPLELEKTAIQNYTVTEFQPLYYVAESFNDAKEKVRNFAATIPRPFSVRYDPYTQR IEVLDNTQQLKILADSINSEIGILCSALQKIK </sequence> </representative. Member> ●Speeding up similarity search ●Reducing bias in homology searches by providing more even sequence space ●Using the clusters for family classification ●Using the clusters to annotate EST and other sequence databases ●Using the clusters to check the consistency of Uni. Prot. KB annotations Swiss Institute of Bioinformatics (SIB) European Bioinformatics Institute (EMBL-EBI) Contact help@uniprot. org Protein Information Resource (PIR) Uni. Prot is mainly supported by the National Institutes of Health (NIH) grant 2 U 01 HG 02712 -04. Additional support for the EBI's involvement in Uni. Prot comes from the European Commission contract FELICS (021902) and from the NIH grant 5 P 41 HG 0227306. Uni. Prot. KB/Swiss-Prot activities at the SIB are supported by the Swiss Federal Government through the Federal Office of Education and Science. PIR activities are also supported by the NIH grants for NIAID proteomic resource (HHSN 266200400061 C) and grid enablement (NCI-ca. BIG-ICR), and National Science Foundation grants for protein ontology (ITR-0205470) and Bio. Tagger (IIS-0430743).