The Genetic Code MathCS Camp 19 07 06


































- Slides: 89

The Genetic Code Math-CS Camp, 19. 07. 06, Singapore Mikhail S. Gelfand Research and Training Center of Bioinformatics, Institute for Information Transmission Problems, Moscow, Russia and Department of Bioengineering and Bioinformatics, Moscow State University

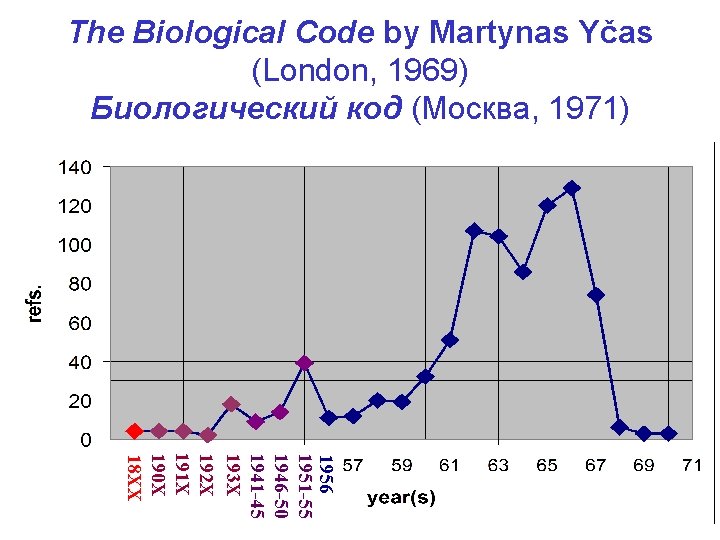
The Biological Code by Martynas Yčas (London, 1969) Биологический код (Mосква, 1971) 1956 1951 -55 1946 -50 1941 -45 193 X 192 X 191 X 190 X 18 XX

To apply mathematics in biology, a mathematician has to understand biology. Israel Gelfand

Plan • Pre-history – Genetics – Evolutionary theory – Chemistry • Cracking the Code • Update

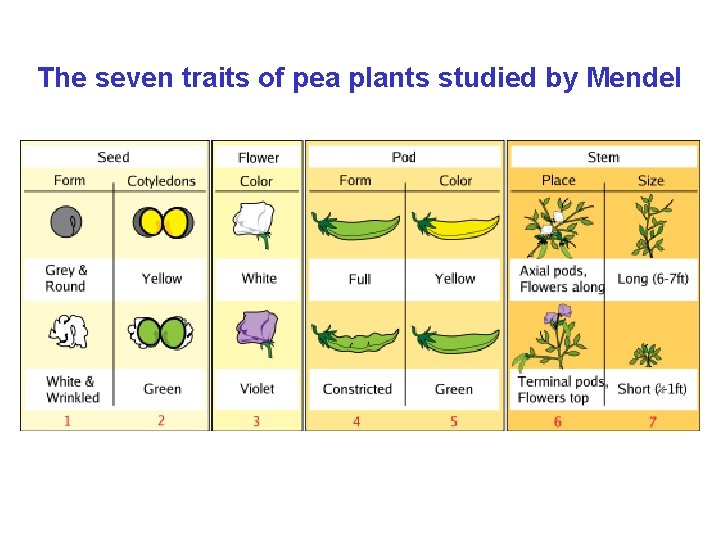
Genetics: Gregor Mendel (1822 -1884) • Attended the Philosophical Institute in Olomouc • Since 1843 – at the Augustinian Abbey of St. Thomas in Brno • 1851 -1853 – studied in the University of Vienna • 1856 -1863 – cultivated 28 thousand pea plants • The Three Laws of Genetics (“Experiments on Plant Hybridization”) – Read to the Natural History Society of Brunn in Bohemia (1865) – Published in Proceedings of the Natural History Society (1866) • Since 1866 – abbot, stopped working in science

The seven traits of pea plants studied by Mendel

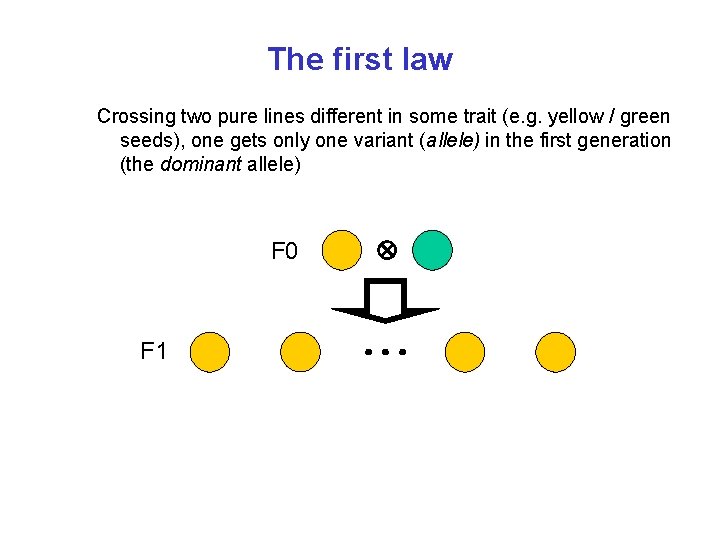
The first law Crossing two pure lines different in some trait (e. g. yellow / green seeds), one gets only one variant (allele) in the first generation (the dominant allele) F 0 F 1

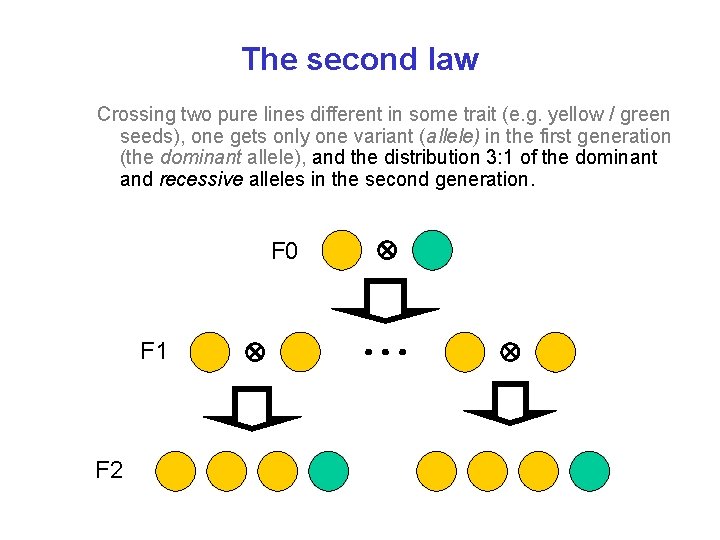
The second law Crossing two pure lines different in some trait (e. g. yellow / green seeds), one gets only one variant (allele) in the first generation (the dominant allele), and the distribution 3: 1 of the dominant and recessive alleles in the second generation. F 0 F 1 F 2

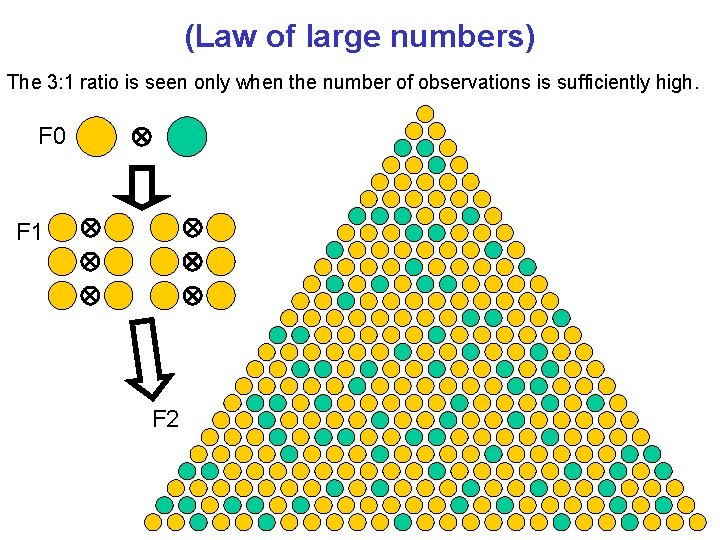
(Law of large numbers) The 3: 1 ratio is seen only when the number of observations is sufficiently high. F 0 F 1 F 2

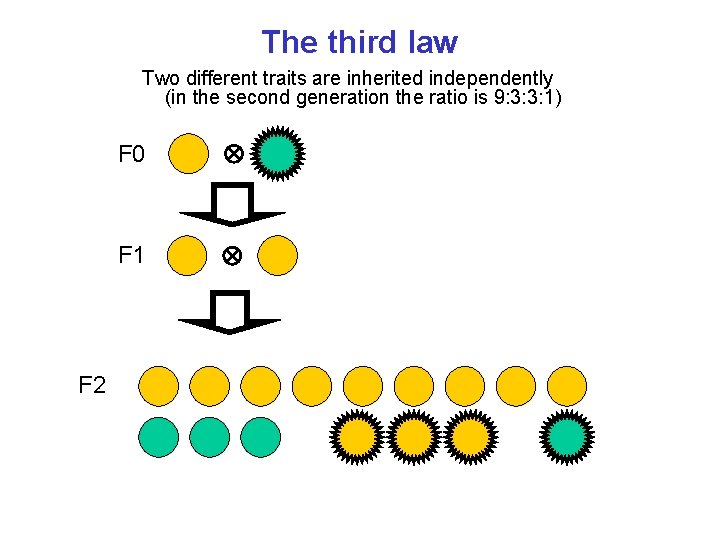
The third law Two different traits are inherited independently (in the second generation the ratio is 9: 3: 3: 1) F 0 F 1 F 2

F 2

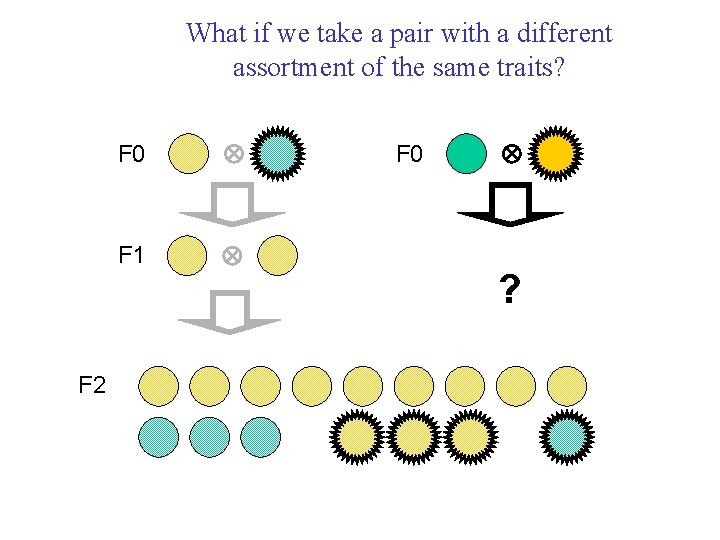
What if we take a pair with a different assortment of the same traits? F 0 F 1 F 2 F 0 ?

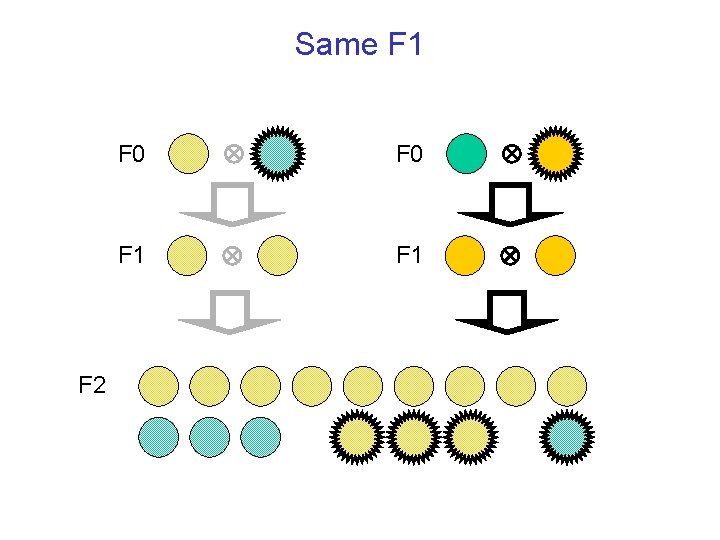
Same F 1 F 2 F 0 F 1

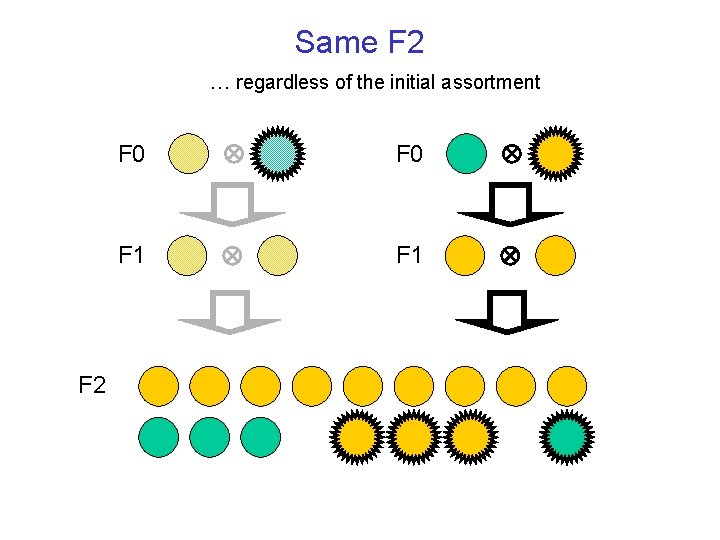
Same F 2 … regardless of the initial assortment F 2 F 0 F 1

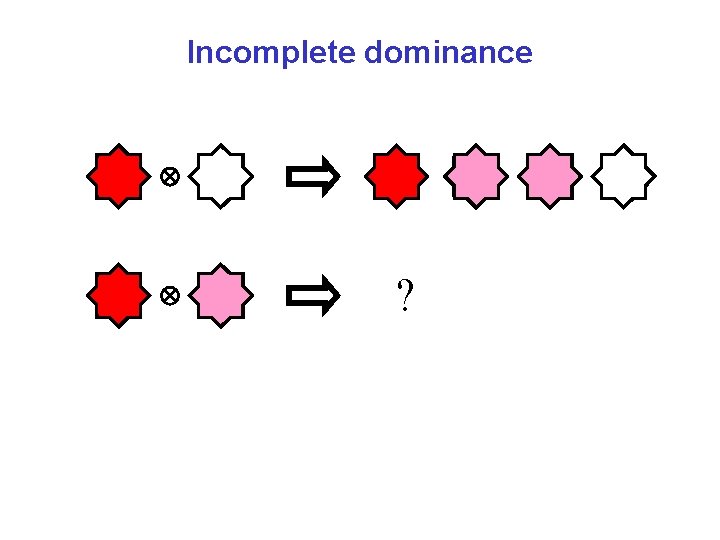
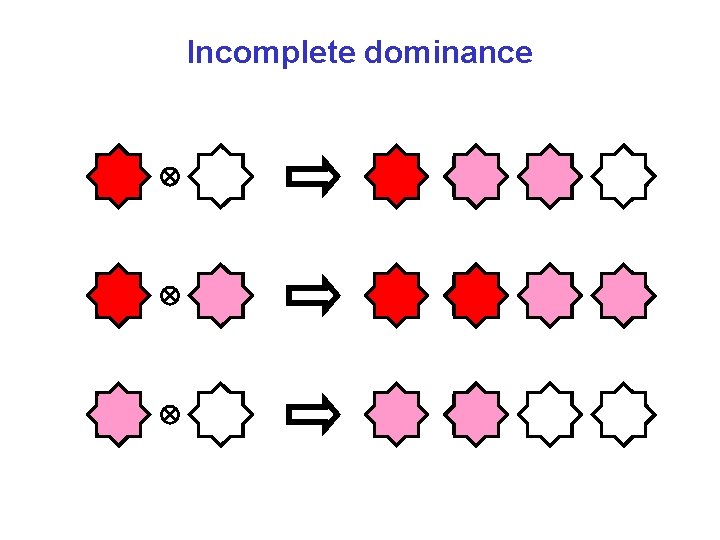
Incomplete dominance

Incomplete dominance ?

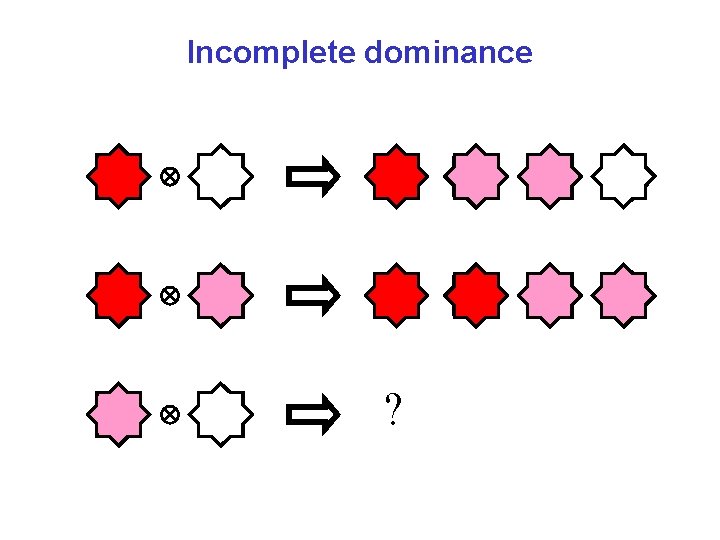
Incomplete dominance ?

Incomplete dominance

Charles Darwin (1809 -1882) • 1825 -27 in Edinburgh University and 182731 in University of Cambridge – natural history, geology, botany • 1831 -1836 – Voyage of the Beagle • Journal of Researches into the Geology and Natural History of the various countries visited by H. M. S. Beagle (1839)

Origin of Species (1859)

The Law of Natural Selection • Species make more offspring than can grow to adulthood. • Populations remain roughly the same size. • Food resources are limited, but are relatively constant most of the time. • In such an environment there will be a struggle for survival among individuals. • In sexually reproducing species, generally no two individuals are identical. • Much of the variation is heritable. • Individuals with the "best" characteristics will be more likely to survive … • … those desirable traits will be passed to their offspring … • … and then inherited by following generations, becoming prevalent and then fixed among the population through time.

Thomas Huxley (1825 -1895) “Darwin’s Bulldog”

Origin of Homo sapiens

Re-discovery of the Mendel laws and emergence of modern genetics • Hugo de Vries (1900) • William Bateson – genetics, gene, allele • Walter Sutton – Link between genes and chromosomes(1902) • Archibald Garrod – Genetic cause of some human disease (1902 -08 -23) • Thomas Morgan, work on Drosophila. – Mutants: spontaneous appearance of new alleles (a fly with white eyes in a population of flies with red eyes) (1908) – Universal acceptance of chromosomes (1915)

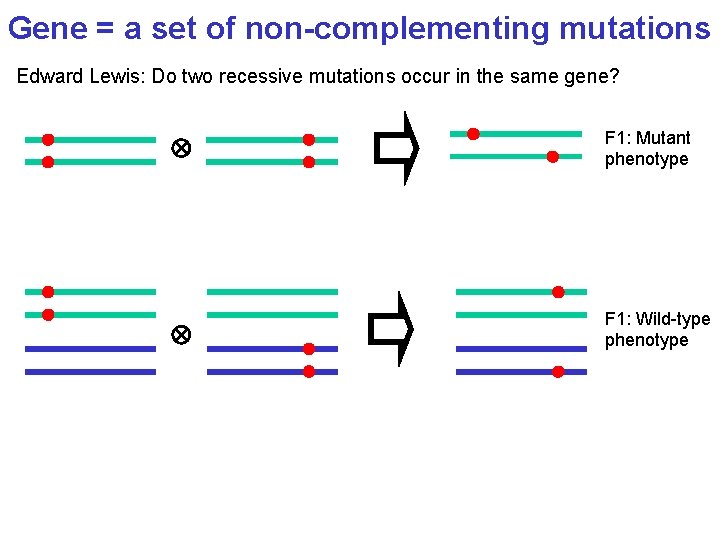
Gene = a set of non-complementing mutations Edward Lewis: Do two recessive mutations occur in the same gene? F 1: Mutant phenotype F 1: Wild-type phenotype

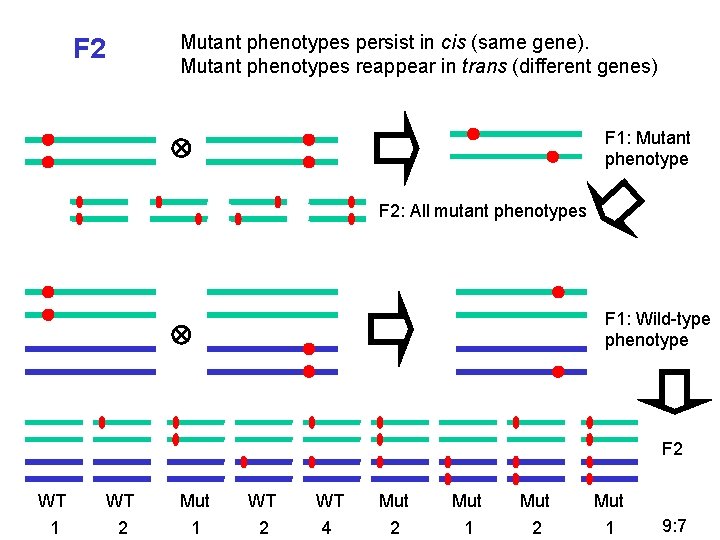
Mutant phenotypes persist in cis (same gene). Mutant phenotypes reappear in trans (different genes) F 2 F 1: Mutant phenotype F 2: All mutant phenotypes F 1: Wild-type phenotype F 2 WT WT Mut Mut Mut 1 2 4 2 1 9: 7

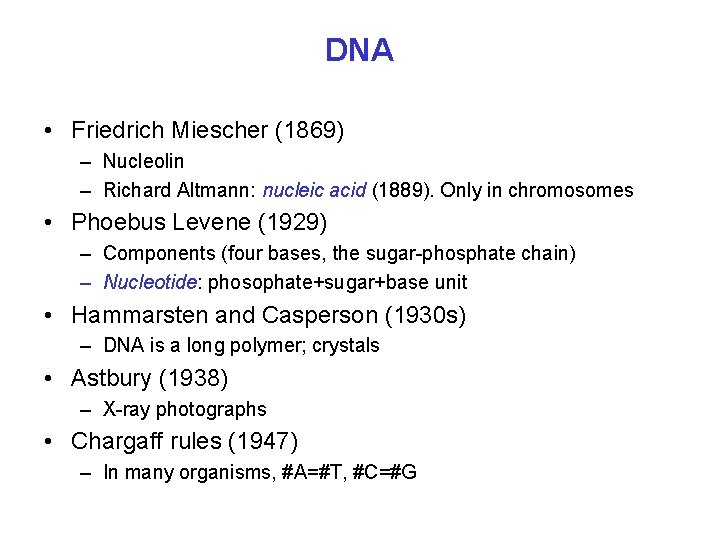
DNA • Friedrich Miescher (1869) – Nucleolin – Richard Altmann: nucleic acid (1889). Only in chromosomes • Phoebus Levene (1929) – Components (four bases, the sugar-phosphate chain) – Nucleotide: phosophate+sugar+base unit • Hammarsten and Casperson (1930 s) – DNA is a long polymer; crystals • Astbury (1938) – X-ray photographs • Chargaff rules (1947) – In many organisms, #A=#T, #C=#G

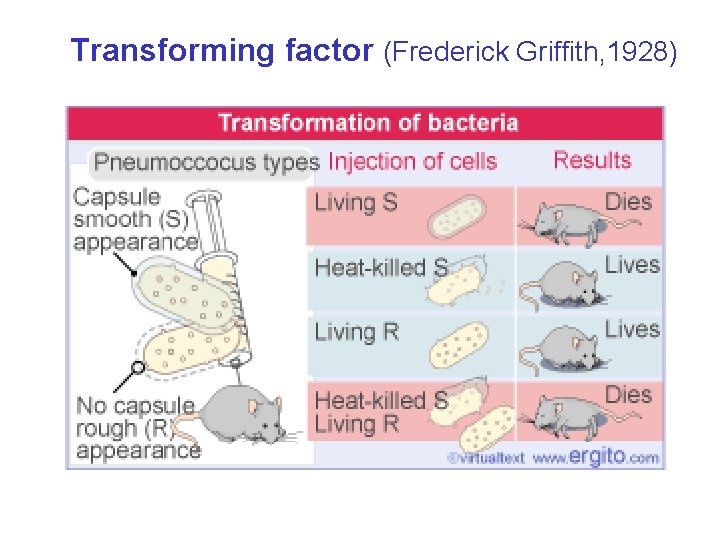
Transforming factor (Frederick Griffith, 1928)

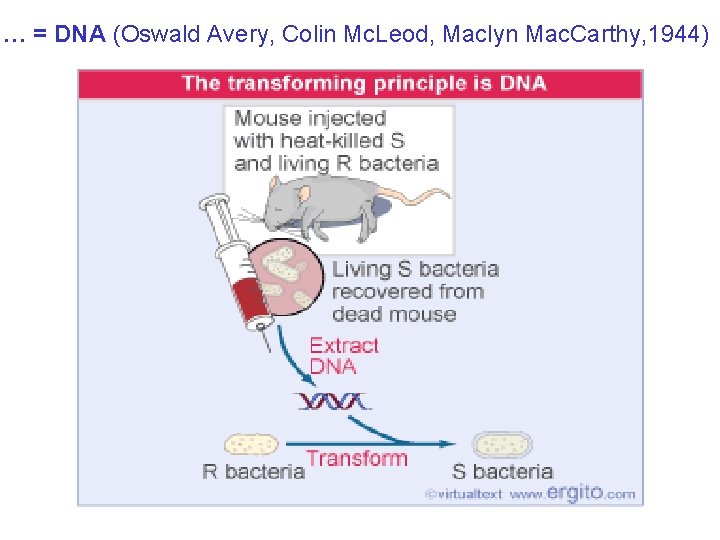
… = DNA (Oswald Avery, Colin Mc. Leod, Maclyn Mac. Carthy, 1944)

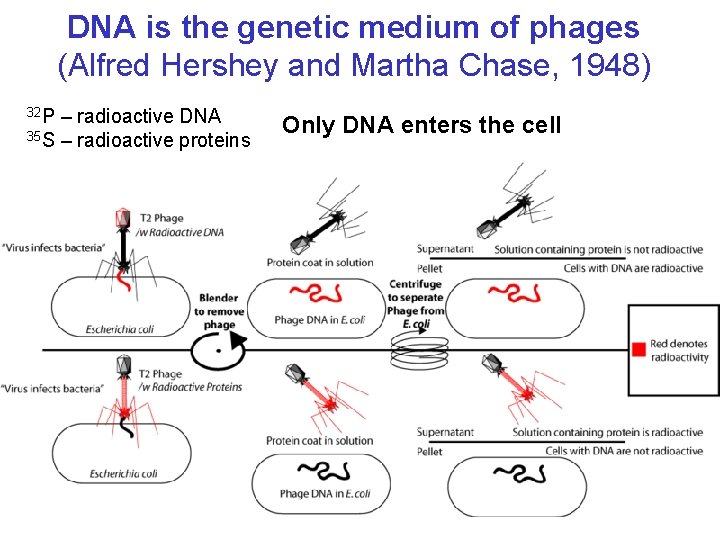
DNA is the genetic medium of phages (Alfred Hershey and Martha Chase, 1948) 32 P – radioactive DNA 35 S – radioactive proteins Only DNA enters the cell

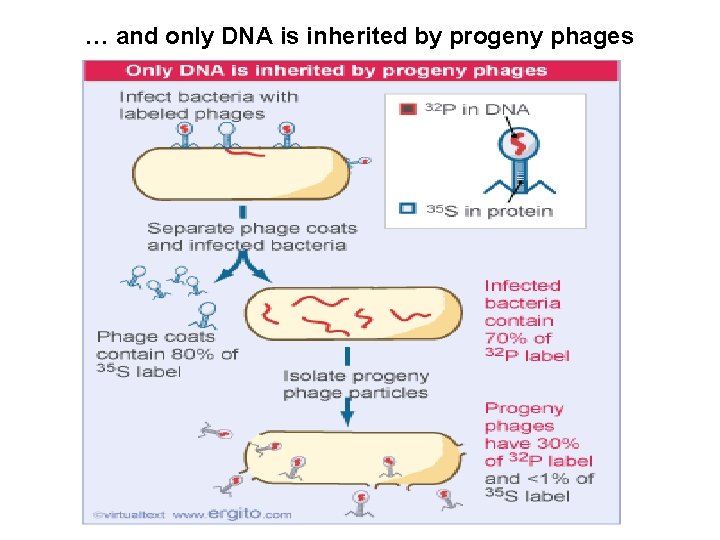
… and only DNA is inherited by progeny phages

Erwin Schrödinger “What is life”, 1946: The gene is an aperiodic crystal

The structure of DNA … • Maurice Wilkins and Rosalind Franklin: high-resolution crystals (1950 -1953)

… is the double helix James Watson and Francis Crick (1953)

The Nature paper: a few lines more than one page

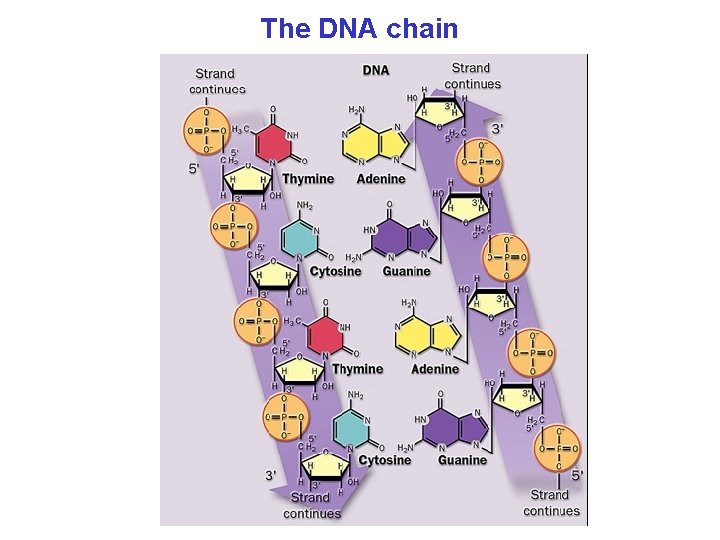
The DNA chain

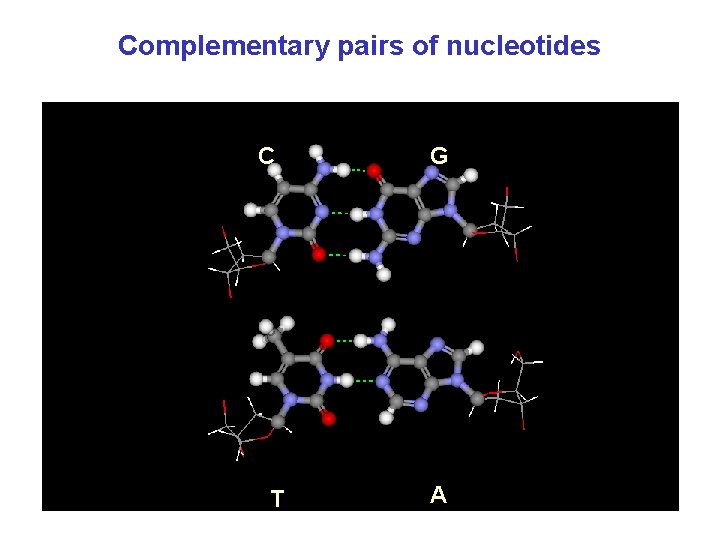
Complementary pairs of nucleotides С Т G A

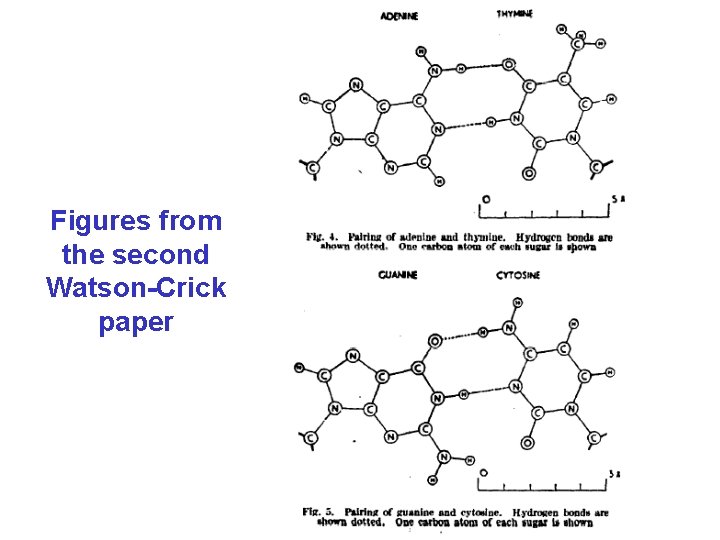
Figures from the second Watson-Crick paper

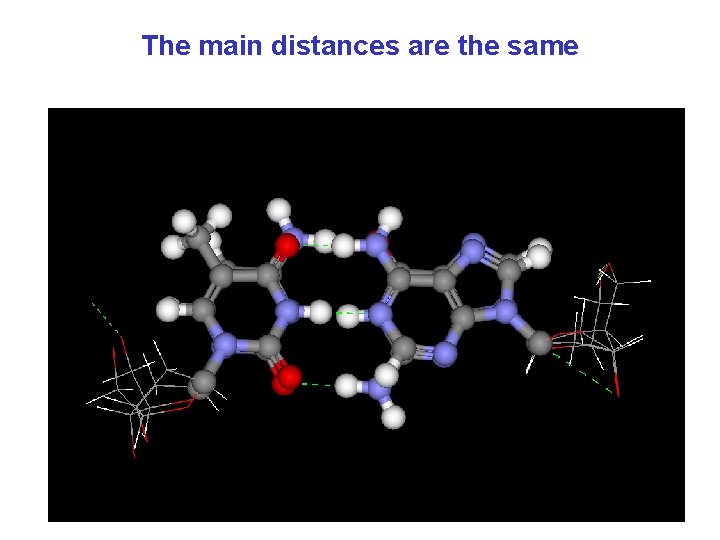
The main distances are the same

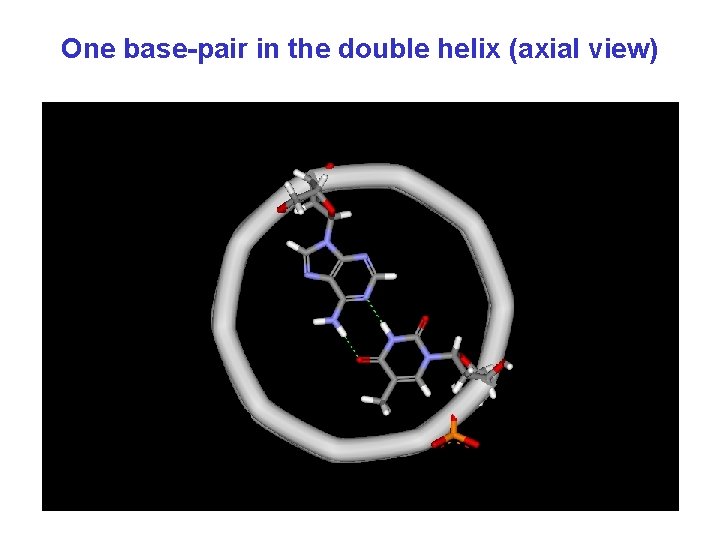
One base-pair in the double helix (axial view)

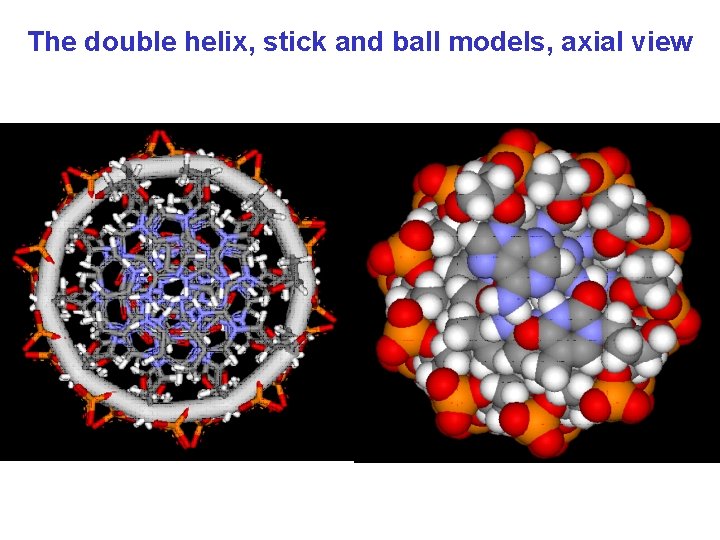
The double helix, stick and ball models, axial view

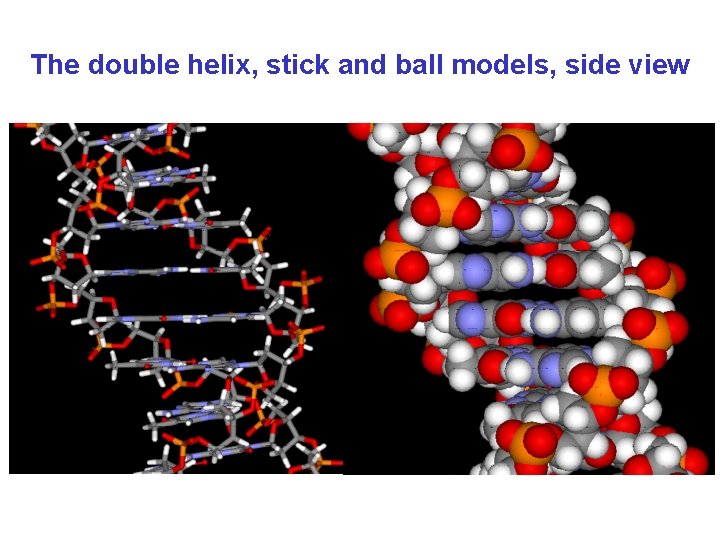
The double helix, stick and ball models, side view

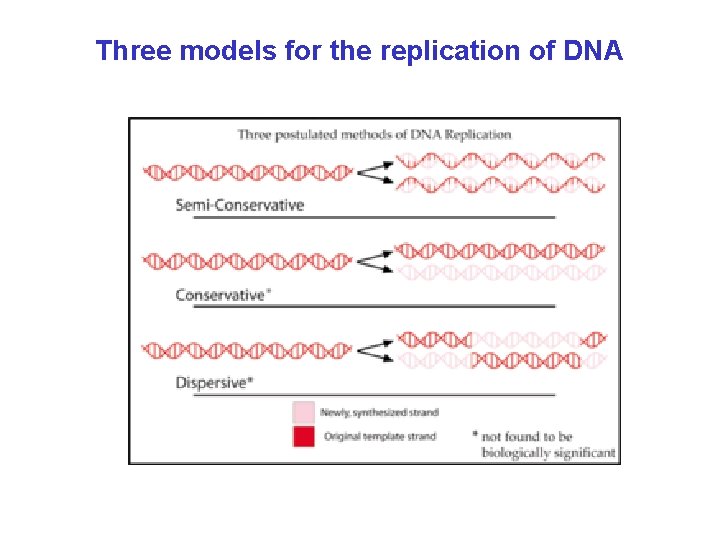
Three models for the replication of DNA

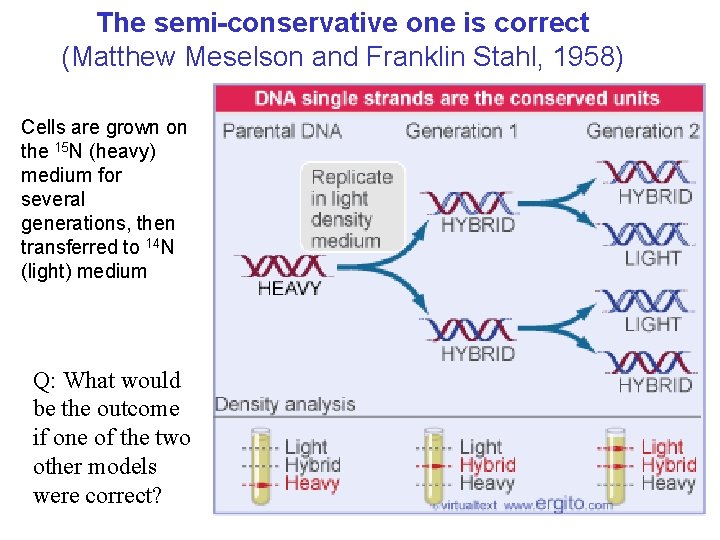
The semi-conservative one is correct (Matthew Meselson and Franklin Stahl, 1958) Cells are grown on the 15 N (heavy) medium for several generations, then transferred to 14 N (light) medium Q: What would be the outcome if one of the two other models were correct?

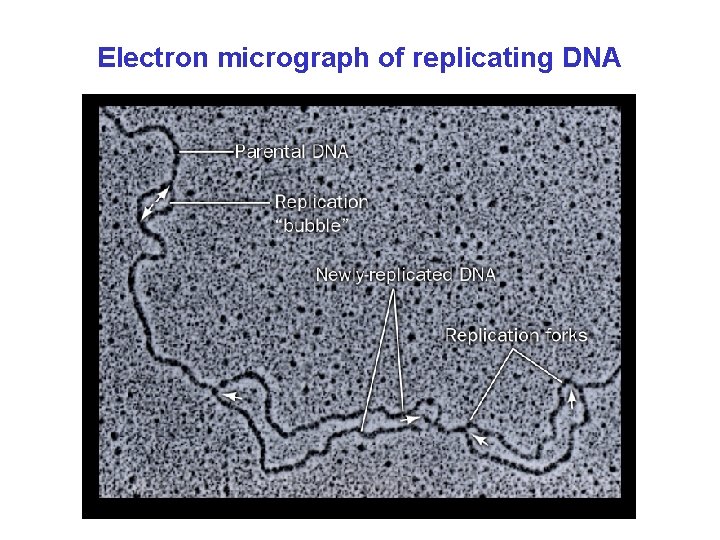
Electron micrograph of replicating DNA

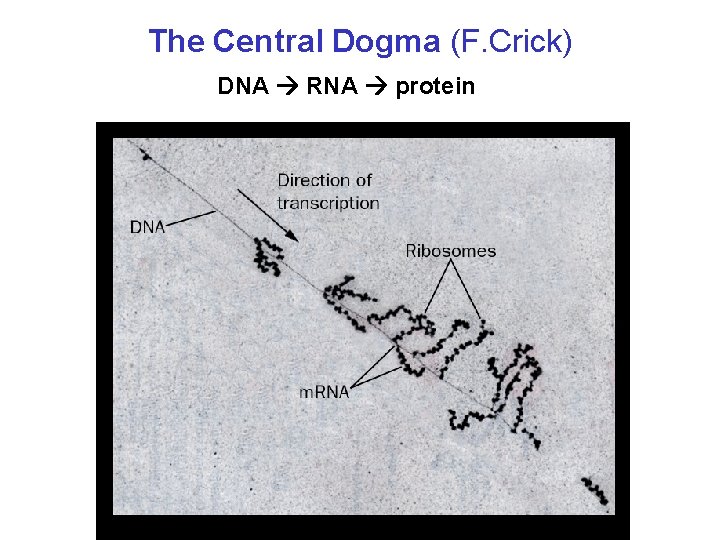
The Central Dogma (F. Crick) DNA RNA protein

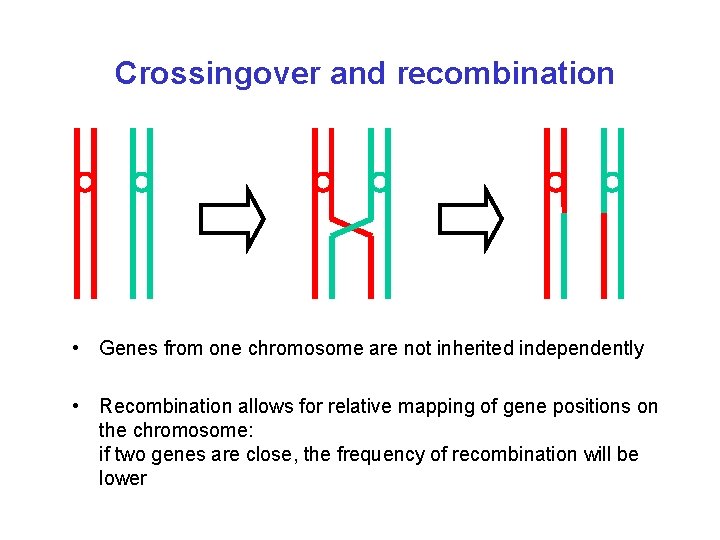
Crossingover and recombination • Genes from one chromosome are not inherited independently • Recombination allows for relative mapping of gene positions on the chromosome: if two genes are close, the frequency of recombination will be lower

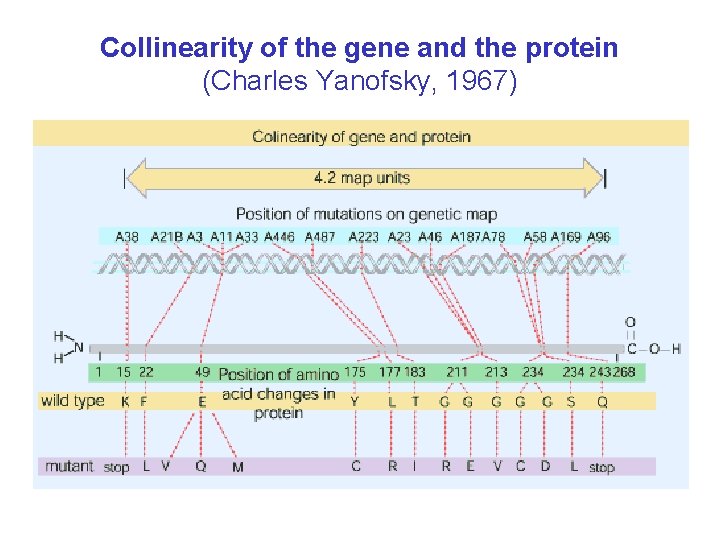
Collinearity of the gene and the protein (Charles Yanofsky, 1967)

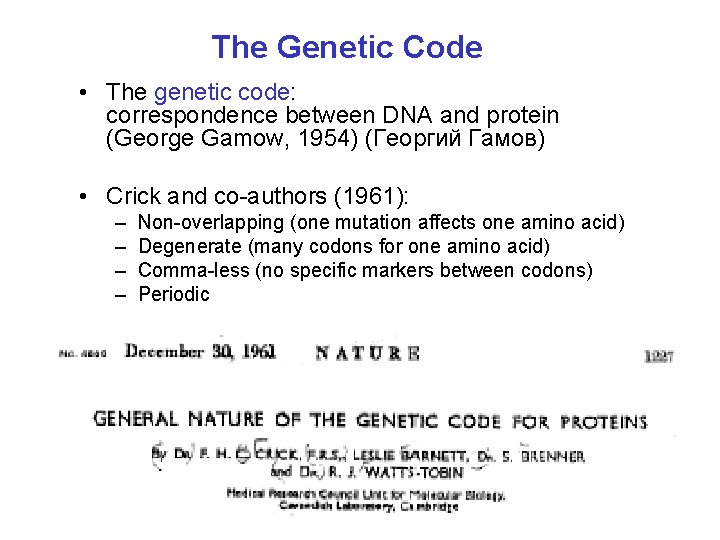
The Genetic Code • The genetic code: correspondence between DNA and protein (George Gamow, 1954) (Георгий Гамов) • Crick and co-authors (1961): – – Non-overlapping (one mutation affects one amino acid) Degenerate (many codons for one amino acid) Comma-less (no specific markers between codons) Periodic

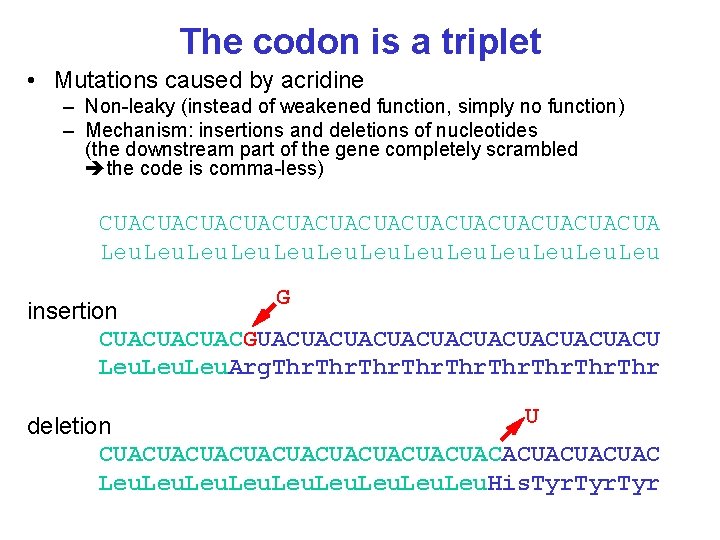
The codon is a triplet • Mutations caused by acridine – Non-leaky (instead of weakened function, simply no function) – Mechanism: insertions and deletions of nucleotides (the downstream part of the gene completely scrambled the code is comma-less) CUACUACUACUACUACUACUA Leu. Leu G insertion CUACUACUACGUACUACUACUACUACU Leu. Arg. Thr U deletion CUACUACUACUACUACACUACUACUAC Leu. His. Tyr

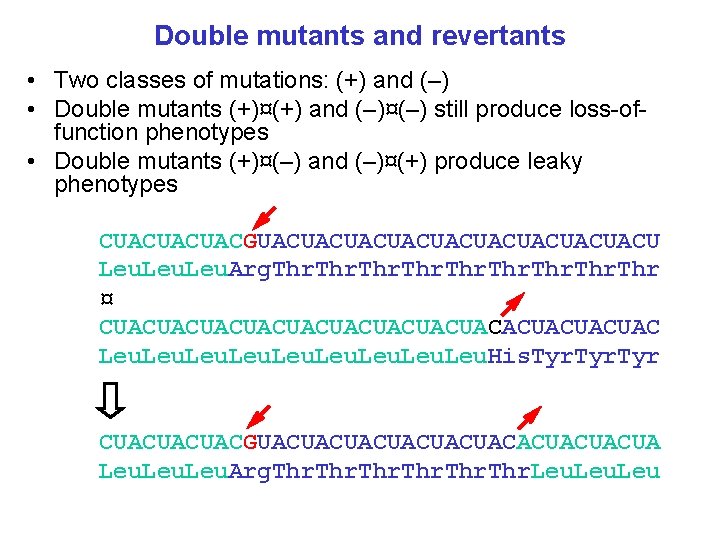
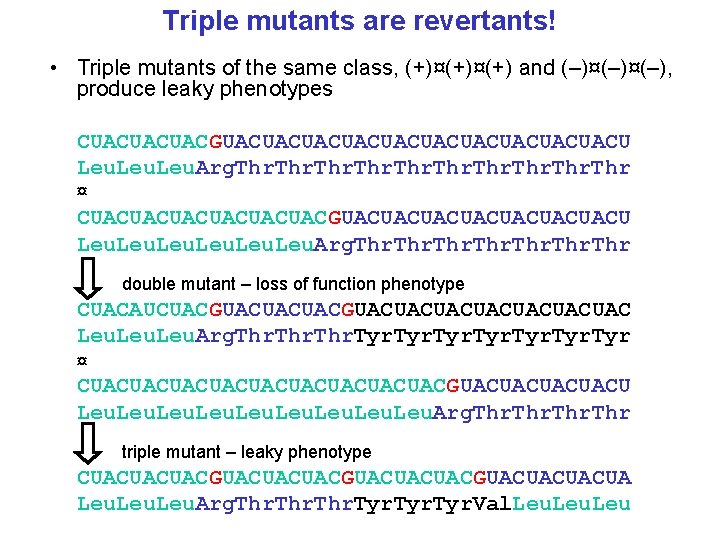
Double mutants and revertants • Two classes of mutations: (+) and (–) • Double mutants (+)¤(+) and (–)¤(–) still produce loss-offunction phenotypes • Double mutants (+)¤(–) and (–)¤(+) produce leaky phenotypes CUACUACUACGUACUACUACUACUACU Leu. Arg. Thr ¤ CUACUACUACUACUACACUACUACUAC Leu. His. Tyr CUACUACUACGUACUACUACACUACUACUA Leu. Arg. Thr. Leu




Triple mutants are revertants! • Triple mutants of the same class, (+)¤(+) and (–)¤(–), produce leaky phenotypes CUACUACUACGUACUACUACUACUACU Leu. Arg. Thr ¤ CUACUACUACGUACUACUACUACU Leu. Arg. Thr double mutant – loss of function phenotype CUACAUCUACGUACUACUACUACUAC Leu. Arg. Thr. Tyr ¤ CUACUACUACUACUACGUACUACU Leu. Arg. Thr triple mutant – leaky phenotype CUACUACUACGUACUACUACGUACUACUACUA Leu. Arg. Thr. Tyr. Val. Leu


Cracking the Code (F. Crick, M. Nirenberg, J. Matthaei, S. Ochoa, G. Khorana, … and you) • Regular oligonucleotides – … UUUUU … – … UCUCUC … – … UCAUCAUCAU … • Random oligonucleotides with known composition • Changes in proteins caused by deaminationcaused mutations: C U, A G • Changes in proteins caused random mutations • (t. RNA binding in the presense of trinucleotides)

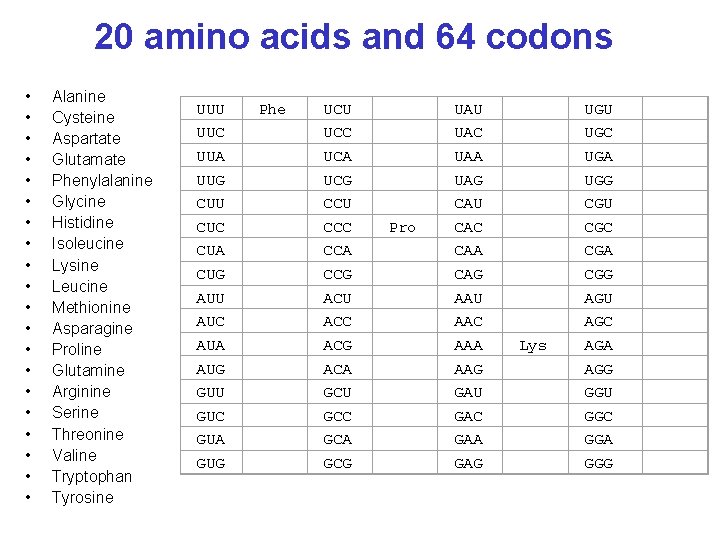
20 amino acids and 64 codons • • • • • Alanine Cysteine Aspartate Glutamate Phenylalanine Glycine Histidine Isoleucine Lysine Leucine Methionine Asparagine Proline Glutamine Arginine Serine Threonine Valine Tryptophan Tyrosine UUU UUC UUA UUG CUU CUC CUA CUG AUU AUC AUA AUG GUU GUC GUA GUG Phe UCU UCC UCA UCG CCU CCC CCA CCG ACU ACC ACG ACA GCU GCC GCA GCG Pro UAU UAC UAA UAG CAU CAC CAA CAG AAU AAC AAA AAG GAU GAC GAA GAG Lys UGU UGC UGA UGG CGU CGC CGA CGG AGU AGC AGA AGG GGU GGC GGA GGG

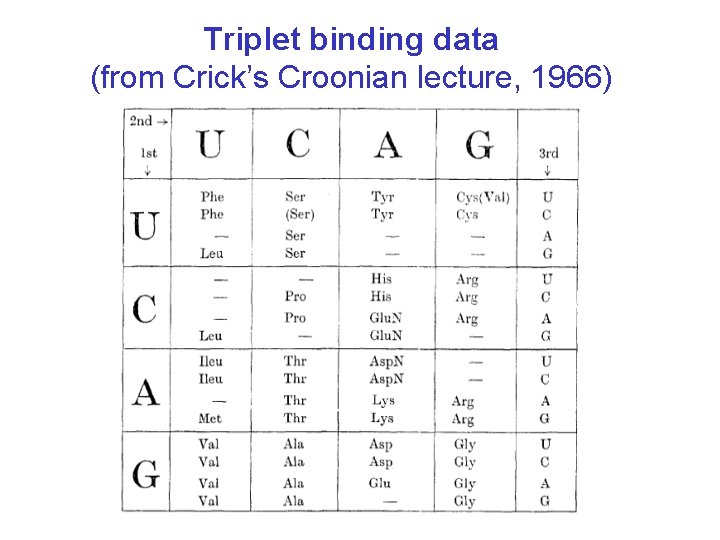
Triplet binding data (from Crick’s Croonian lecture, 1966)

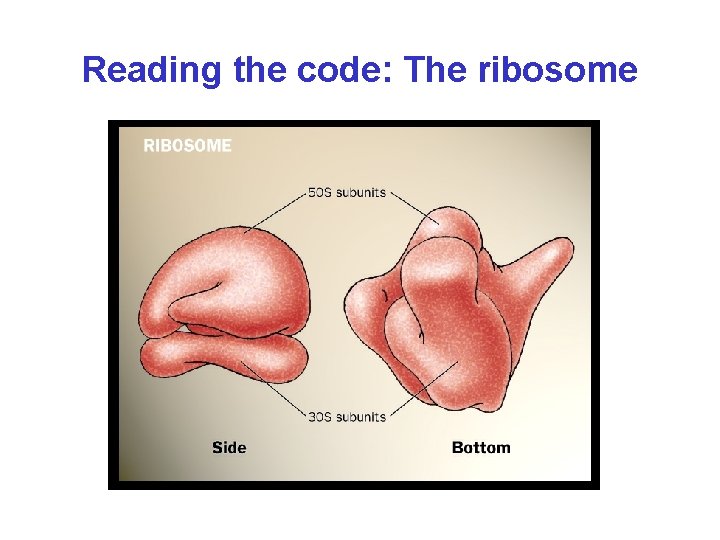
Reading the code: The ribosome

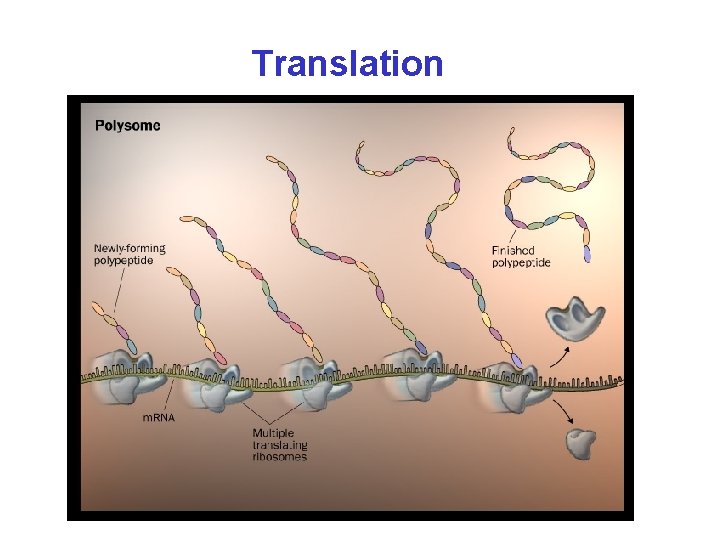
Translation

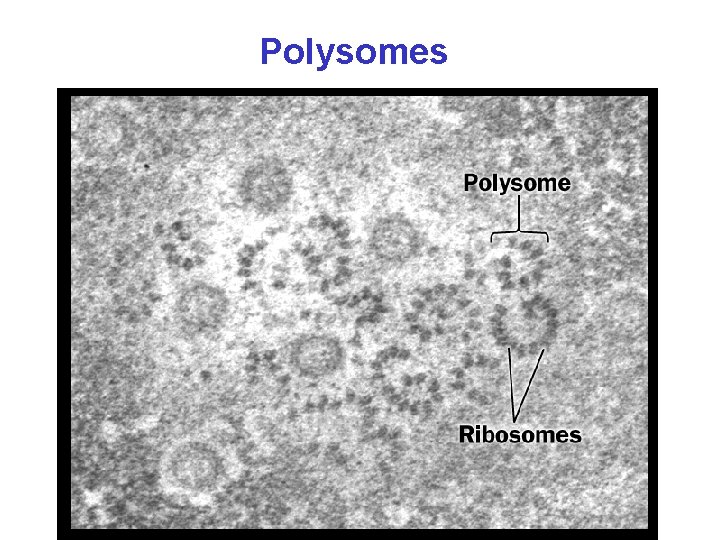
Polysomes

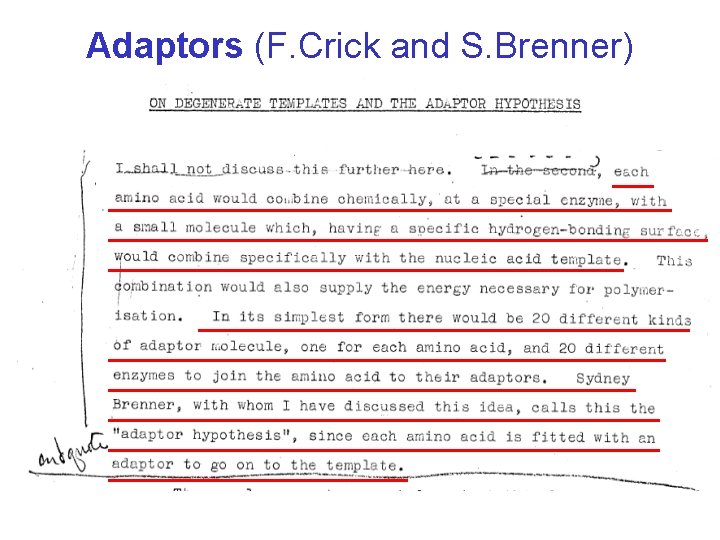
Adaptors (F. Crick and S. Brenner)

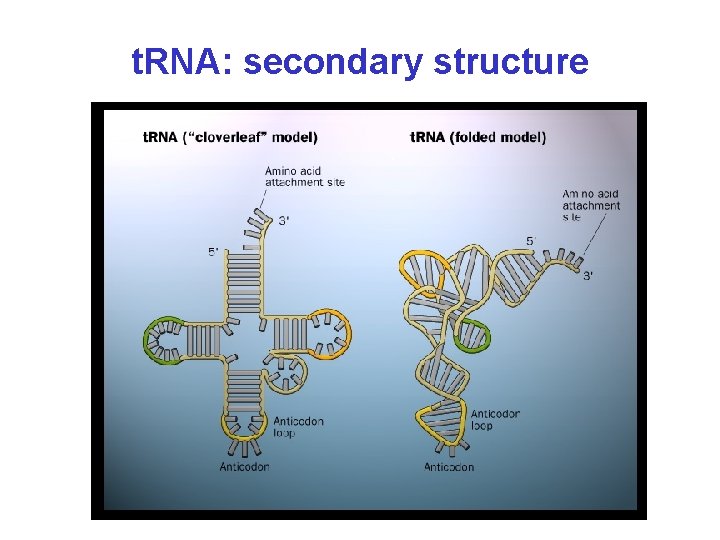
t. RNA: secondary structure

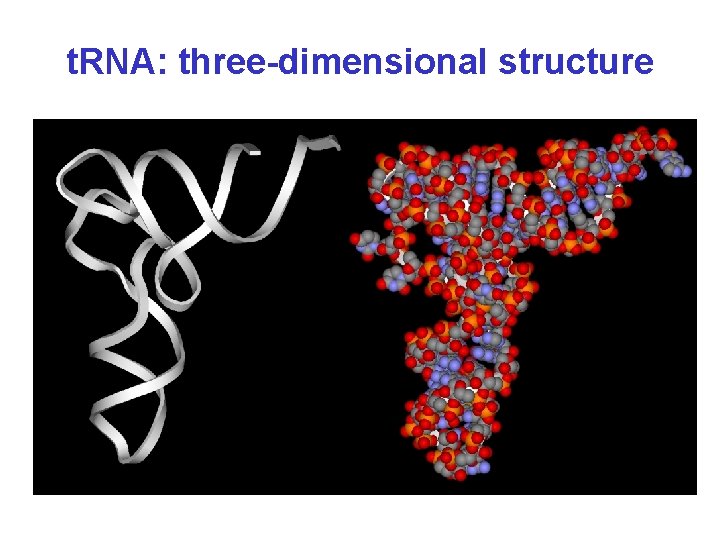
t. RNA: three-dimensional structure

t. RNA and aminoacid-t. RNA-synthetase

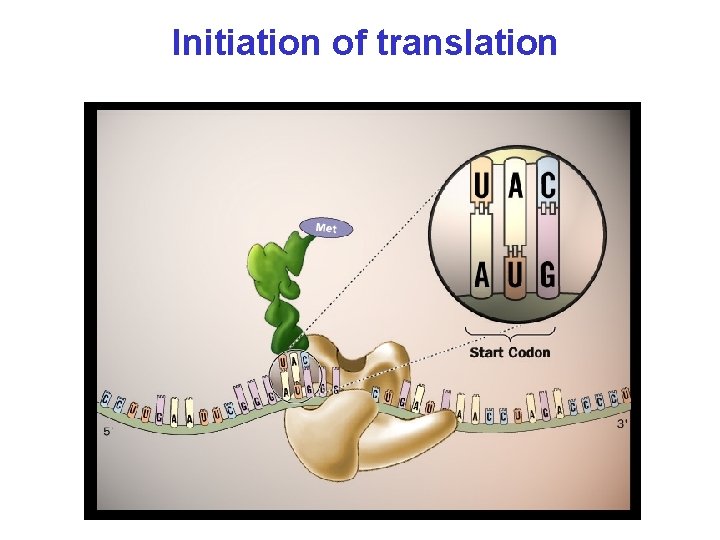
Initiation of translation

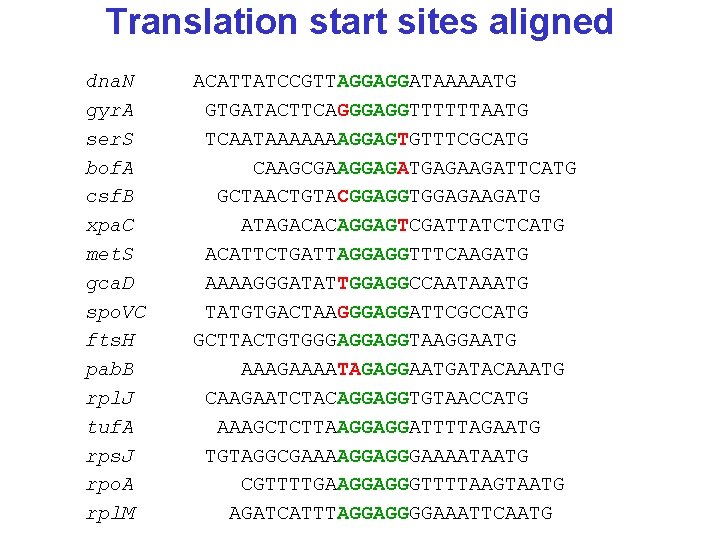
Translation start sites dna. N gyr. A ser. S bof. A csf. B xpa. C met. S gca. D spo. VC fts. H pab. B rpl. J tuf. A rps. J rpo. A rpl. M ACATTATCCGTTAGGAGGATAAAAATG GTGATACTTCAGGGAGGTTTTTTAATG TCAATAAAAAAAGGAGTGTTTCGCATG CAAGCGAAGGAGATGAGAAGATTCATG GCTAACTGTACGGAGGTGGAGAAGATG ATAGACACAGGAGTCGATTATCTCATG ACATTCTGATTAGGAGGTTTCAAGATG AAAAGGGATATTGGAGGCCAATAAATG TATGTGACTAAGGGAGGATTCGCCATG GCTTACTGTGGGAGGAGGTAAGGAATG AAAGAAAATAGAGGAATGATACAAATG CAAGAATCTACAGGAGGTGTAACCATG AAAGCTCTTAAGGAGGATTTTAGAATG TGTAGGCGAAAAGGAGGGAAAATAATG CGTTTTGAAGGAGGGTTTTAAGTAATG AGATCATTTAGGAGGGGAAATTCAATG

Translation start sites aligned dna. N gyr. A ser. S bof. A csf. B xpa. C met. S gca. D spo. VC fts. H pab. B rpl. J tuf. A rps. J rpo. A rpl. M ACATTATCCGTTAGGAGGATAAAAATG GTGATACTTCAGGGAGGTTTTTTAATG TCAATAAAAAAAGGAGTGTTTCGCATG CAAGCGAAGGAGATGAGAAGATTCATG GCTAACTGTACGGAGGTGGAGAAGATG ATAGACACAGGAGTCGATTATCTCATG ACATTCTGATTAGGAGGTTTCAAGATG AAAAGGGATATTGGAGGCCAATAAATG TATGTGACTAAGGGAGGATTCGCCATG GCTTACTGTGGGAGGAGGTAAGGAATG AAAGAAAATAGAGGAATGATACAAATG CAAGAATCTACAGGAGGTGTAACCATG AAAGCTCTTAAGGAGGATTTTAGAATG TGTAGGCGAAAAGGAGGGAAAATAATG CGTTTTGAAGGAGGGTTTTAAGTAATG AGATCATTTAGGAGGGGAAATTCAATG

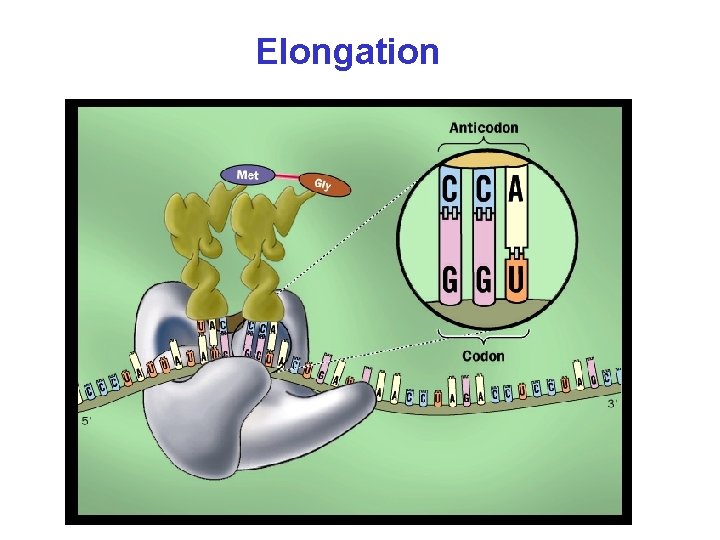
Elongation

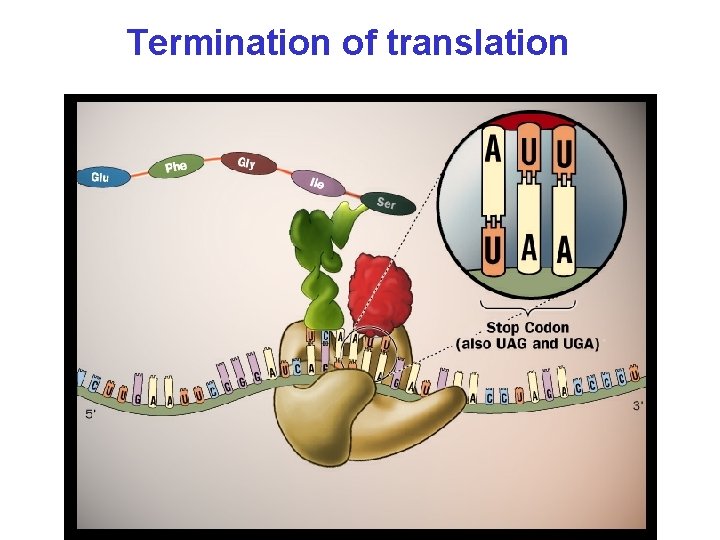
Termination of translation

Dialects • • • The genetic code is not universal … but the differences are relatively minor … occur mainly in small genomes of organelles … and involve specific codon families. In many cases symmetry is increased, or entire families reassigned. • Many changes involve stop codons

Reassignment CUN (=CUU, CUC, CUA, CUG): Leu Thr Possible initiation codons in addition to AUG (Met): NUG (=GUG, UUG, CUG), AUN (=AUU, AUC, AUA) UAA, UAG: stop Gln

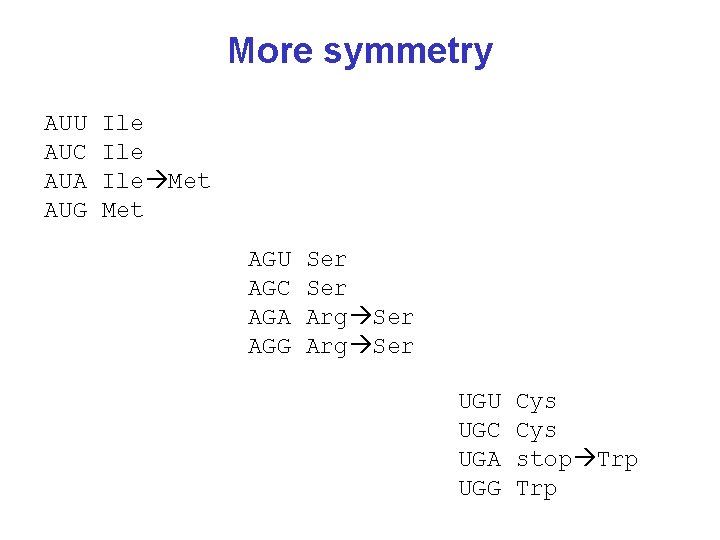
More symmetry AUU AUC AUA AUG Ile Ile Met AGU AGC AGA AGG Ser Arg Ser UGU UGC UGA UGG Cys stop Trp

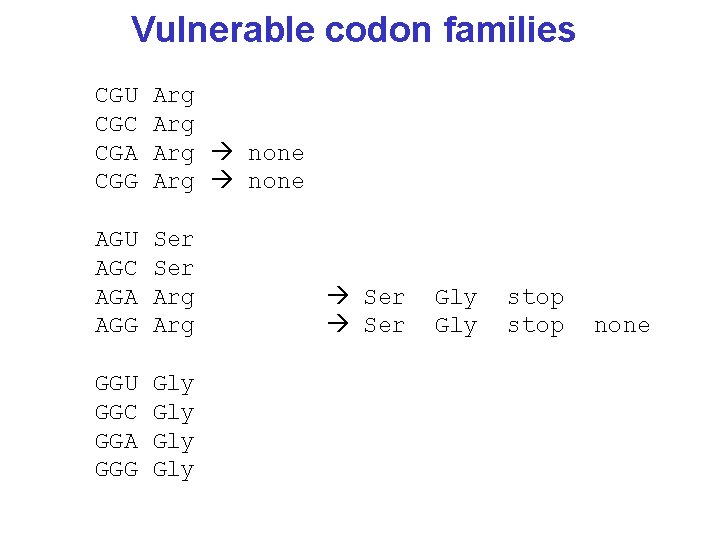
Vulnerable codon families CGU CGC CGA CGG Arg Arg none AGU AGC AGA AGG Ser Arg GGU GGC GGA GGG Gly Gly Ser Gly stop none

Stop-containing families UGU UGC UGA UGG Cys stop Trp UAU UAC UAA UAG Tyr stop Cys Sec Gln (Pyl)

How many letters are there in the English alphabet?

How many letters are there in the English alphabet? • 26 (everybody knows) …

How many letters are there in the English alphabet? • 26 (everybody knows) … • … but we are discussing the book by Yčas …

How many letters are there in the English alphabet? • 26 (everybody knows) … • … but we are discussing the book by Yčas … • … so everybody are naïve

How many amino acids? • Chemists: hundreds – many occur in proteins: post-translation modifications • How many amino acids are encoded by DNA?

Crick:

Is formyl-methionine a “standard” amino acid? • Occurs in bacteria at N-termini of all recently synthesized proteins (may be enzymatically removed later on) • Has three codons: AUG, GUG, UUG – unlike “inernal” methionine encoded only by AUG – by the way, internal GUG encodes Valine and internal UUG encodes Leucine

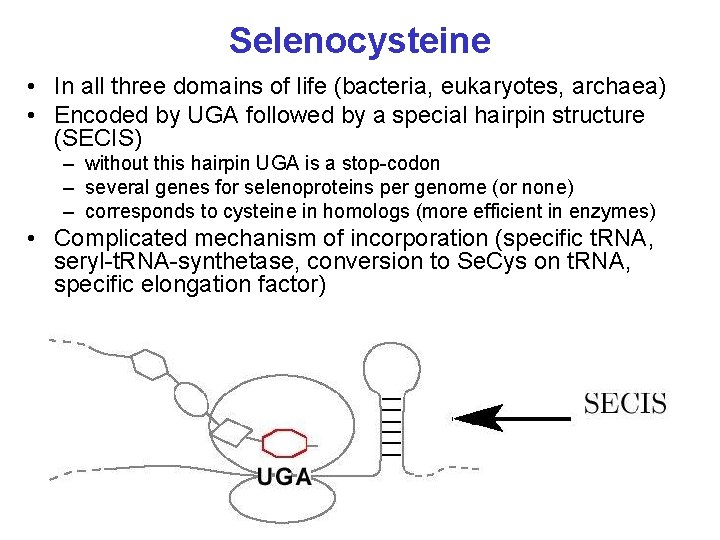
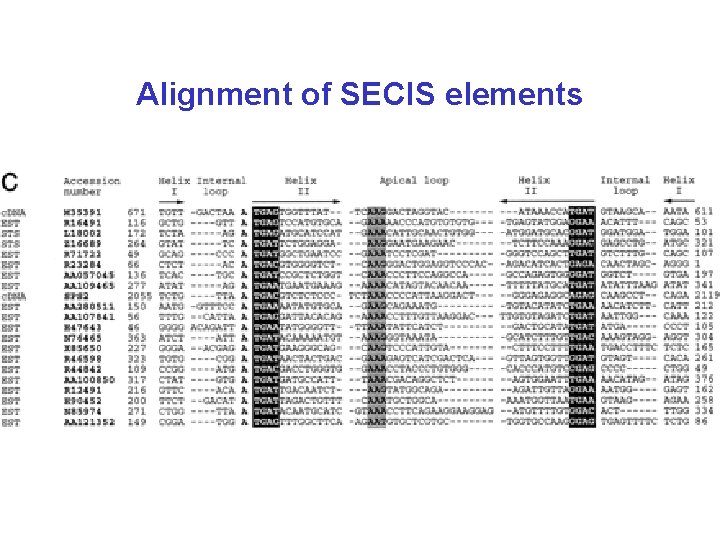
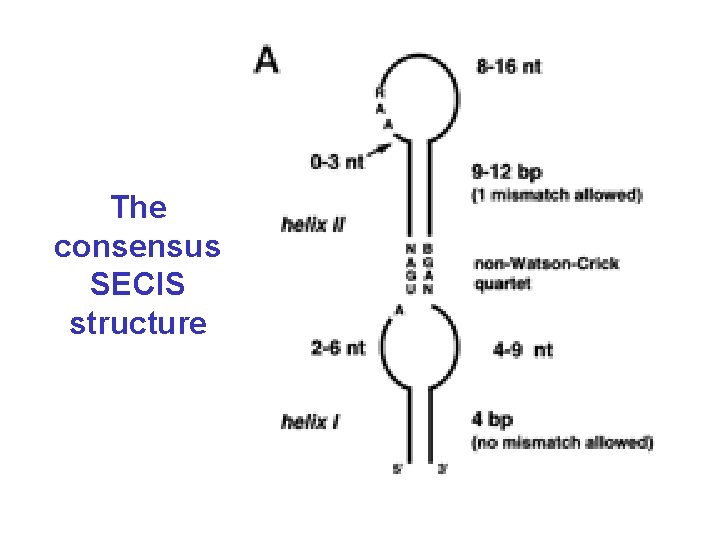
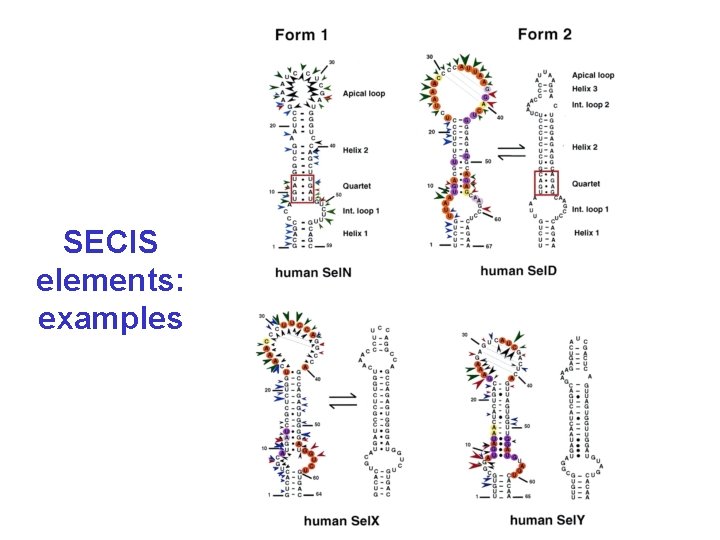
Selenocysteine • In all three domains of life (bacteria, eukaryotes, archaea) • Encoded by UGA followed by a special hairpin structure (SECIS) – without this hairpin UGA is a stop-codon – several genes for selenoproteins per genome (or none) – corresponds to cysteine in homologs (more efficient in enzymes) • Complicated mechanism of incorporation (specific t. RNA, seryl-t. RNA-synthetase, conversion to Se. Cys on t. RNA, specific elongation factor)

Alignment of SECIS elements

The consensus SECIS structure

SECIS elements: examples

Pyrrolysine • In methanogenic archaea • A derivative of lysine • Directly encoded (unlike selenocysteine). Standard mechanism: – UAG codon – specific t. RNA – aminoacyl-t. RNA • UAG rarely used as a stop codon – never as the only stop of a gene

Thanks • Wikipedia • Ergito • Authors of papers, photographs and Internet resources • • Professor Leong Hon Wai The organizers The assistants The students