Methods of Genome Mapping linkage maps physical maps

- Slides: 14

Methods of Genome Mapping linkage maps, physical maps, QTL analysis The focus of the course should be on analytical (bioinformatic) tools for genome mapping, i. e. , relevant background from (a) statistics, (b) appl. math. (c) software

A few elementary genetic and molecular-genetic notions (subjects) you are supposed to know General Genetics: meiosis, syngamy, gamete, zygote, DNA, genome, nucleus, chromosome, centromere, bivalent, hybrid, homozygote, F 1, F 2, heterozygote, inbred, haploid, diploid, mutant, gene, allele, locus, phenotype, Mendelian segregation (single-, two-, multilocus), dominant, co-dominant, recessive, additive, linkage, recombination, epistasis, quantitative variation, heritability, test-cross, backcross, intercross, linkage phase (coupling, repulsion), multiple crossovers, interference, polymorphism, linkage disequilibrium, haplotype Molecular Genetics: restriction fragment, DNA hybridization, Southern blot analysis, PCR, tandem repeats, microsatellite, SNP, DNA cloning, BAC-clone, genomic library, DNA fingerprinting, overlapping clones, contig, radiation hybrid, candidate gene, microarray

Mendelian (qualitative) vs qualitative traits Simple Mendelian traits - discrete (discontinuous) traits - • One gene = one trait • Finite number of genotypes • One gene = 3 genotypes = 2 or 3 phenotypes (folding hands, blood type, fruit color, wing shape) Complex non-Mendelian traits – continuous distribution • Quantitative or continuous traits - controlled by several loci Each quantitative trait locus (QTL) contributes to phenotype QTL(s) + Environment (e. g. , climate) + Culture = Phenotype A fundamental question: Do QTLs represent the same Mendelian genes, or these are a specific class of elements ?

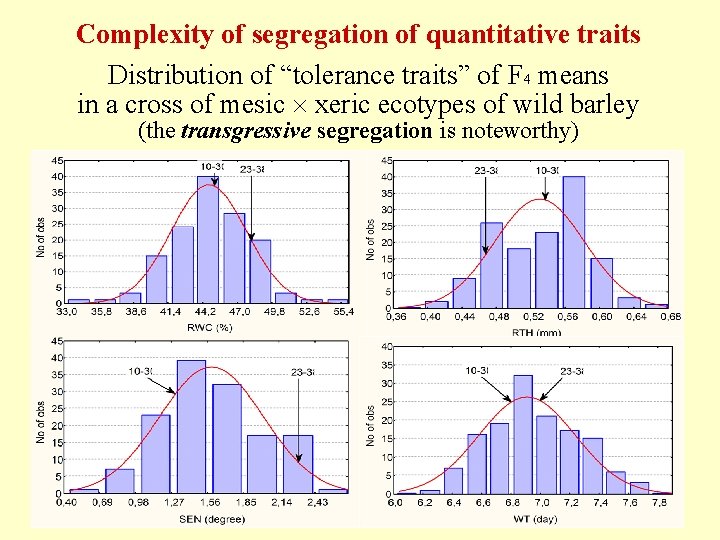
Complexity of segregation of quantitative traits Distribution of “tolerance traits” of F 4 means in a cross of mesic xeric ecotypes of wild barley (the transgressive segregation is noteworthy)

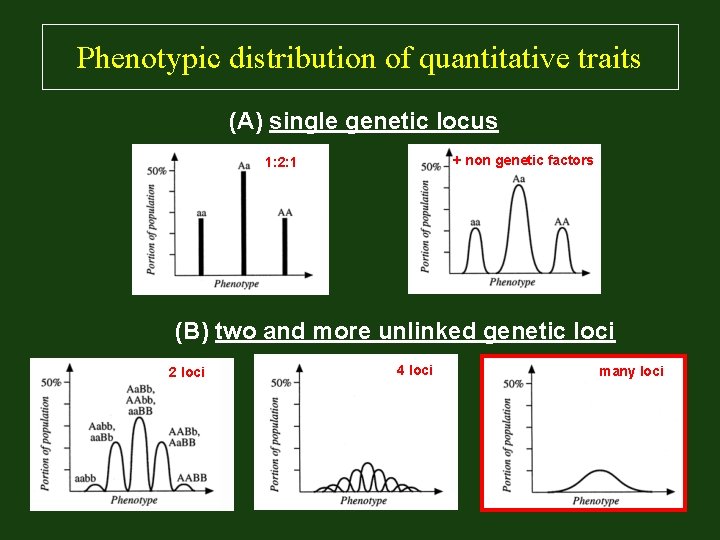
Phenotypic distribution of quantitative traits (A) single genetic locus + non genetic factors 1: 2: 1 (B) two and more unlinked genetic loci 2 loci 4 loci many loci

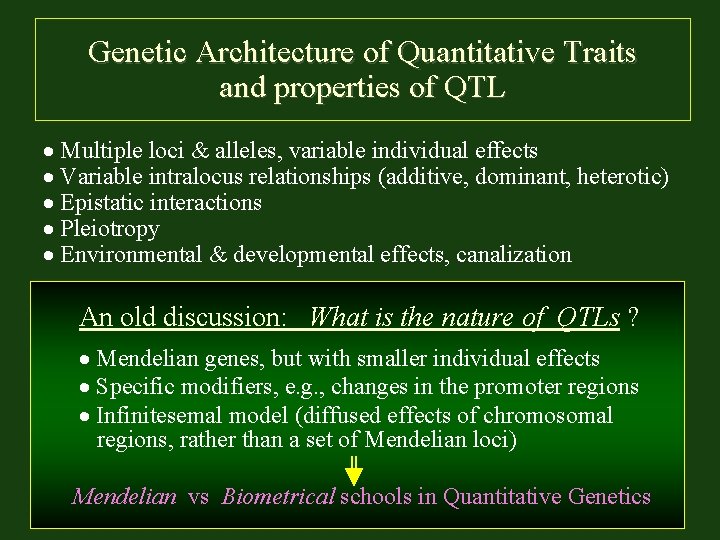
Genetic Architecture of Quantitative Traits and properties of QTL Multiple loci & alleles, variable individual effects Variable intralocus relationships (additive, dominant, heterotic) Epistatic interactions Pleiotropy Environmental & developmental effects, canalization An old discussion: What is the nature of QTLs ? Mendelian genes, but with smaller individual effects Specific modifiers, e. g. , changes in the promoter regions Infinitesemal model (diffused effects of chromosomal regions, rather than a set of Mendelian loci) Mendelian vs Biometrical schools in Quantitative Genetics

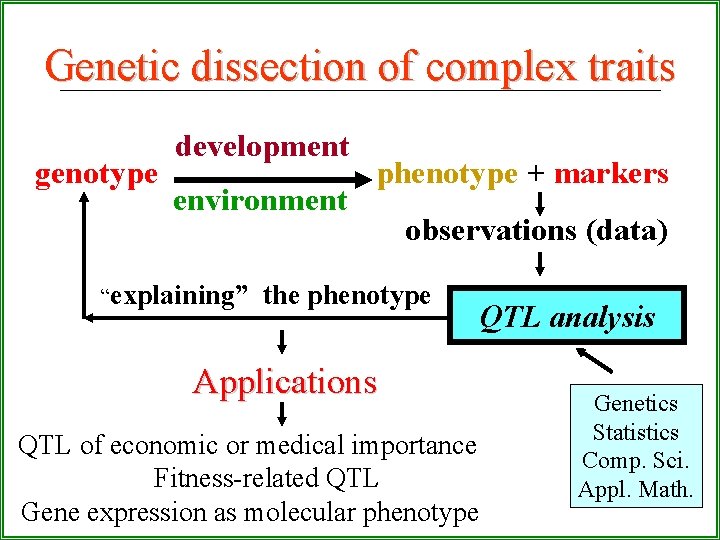
Genetic dissection of complex traits development genotype phenotype + markers environment observations (data) “explaining” the phenotype QTL analysis Applications Genetics QTL of economic or medical importance Fitness-related QTL Gene expression as molecular phenotype Statistics Comp. Sci. Appl. Math.

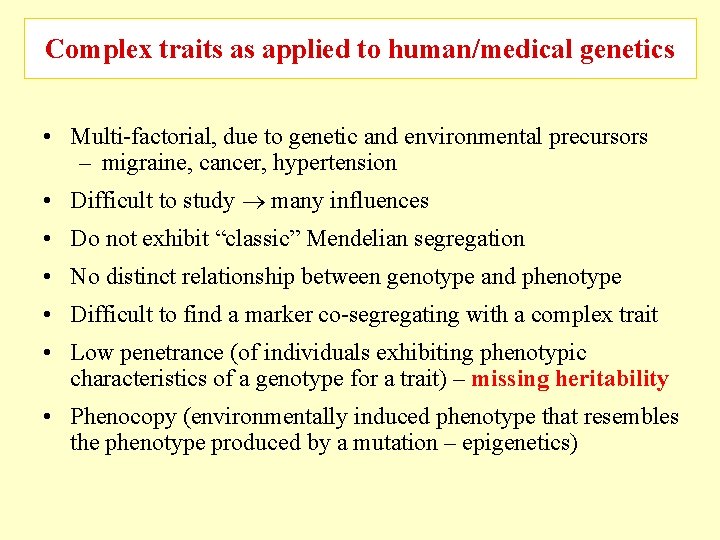
Complex traits as applied to human/medical genetics • Multi-factorial, due to genetic and environmental precursors – migraine, cancer, hypertension • Difficult to study many influences • Do not exhibit “classic” Mendelian segregation • No distinct relationship between genotype and phenotype • Difficult to find a marker co-segregating with a complex trait • Low penetrance (of individuals exhibiting phenotypic characteristics of a genotype for a trait) – missing heritability • Phenocopy (environmentally induced phenotype that resembles the phenotype produced by a mutation – epigenetics)

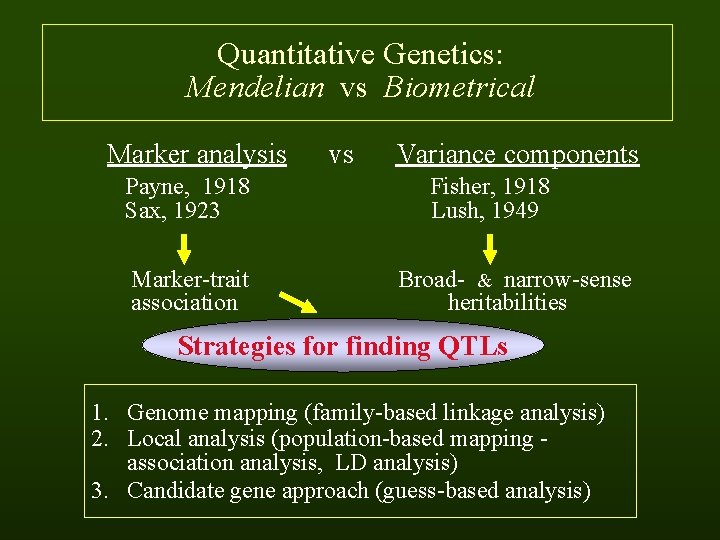
Quantitative Genetics: Mendelian vs Biometrical Marker analysis vs Variance components Payne, 1918 Fisher, 1918 Sax, 1923 Lush, 1949 Marker-trait Broad- & narrow-sense association heritabilities Strategies for finding QTLs 1. Genome mapping (family-based linkage analysis) 2. Local analysis (population-based mapping - association analysis, LD analysis) 3. Candidate gene approach (guess-based analysis)

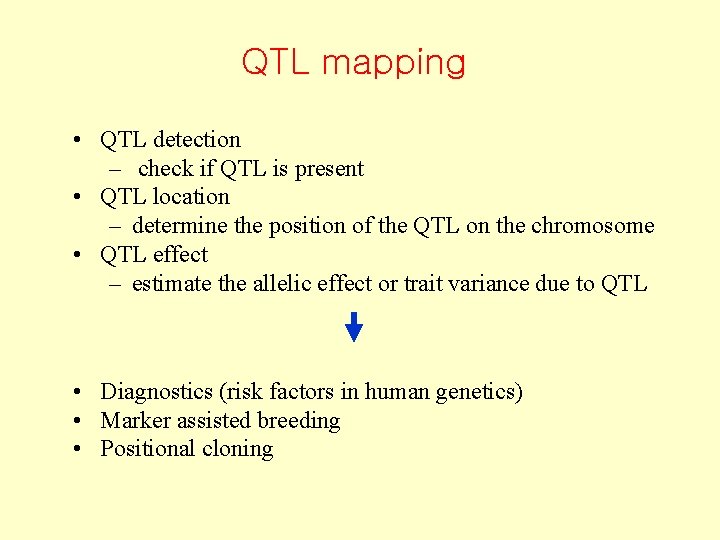
QTL mapping • QTL detection – check if QTL is present • QTL location – determine the position of the QTL on the chromosome • QTL effect – estimate the allelic effect or trait variance due to QTL • Diagnostics (risk factors in human genetics) • Marker assisted breeding • Positional cloning

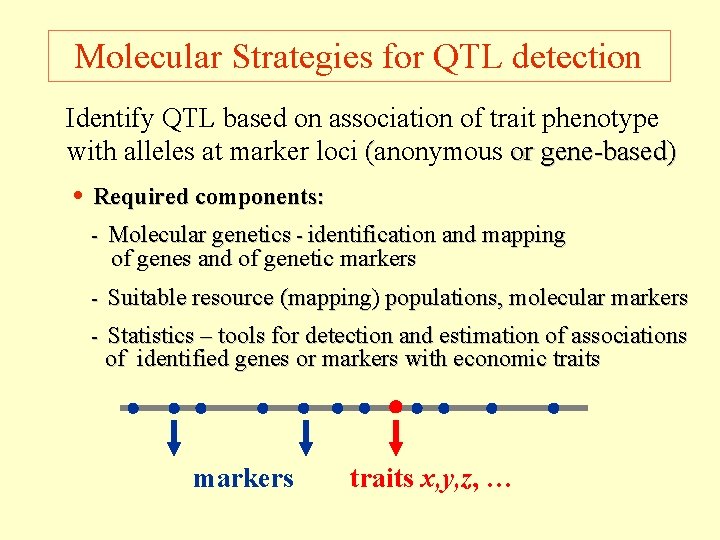
Molecular Strategies for QTL detection Identify QTL based on association of trait phenotype with alleles at marker loci (anonymous or gene-based) • Required components: - Molecular genetics - identification and mapping of genes and of genetic markers - Suitable resource (mapping) populations, molecular markers - Statistics – tools for detection and estimation of associations of identified genes or markers with economic traits markers traits x, y, z, …

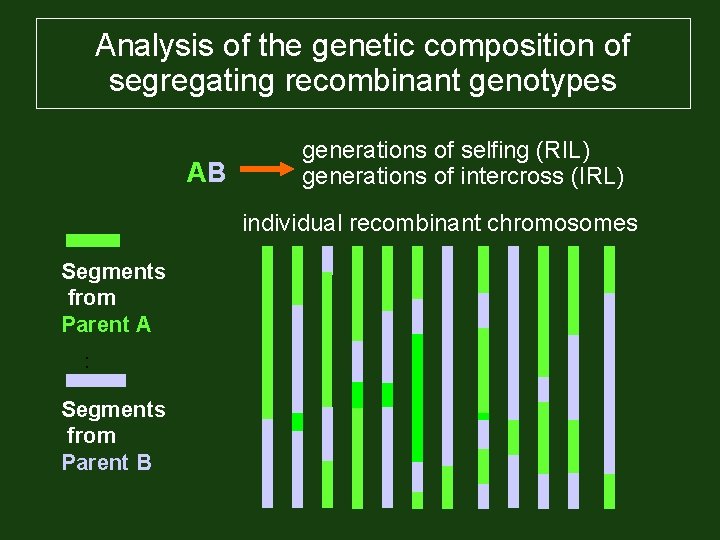
Analysis of the genetic composition of segregating recombinant genotypes AB generations of selfing (RIL) generations of intercross (IRL) individual recombinant chromosomes Segments from Parent A : Segments from Parent B

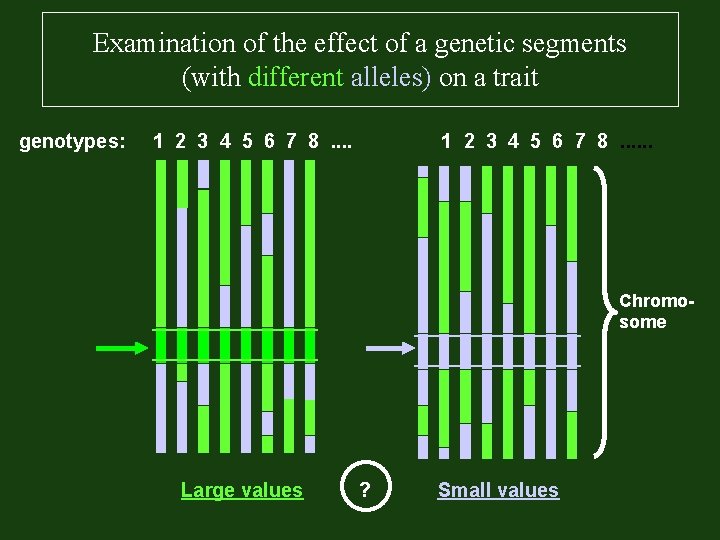
Examination of the effect of a genetic segments (with different alleles) on a trait genotypes: 1 2 3 4 5 6 7 8. . . Chromosome Large values ? Small values

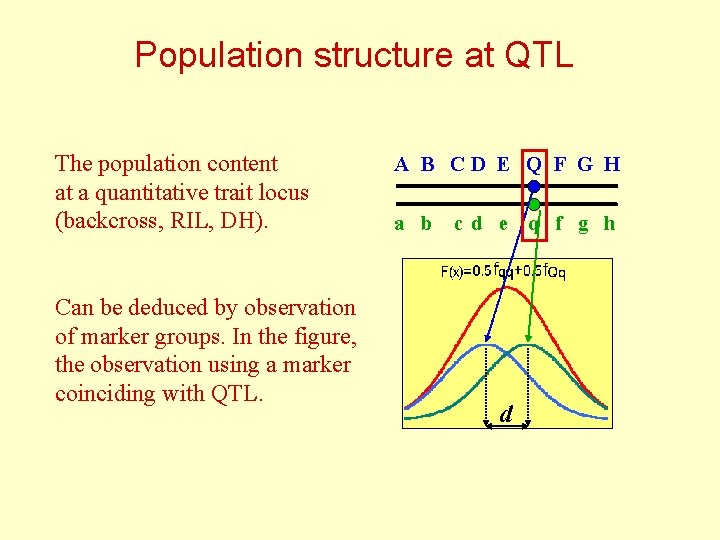
Population structure at QTL The population content at a quantitative trait locus (backcross, RIL, DH). Can be deduced by observation of marker groups. In the figure, the observation using a marker coinciding with QTL. A B CD E Q F G H a b c d e q f g h d