1 2 3 4 5 Phusion PCR mix








- Slides: 89

1. 2. 3. 4. 5. Phusion PCR mix: for 20 µl On ICE! Prepare 100 p. Mol/µl solutions of each of your primers with molecular grade water prepare 10 x dilution of F primer and add 2 µl to your PCR tube prepare 10 x dilution of R primer and add 2 µl to your PCR tube Add 1 µl of suitable genomic DNA Add 15 µl Phusion master mix (for 140 µl total volume) 1. 28 µl 5 x HF buffer 2. 2. 8 µl 10 m. M d. NTP 3. 1. 4 µl Phusion 4. 72. 8 µl molecular grade water

Cycle 1 x 60” @ 98˚ C 35 x: 30” @ 98˚ C : (55 -cycle #)” @ Topt : 1 min/1000 bp@ 72˚ C 1 x 5 ‘ @ 72˚ C Transfer 5 µl to fresh tube, add 1 µl dye & run on 2% gel

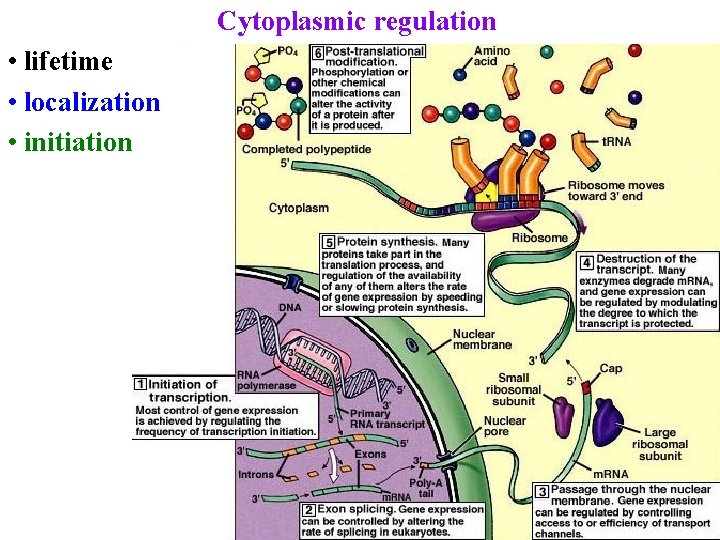
Cytoplasmic regulation • lifetime • localization • initiation

Post-transcriptional regulation Nearly ½ of human genome is transcribed, only 1% is CDS • 98% of RNA made is non-coding • ~1/3 intron • ~2/3 “independently transcribed” • Polymerases II & III (+ IV & V in plants) all help • many are from transposons or gene fragments made by transposons (pack-MULES) • ~ 10 -25% is anti-sense: same region is transcribed off both strands

Hypotheses 1. Accident: transcription unveils “cryptic promoters” on opposite strand (Zilberman et al) 2. Functional a. si. RNA b. mi. RNA c. Silencing d. Priming: chromatin remodeling requires transcription!

Post-transcriptional regulation RNA degradation is crucial with so much “extra” RNA

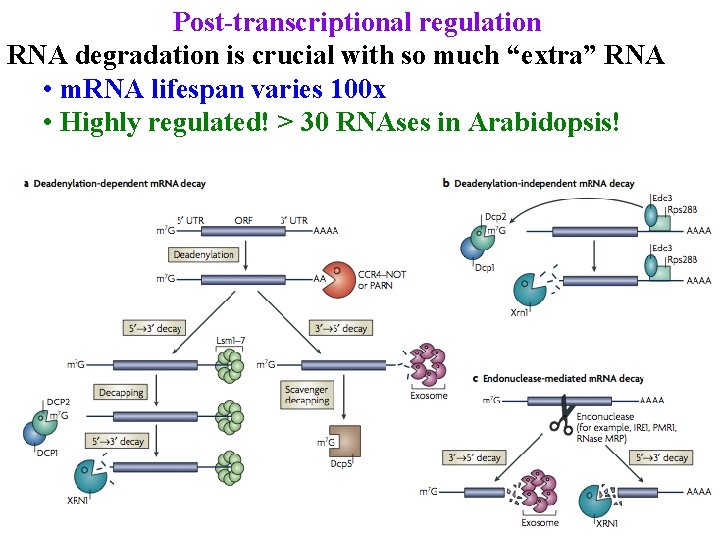
Post-transcriptional regulation RNA degradation is crucial with so much “extra” RNA • m. RNA lifespan varies 100 x • Highly regulated! > 30 RNAses in Arabidopsis!

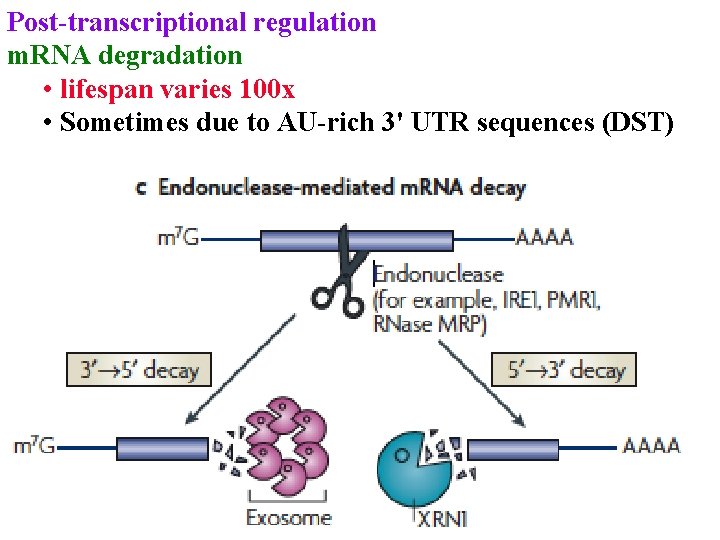
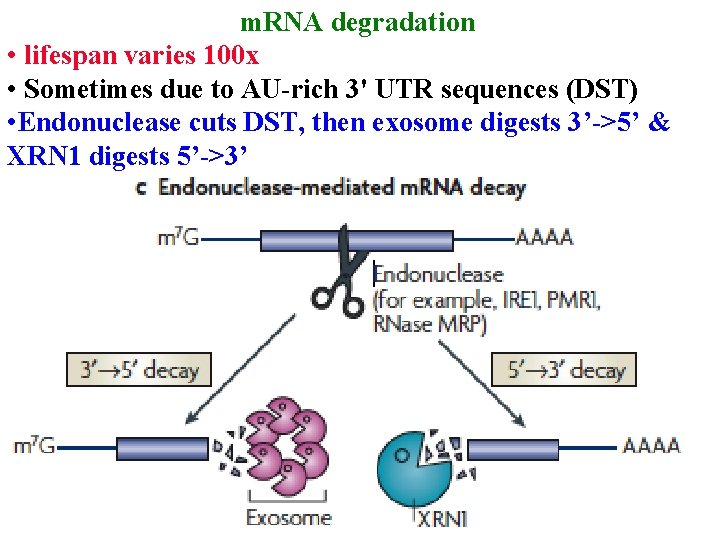
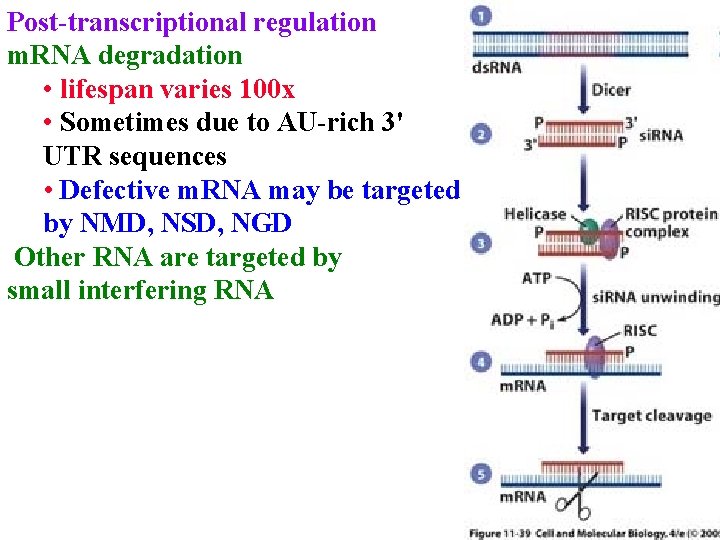
Post-transcriptional regulation m. RNA degradation • lifespan varies 100 x • Sometimes due to AU-rich 3' UTR sequences (DST)

m. RNA degradation • lifespan varies 100 x • Sometimes due to AU-rich 3' UTR sequences (DST) • Endonuclease cuts DST, then exosome digests 3’->5’ & XRN 1 digests 5’->3’

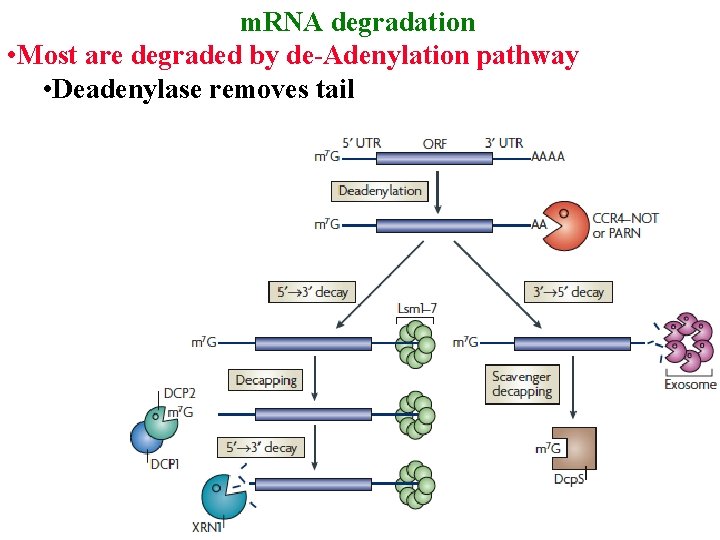
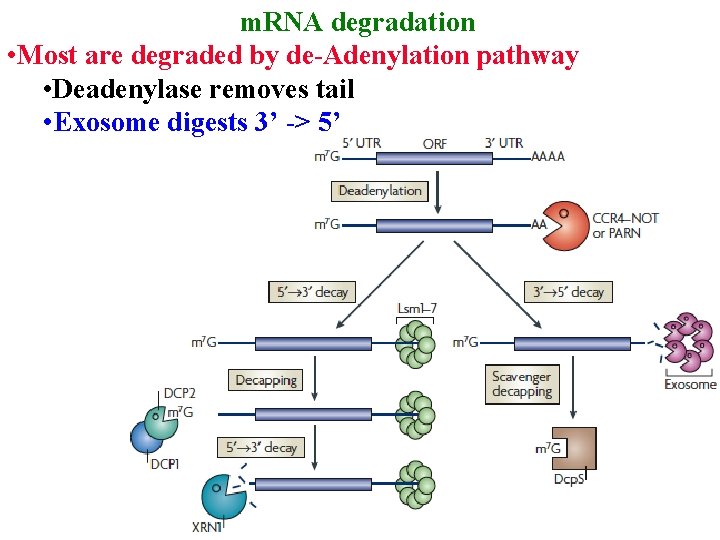
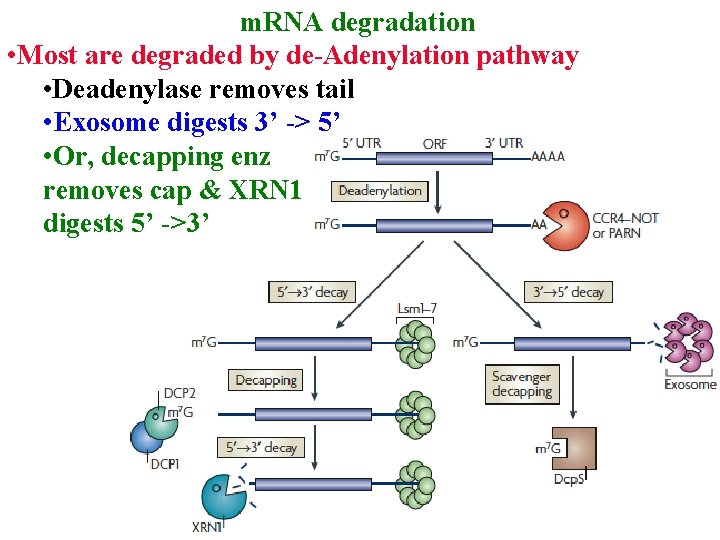
m. RNA degradation • Most are degraded by de-Adenylation pathway • Deadenylase removes tail

m. RNA degradation • Most are degraded by de-Adenylation pathway • Deadenylase removes tail • Exosome digests 3’ -> 5’

m. RNA degradation • Most are degraded by de-Adenylation pathway • Deadenylase removes tail • Exosome digests 3’ -> 5’ • Or, decapping enz removes cap & XRN 1 digests 5’ ->3’

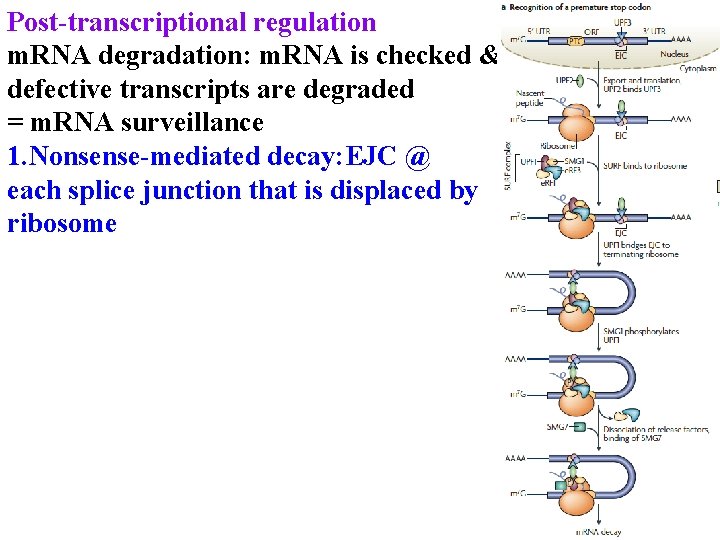
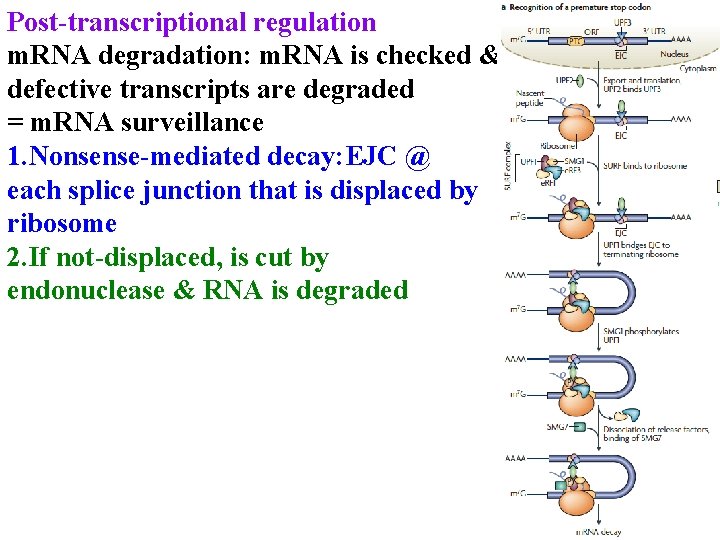
Post-transcriptional regulation m. RNA degradation: m. RNA is checked & defective transcripts are degraded = m. RNA surveillance 1. Nonsense-mediated decay: EJC @ each splice junction that is displaced by ribosome

Post-transcriptional regulation m. RNA degradation: m. RNA is checked & defective transcripts are degraded = m. RNA surveillance 1. Nonsense-mediated decay: EJC @ each splice junction that is displaced by ribosome 2. If not-displaced, is cut by endonuclease & RNA is degraded

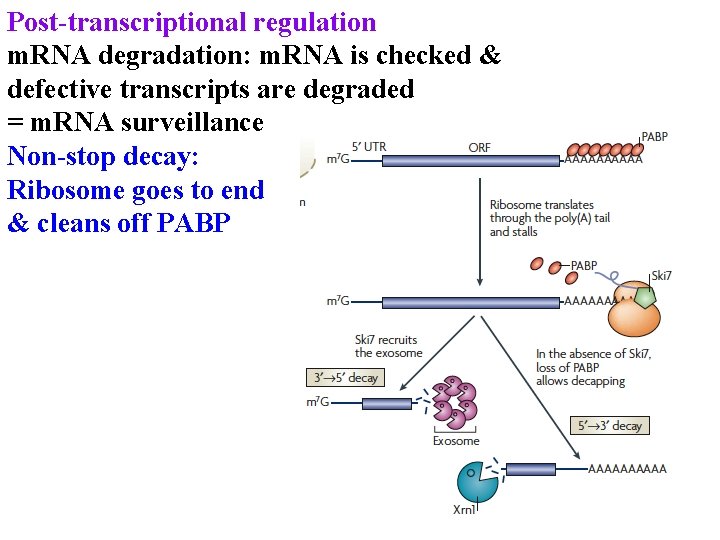
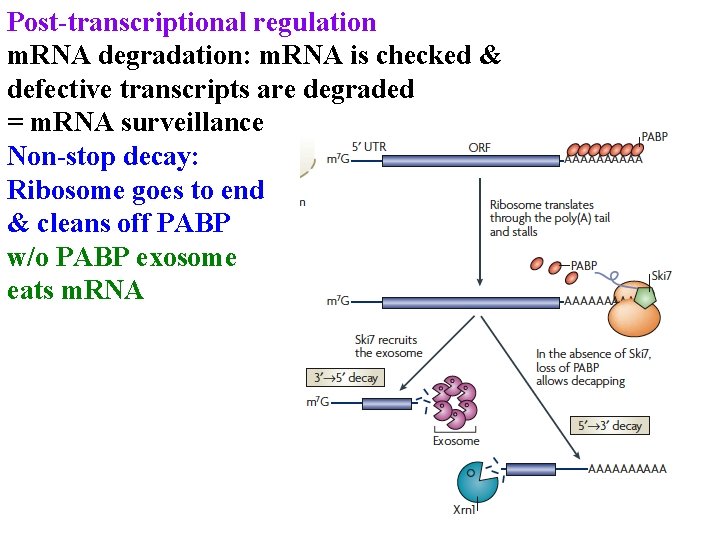
Post-transcriptional regulation m. RNA degradation: m. RNA is checked & defective transcripts are degraded = m. RNA surveillance Non-stop decay: Ribosome goes to end & cleans off PABP

Post-transcriptional regulation m. RNA degradation: m. RNA is checked & defective transcripts are degraded = m. RNA surveillance Non-stop decay: Ribosome goes to end & cleans off PABP w/o PABP exosome eats m. RNA

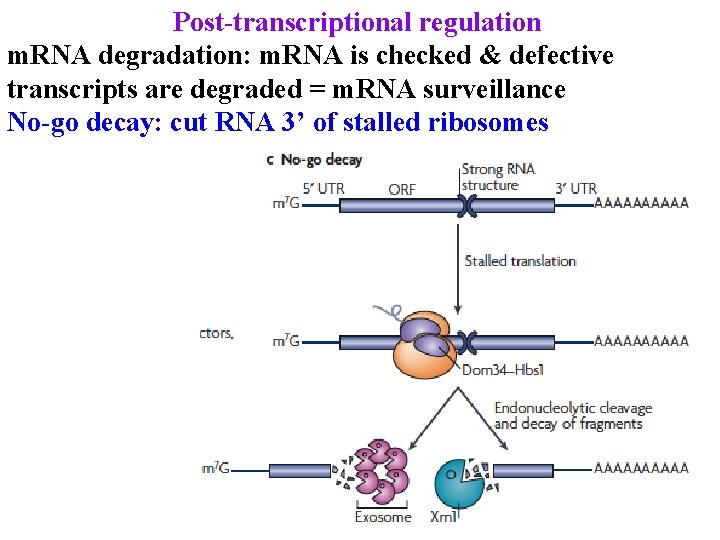
Post-transcriptional regulation m. RNA degradation: m. RNA is checked & defective transcripts are degraded = m. RNA surveillance No-go decay: cut RNA 3’ of stalled ribosomes

Post-transcriptional regulation m. RNA degradation • lifespan varies 100 x • Sometimes due to AU-rich 3' UTR sequences • Defective m. RNA may be targeted by NMD, NSD, NGD Other RNA are targeted by small interfering RNA

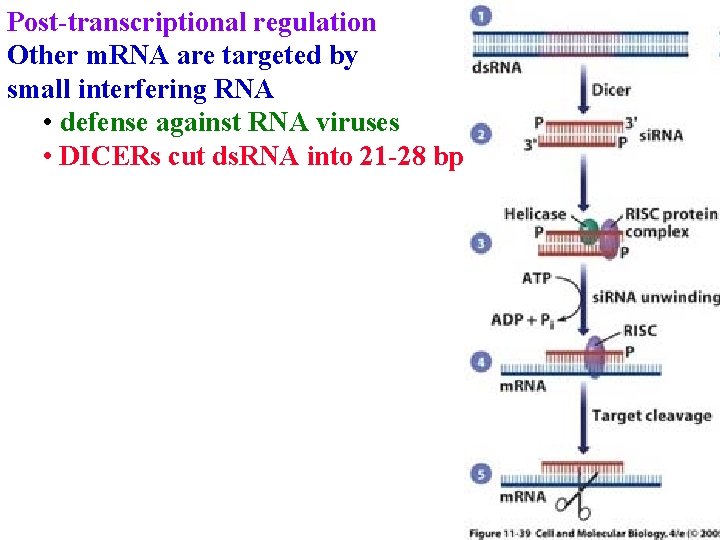
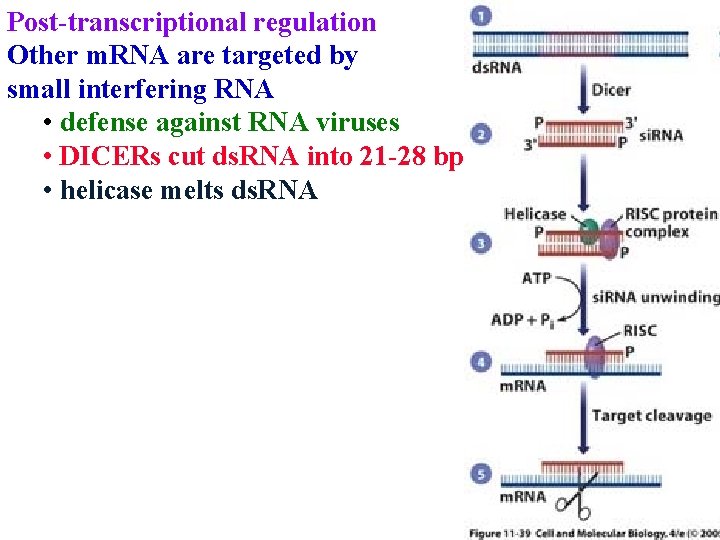
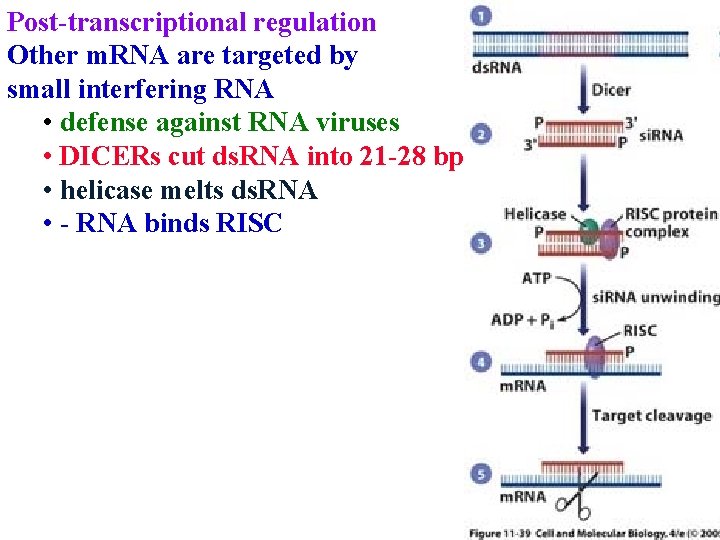
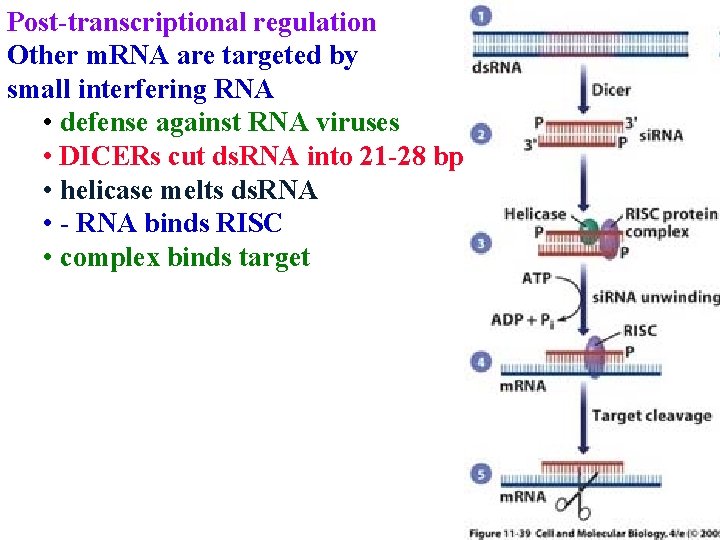
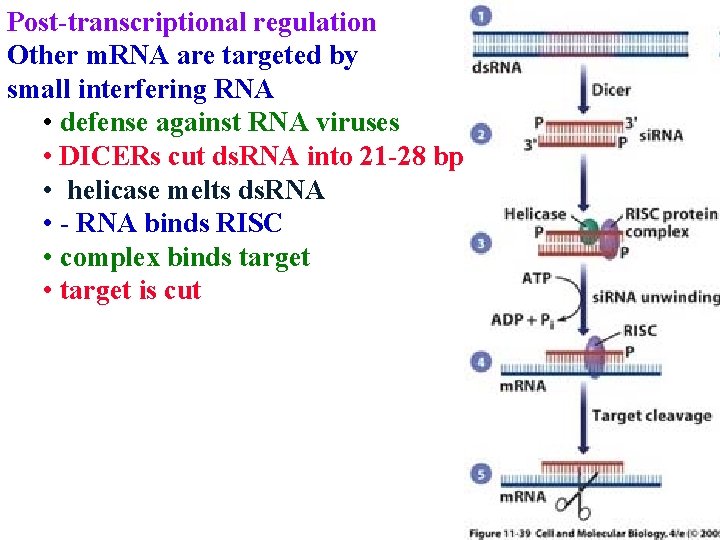
Post-transcriptional regulation Other m. RNA are targeted by small interfering RNA • defense against RNA viruses • DICERs cut ds. RNA into 21 -28 bp

Post-transcriptional regulation Other m. RNA are targeted by small interfering RNA • defense against RNA viruses • DICERs cut ds. RNA into 21 -28 bp • helicase melts ds. RNA

Post-transcriptional regulation Other m. RNA are targeted by small interfering RNA • defense against RNA viruses • DICERs cut ds. RNA into 21 -28 bp • helicase melts ds. RNA • - RNA binds RISC

Post-transcriptional regulation Other m. RNA are targeted by small interfering RNA • defense against RNA viruses • DICERs cut ds. RNA into 21 -28 bp • helicase melts ds. RNA • - RNA binds RISC • complex binds target

Post-transcriptional regulation Other m. RNA are targeted by small interfering RNA • defense against RNA viruses • DICERs cut ds. RNA into 21 -28 bp • helicase melts ds. RNA • - RNA binds RISC • complex binds target • target is cut

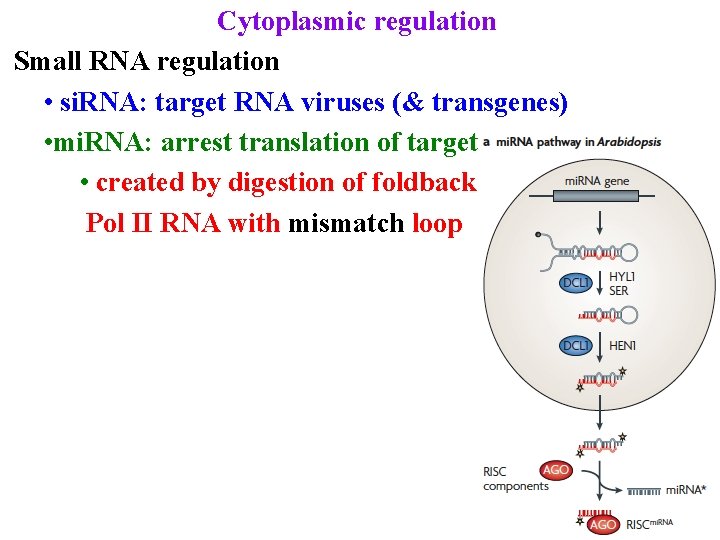
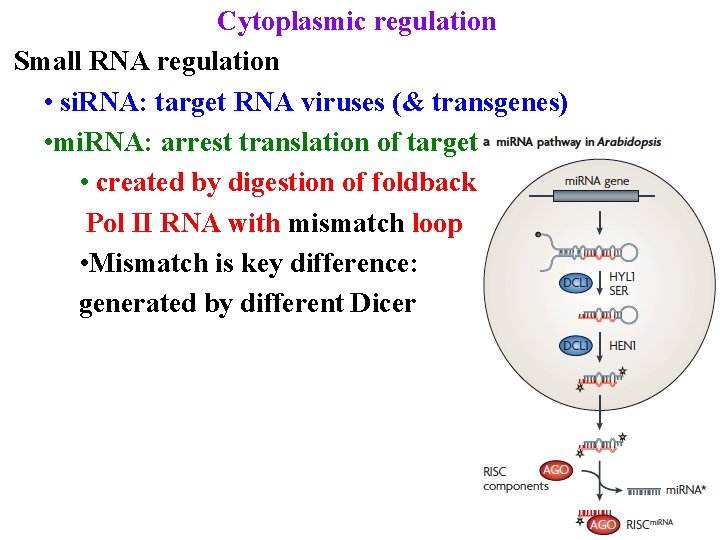
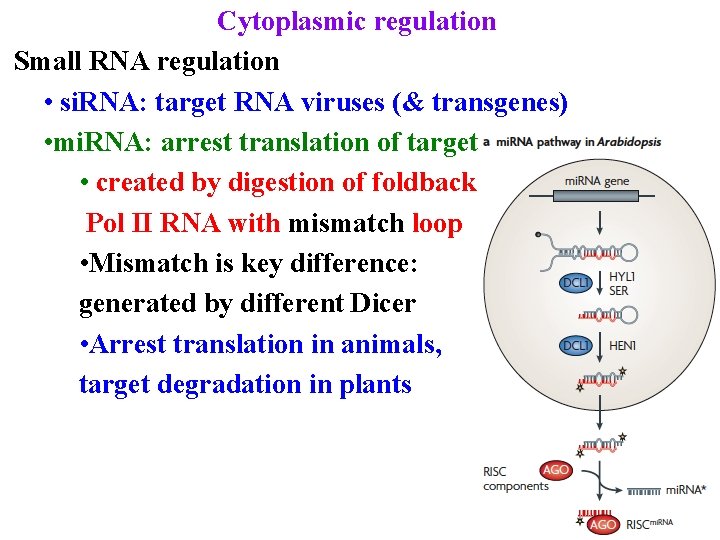
Cytoplasmic regulation Small RNA regulation • si. RNA: target RNA viruses (& transgenes) • mi. RNA: arrest translation of targets • created by digestion of foldback Pol II RNA with mismatch loop

Cytoplasmic regulation Small RNA regulation • si. RNA: target RNA viruses (& transgenes) • mi. RNA: arrest translation of targets • created by digestion of foldback Pol II RNA with mismatch loop • Mismatch is key difference: generated by different Dicer

Cytoplasmic regulation Small RNA regulation • si. RNA: target RNA viruses (& transgenes) • mi. RNA: arrest translation of targets • created by digestion of foldback Pol II RNA with mismatch loop • Mismatch is key difference: generated by different Dicer • Arrest translation in animals, target degradation in plants

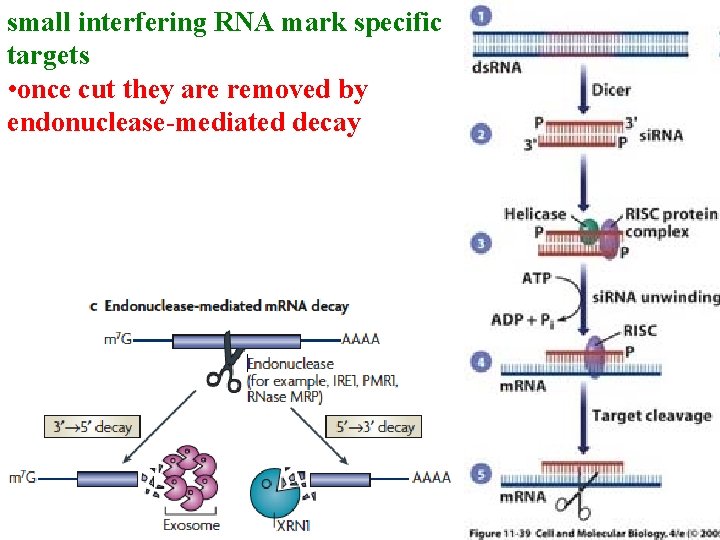
small interfering RNA mark specific targets • once cut they are removed by endonuclease-mediated decay


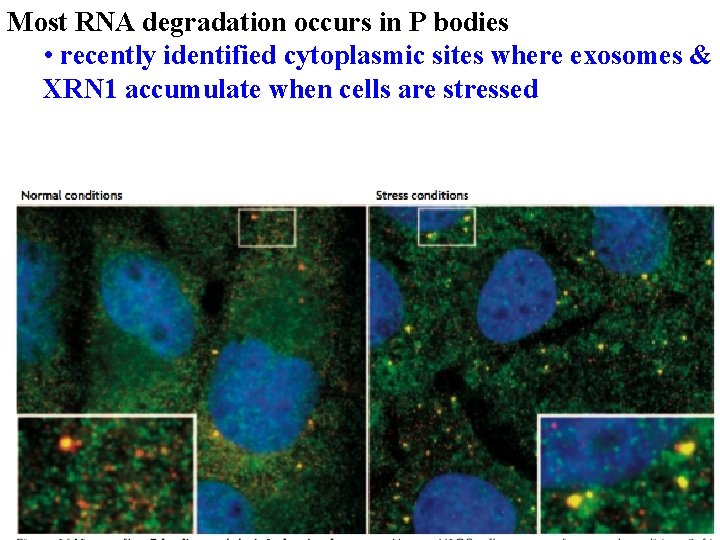
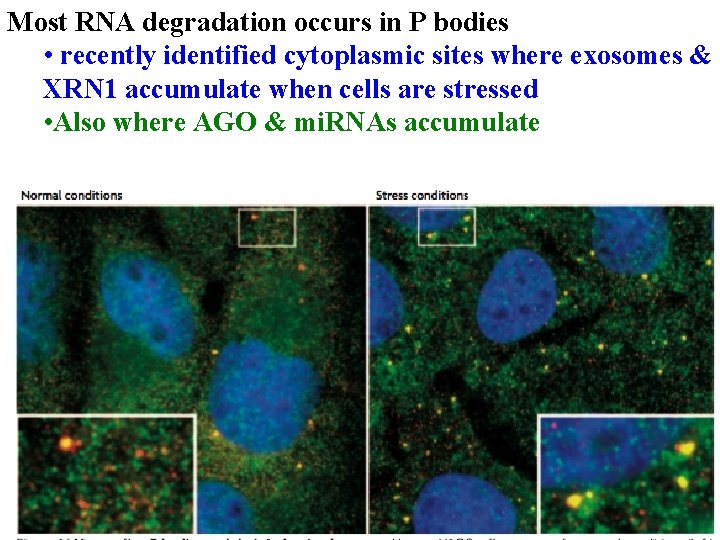
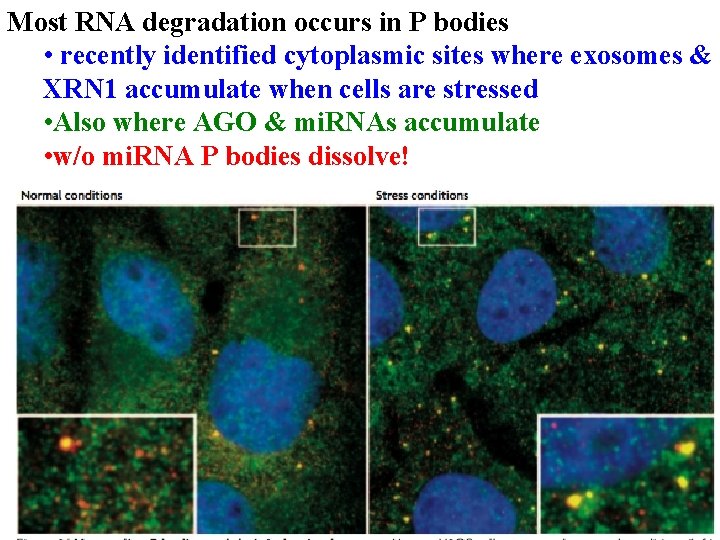
Most RNA degradation occurs in P bodies • recently identified cytoplasmic sites where exosomes & XRN 1 accumulate when cells are stressed

Most RNA degradation occurs in P bodies • recently identified cytoplasmic sites where exosomes & XRN 1 accumulate when cells are stressed • Also where AGO & mi. RNAs accumulate

Most RNA degradation occurs in P bodies • recently identified cytoplasmic sites where exosomes & XRN 1 accumulate when cells are stressed • Also where AGO & mi. RNAs accumulate • w/o mi. RNA P bodies dissolve!

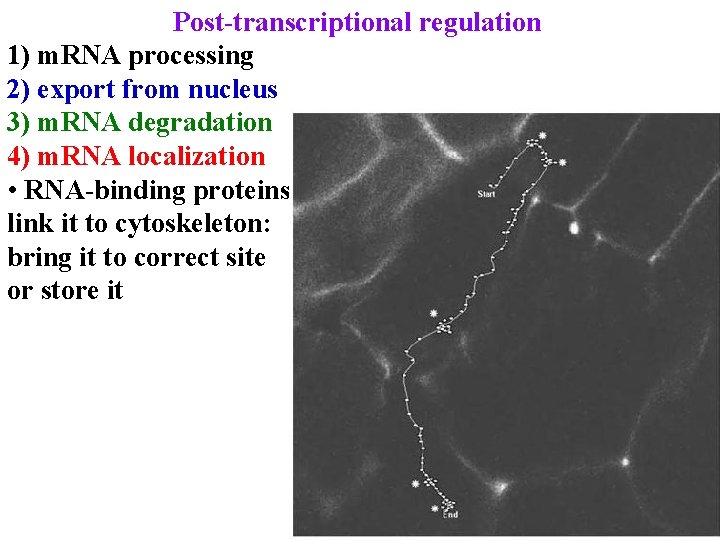
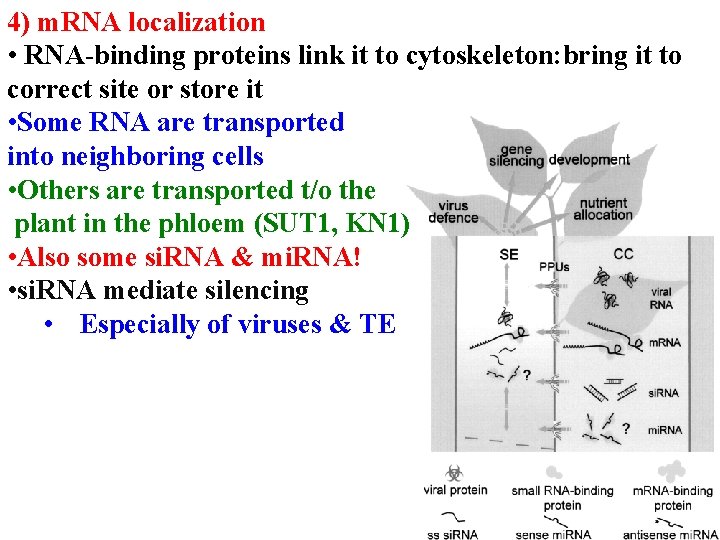
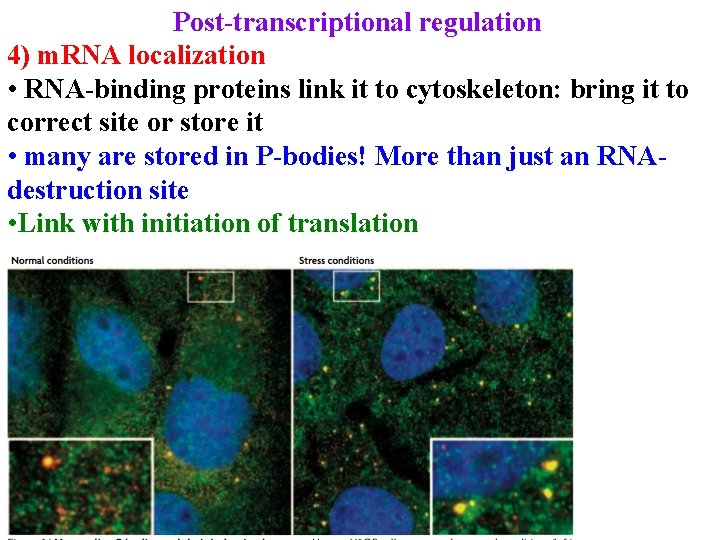
Post-transcriptional regulation 1) m. RNA processing 2) export from nucleus 3) m. RNA degradation 4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it

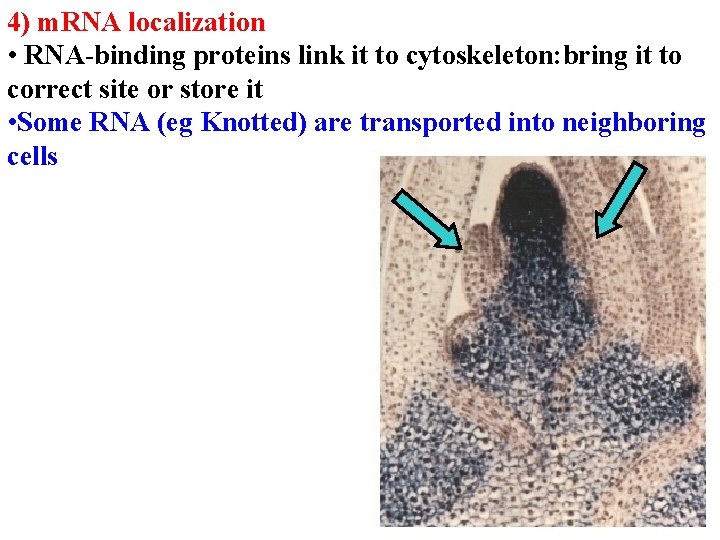
4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • Some RNA (eg Knotted) are transported into neighboring cells

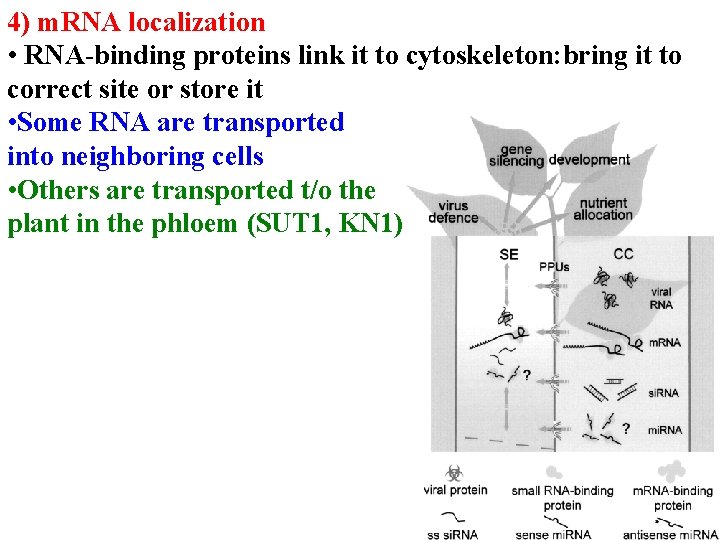
4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • Some RNA are transported into neighboring cells • Others are transported t/o the plant in the phloem (SUT 1, KN 1)

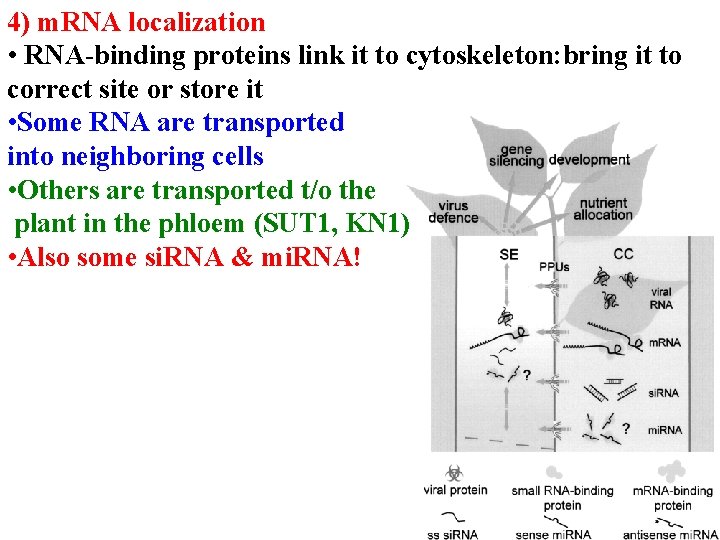
4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • Some RNA are transported into neighboring cells • Others are transported t/o the plant in the phloem (SUT 1, KN 1) • Also some si. RNA & mi. RNA!

4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • Some RNA are transported into neighboring cells • Others are transported t/o the plant in the phloem (SUT 1, KN 1) • Also some si. RNA & mi. RNA! • si. RNA mediate silencing • Especially of viruses & TE

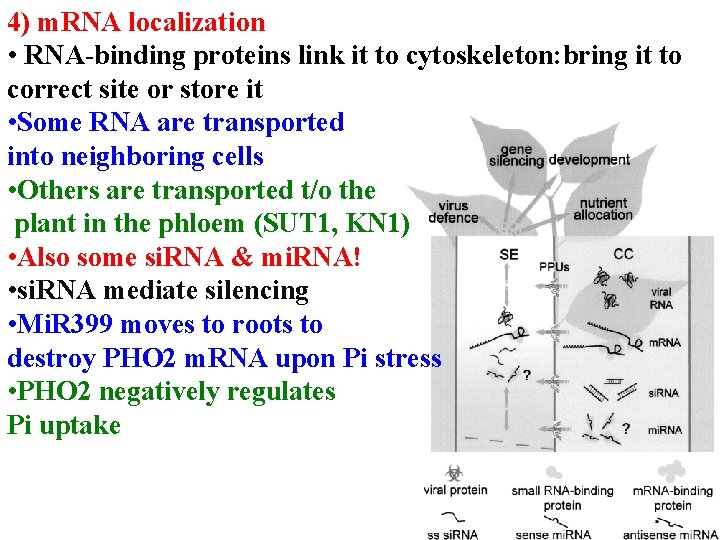
4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • Some RNA are transported into neighboring cells • Others are transported t/o the plant in the phloem (SUT 1, KN 1) • Also some si. RNA & mi. RNA! • si. RNA mediate silencing • Mi. R 399 moves to roots to destroy PHO 2 m. RNA upon Pi stress • PHO 2 negatively regulates Pi uptake

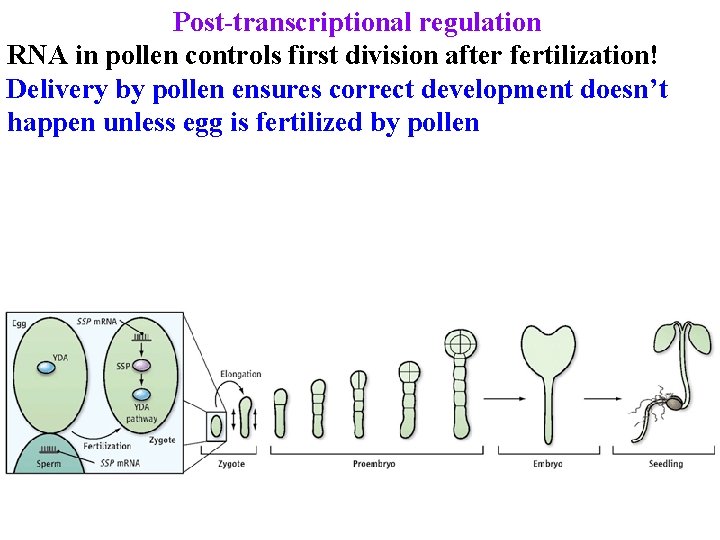
Post-transcriptional regulation RNA in pollen controls first division after fertilization!

Post-transcriptional regulation RNA in pollen controls first division after fertilization! Delivery by pollen ensures correct development doesn’t happen unless egg is fertilized by pollen

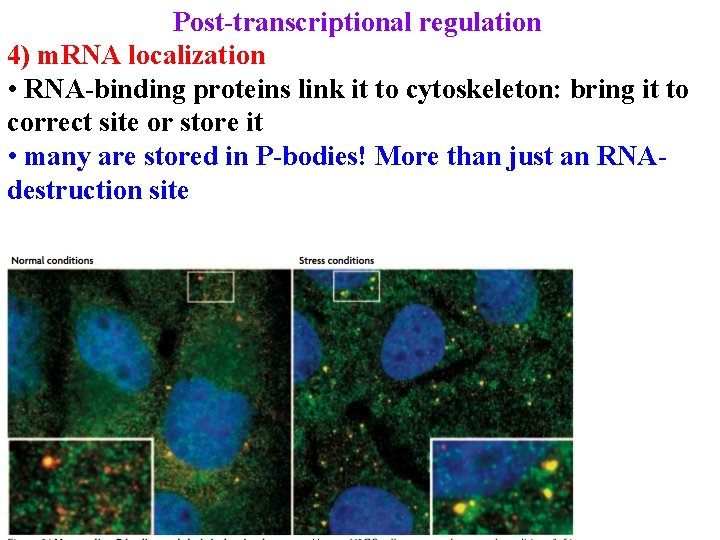
Post-transcriptional regulation 4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • many are stored in P-bodies! More than just an RNAdestruction site

Post-transcriptional regulation 4) m. RNA localization • RNA-binding proteins link it to cytoskeleton: bring it to correct site or store it • many are stored in P-bodies! More than just an RNAdestruction site • Link with initiation of translation

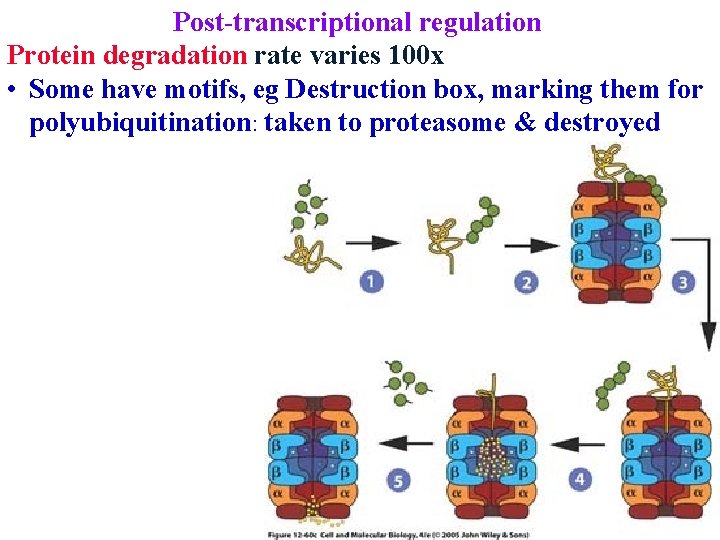
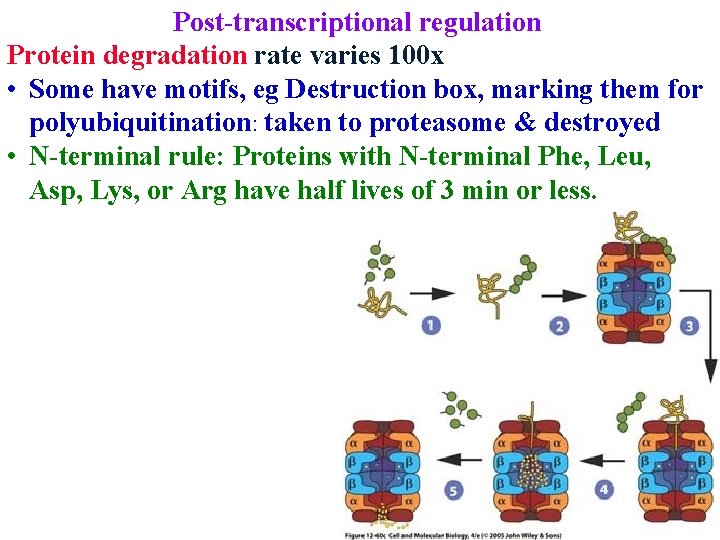
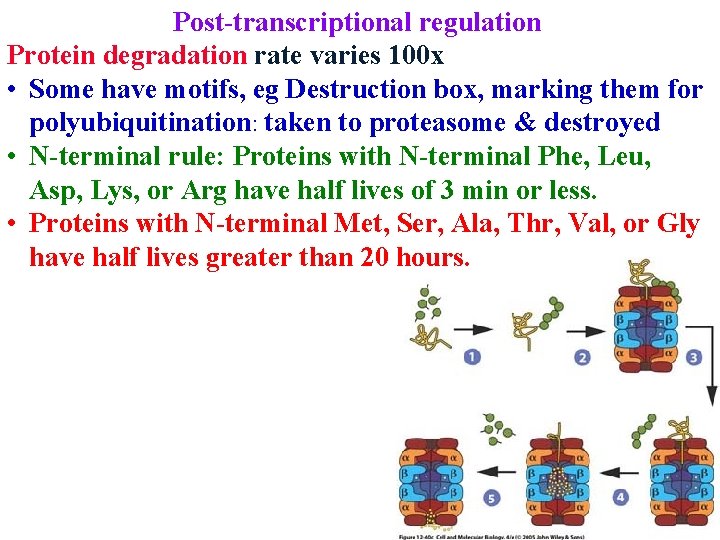
Post-transcriptional regulation Protein degradation rate varies 100 x • Some have motifs, eg Destruction box, marking them for polyubiquitination: taken to proteasome & destroyed

Post-transcriptional regulation Protein degradation rate varies 100 x • Some have motifs, eg Destruction box, marking them for polyubiquitination: taken to proteasome & destroyed • N-terminal rule: Proteins with N-terminal Phe, Leu, Asp, Lys, or Arg have half lives of 3 min or less.

Post-transcriptional regulation Protein degradation rate varies 100 x • Some have motifs, eg Destruction box, marking them for polyubiquitination: taken to proteasome & destroyed • N-terminal rule: Proteins with N-terminal Phe, Leu, Asp, Lys, or Arg have half lives of 3 min or less. • Proteins with N-terminal Met, Ser, Ala, Thr, Val, or Gly have half lives greater than 20 hours.

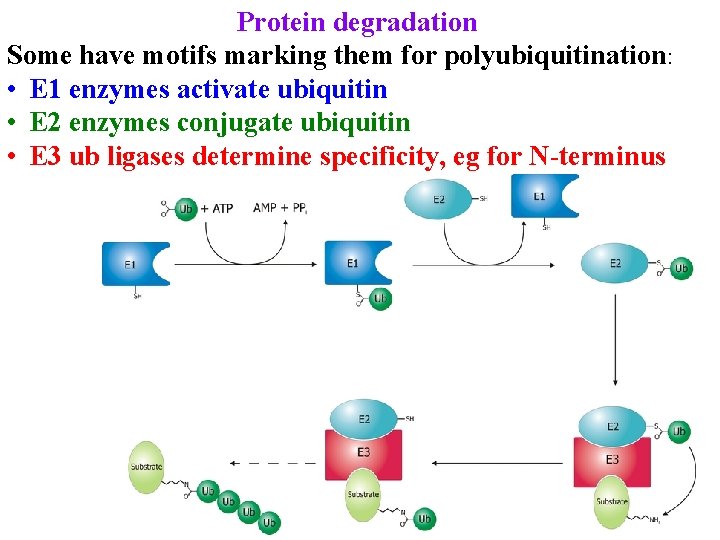
Protein degradation Some have motifs marking them for polyubiquitination: • E 1 enzymes activate ubiquitin • E 2 enzymes conjugate ubiquitin • E 3 ub ligases determine specificity, eg for N-terminus

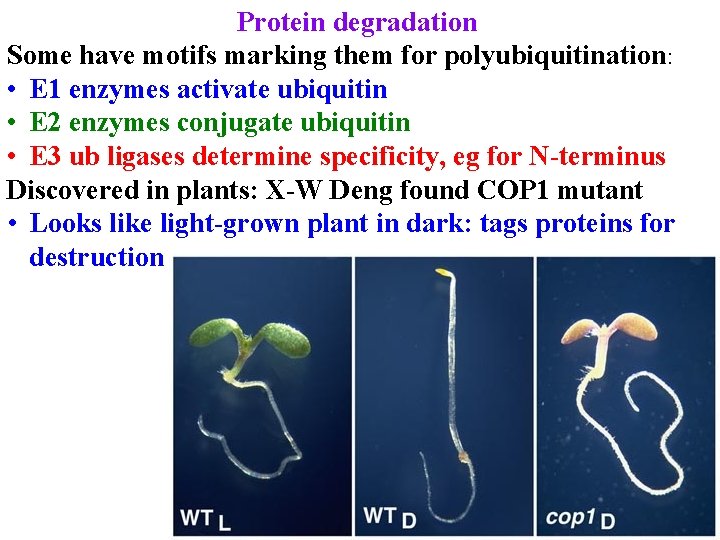
Protein degradation Some have motifs marking them for polyubiquitination: • E 1 enzymes activate ubiquitin • E 2 enzymes conjugate ubiquitin • E 3 ub ligases determine specificity, eg for N-terminus Discovered in plants: X-W Deng found COP 1 mutant • Looks like light-grown plant in dark: tags proteins for destruction

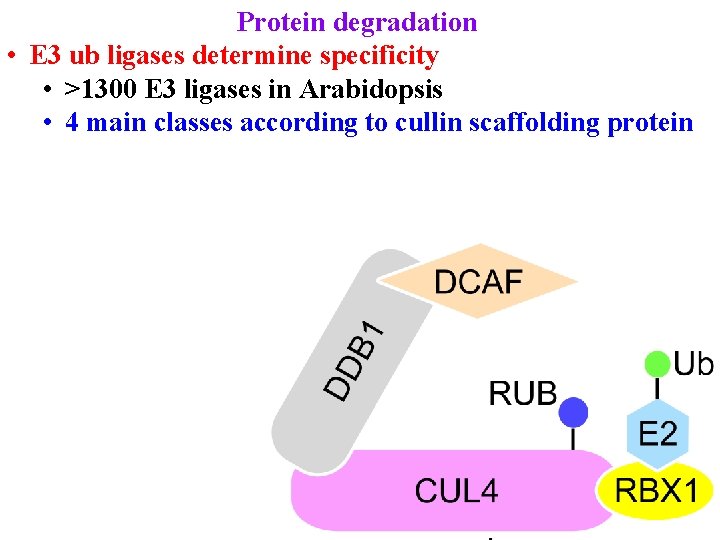
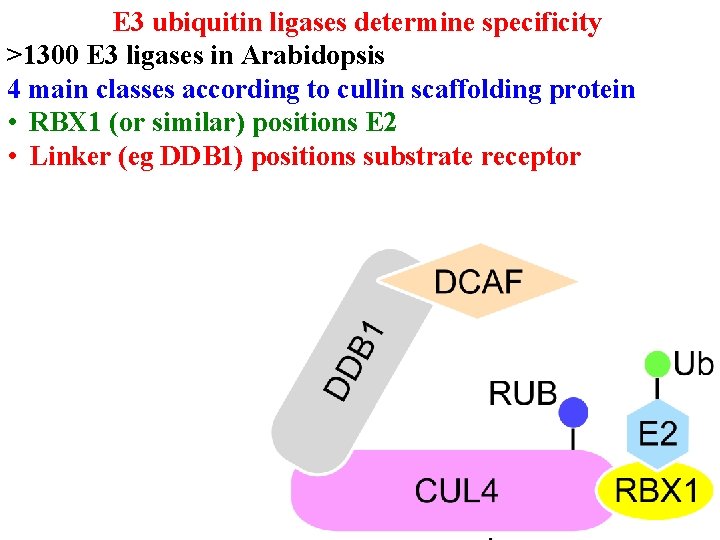
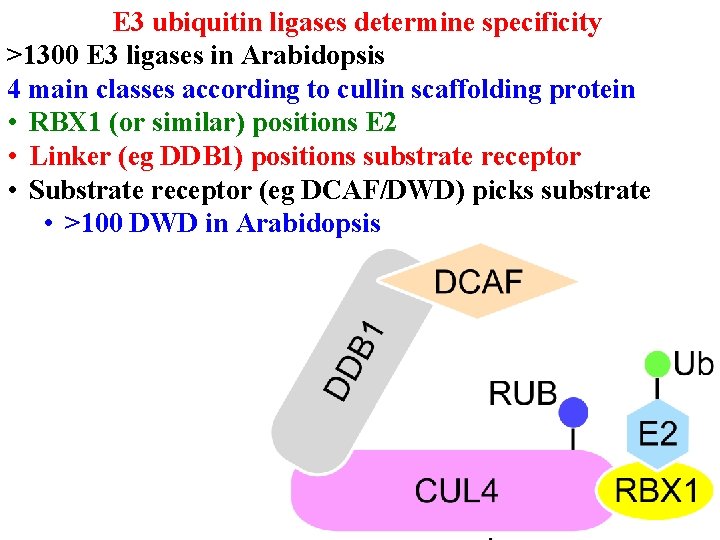
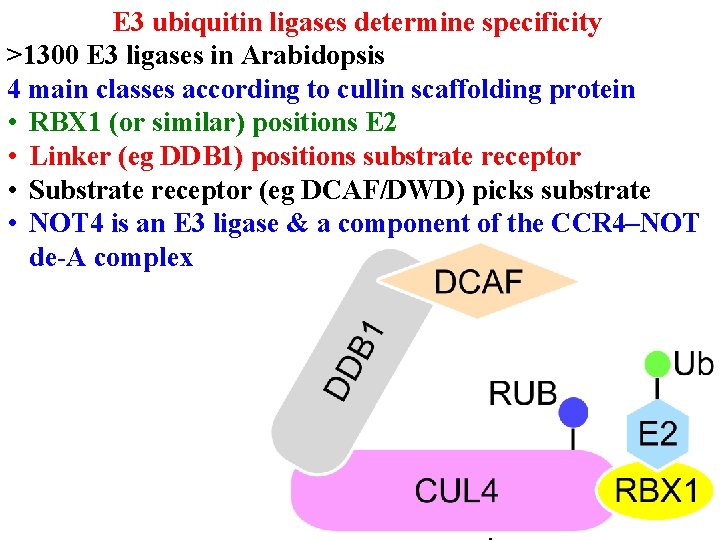
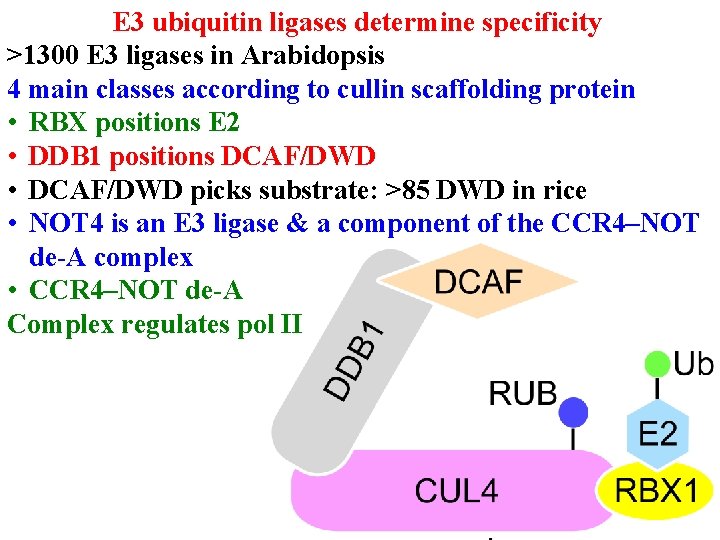
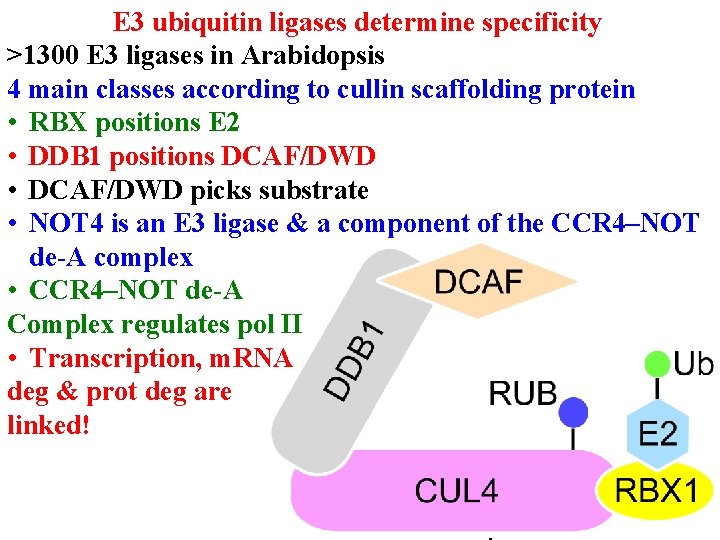
Protein degradation • E 3 ub ligases determine specificity • >1300 E 3 ligases in Arabidopsis • 4 main classes according to cullin scaffolding protein

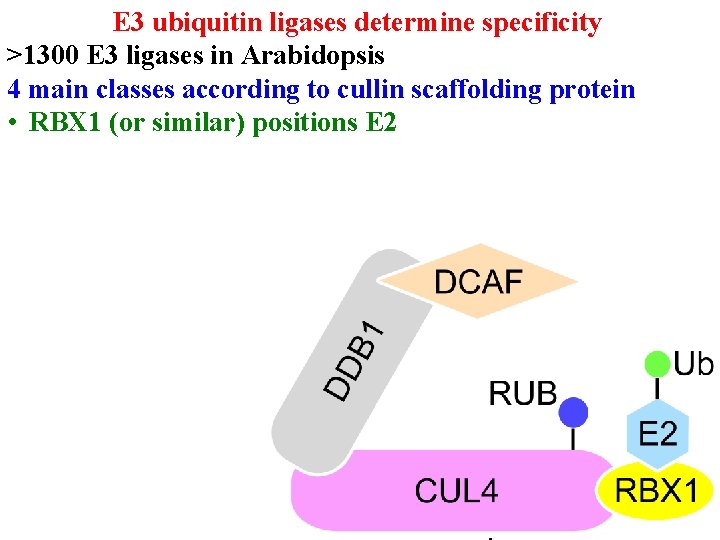
E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX 1 (or similar) positions E 2

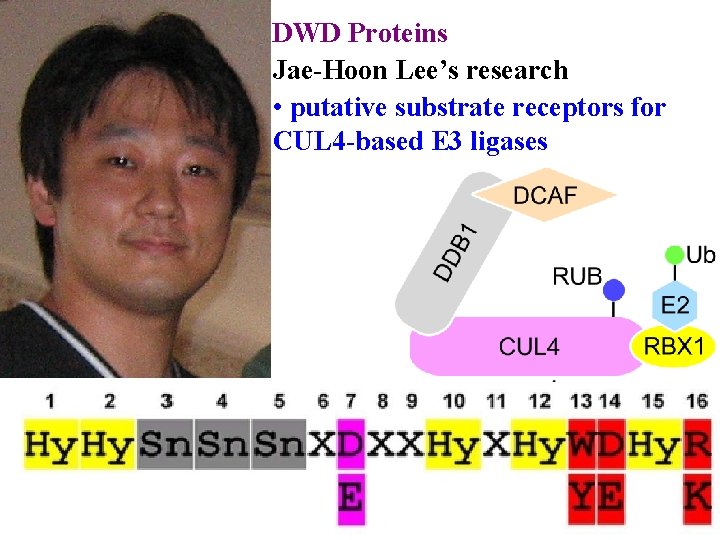
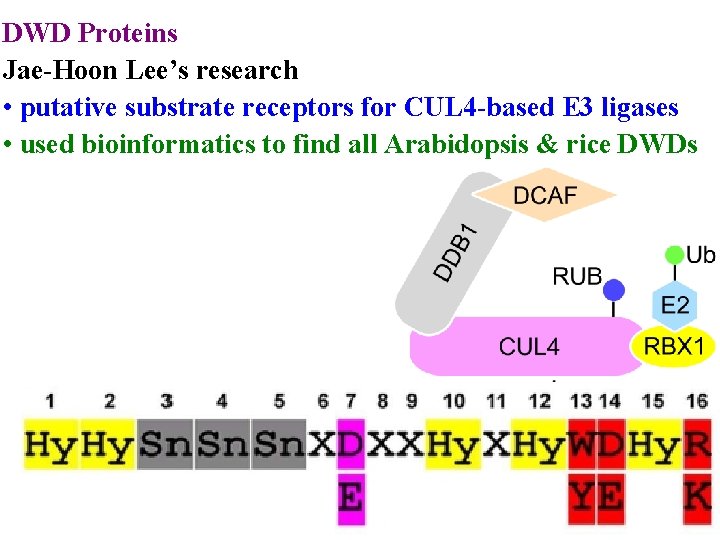
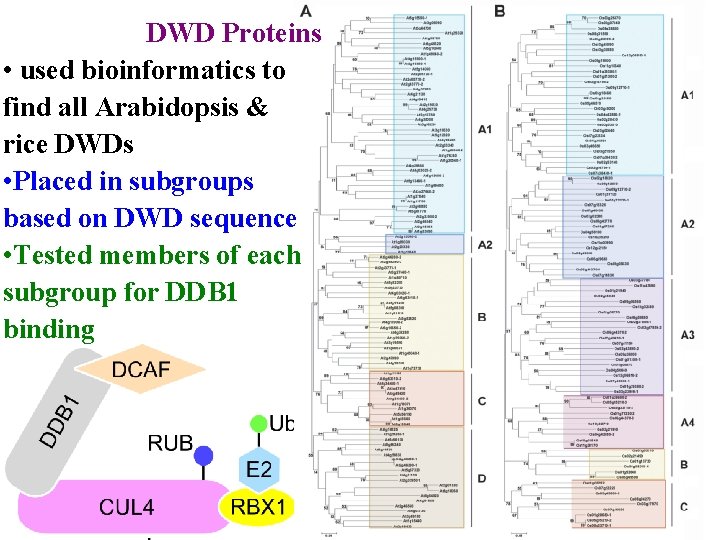
E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX 1 (or similar) positions E 2 • Linker (eg DDB 1) positions substrate receptor

E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX 1 (or similar) positions E 2 • Linker (eg DDB 1) positions substrate receptor • Substrate receptor (eg DCAF/DWD) picks substrate • >100 DWD in Arabidopsis

E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX 1 (or similar) positions E 2 • Linker (eg DDB 1) positions substrate receptor • Substrate receptor (eg DCAF/DWD) picks substrate • NOT 4 is an E 3 ligase & a component of the CCR 4–NOT de-A complex

E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX positions E 2 • DDB 1 positions DCAF/DWD • DCAF/DWD picks substrate: >85 DWD in rice • NOT 4 is an E 3 ligase & a component of the CCR 4–NOT de-A complex • CCR 4–NOT de-A Complex regulates pol II

E 3 ubiquitin ligases determine specificity >1300 E 3 ligases in Arabidopsis 4 main classes according to cullin scaffolding protein • RBX positions E 2 • DDB 1 positions DCAF/DWD • DCAF/DWD picks substrate • NOT 4 is an E 3 ligase & a component of the CCR 4–NOT de-A complex • CCR 4–NOT de-A Complex regulates pol II • Transcription, m. RNA deg & prot deg are linked!

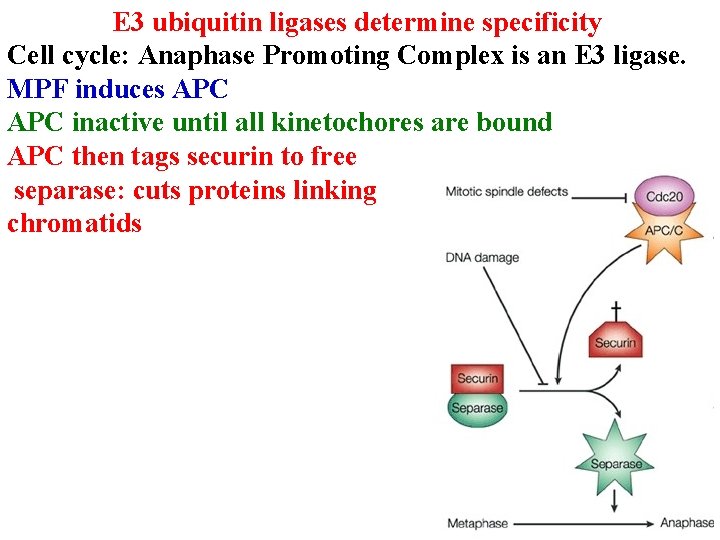
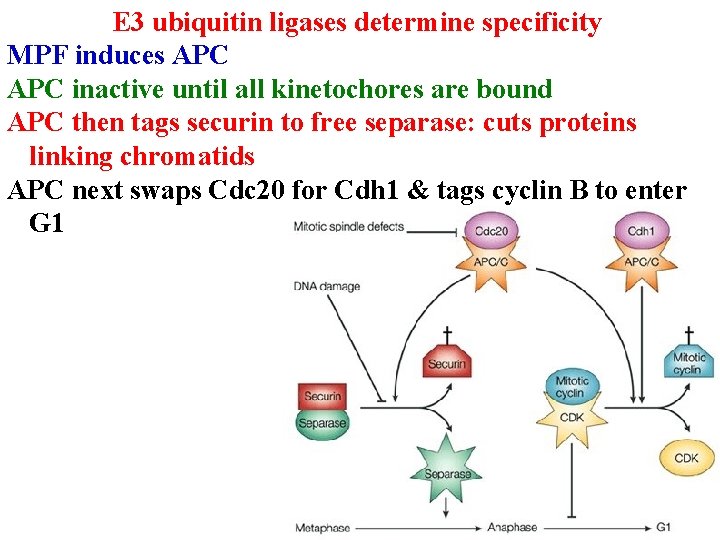
E 3 ubiquitin ligases determine specificity Cell cycle: Anaphase Promoting Complex is an E 3 ligase. MPF induces APC inactive until all kinetochores are bound APC then tags securin to free separase: cuts proteins linking chromatids

E 3 ubiquitin ligases determine specificity MPF induces APC inactive until all kinetochores are bound APC then tags securin to free separase: cuts proteins linking chromatids APC next swaps Cdc 20 for Cdh 1 & tags cyclin B to enter G 1

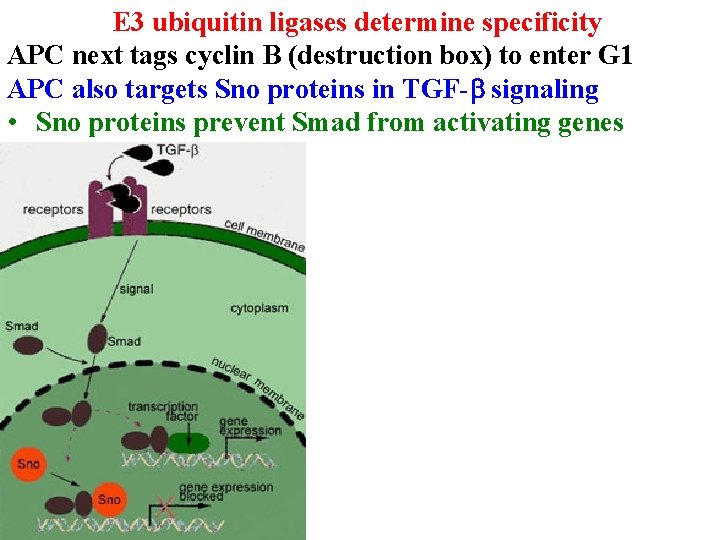
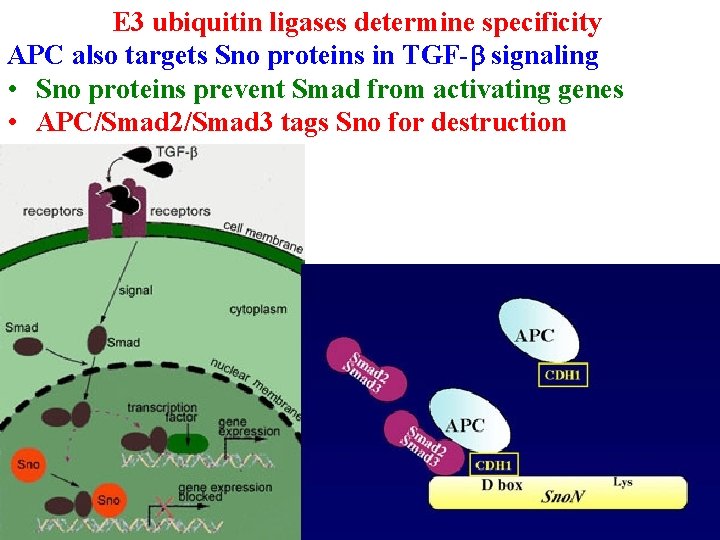
E 3 ubiquitin ligases determine specificity APC next tags cyclin B (destruction box) to enter G 1 APC also targets Sno proteins in TGF-b signaling • Sno proteins prevent Smad from activating genes

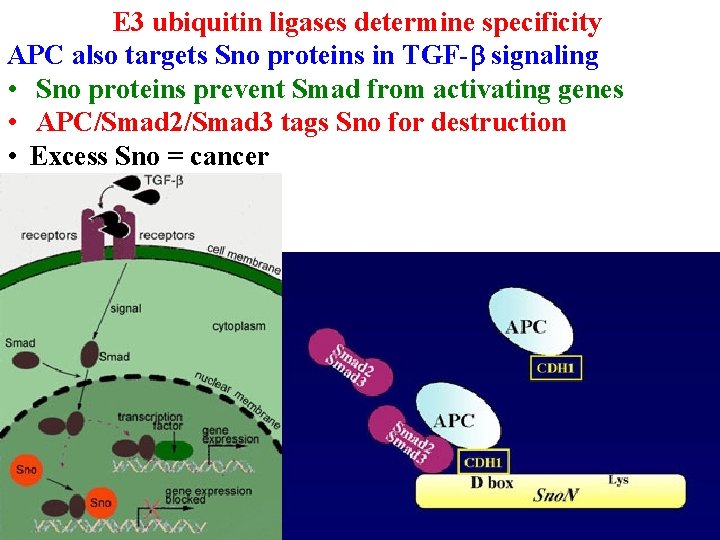
E 3 ubiquitin ligases determine specificity APC also targets Sno proteins in TGF-b signaling • Sno proteins prevent Smad from activating genes • APC/Smad 2/Smad 3 tags Sno for destruction

E 3 ubiquitin ligases determine specificity APC also targets Sno proteins in TGF-b signaling • Sno proteins prevent Smad from activating genes • APC/Smad 2/Smad 3 tags Sno for destruction • Excess Sno = cancer

E 3 ubiquitin ligases determine specificity APC also targets Sno proteins in TGF-b signaling • Sno proteins prevent Smad from activating genes • APC/Smad 2/Smad 3 tags Sno for destruction • Excess Sno = cancer Angelman syndrome = bad UBE 3 A • Only express maternal allele because paternal allele is methylated

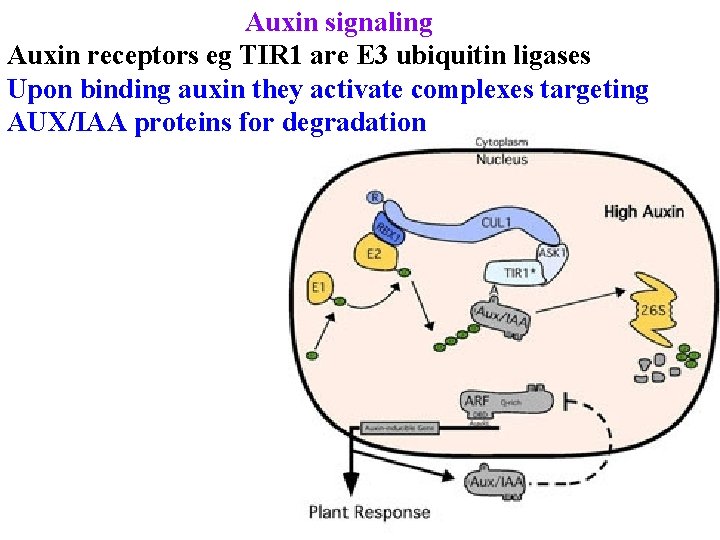
Auxin signaling Auxin receptors eg TIR 1 are E 3 ubiquitin ligases Upon binding auxin they activate complexes targeting AUX/IAA proteins for degradation

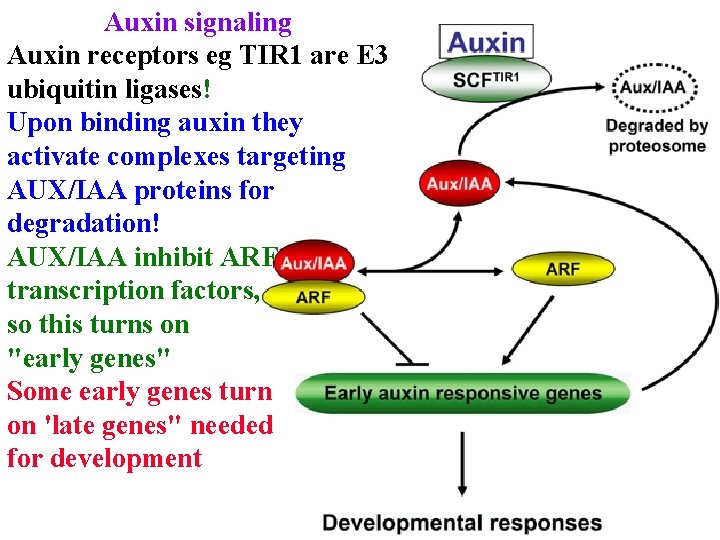
Auxin signaling Auxin receptors eg TIR 1 are E 3 ubiquitin ligases! Upon binding auxin they activate complexes targeting AUX/IAA proteins for degradation AUX/IAA inhibit ARF transcription factors, so this turns on "early genes"

Auxin signaling Auxin receptors eg TIR 1 are E 3 ubiquitin ligases! Upon binding auxin they activate complexes targeting AUX/IAA proteins for degradation! AUX/IAA inhibit ARF transcription factors, so this turns on "early genes" Some early genes turn on 'late genes" needed for development

DWD Proteins Jae-Hoon Lee’s research • putative substrate receptors for CUL 4 -based E 3 ligases

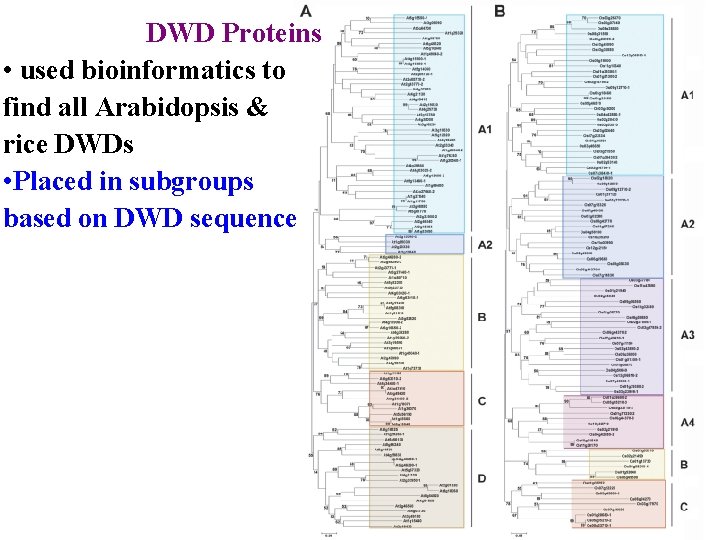
DWD Proteins Jae-Hoon Lee’s research • putative substrate receptors for CUL 4 -based E 3 ligases • used bioinformatics to find all Arabidopsis & rice DWDs

DWD Proteins • used bioinformatics to find all Arabidopsis & rice DWDs • Placed in subgroups based on DWD sequence

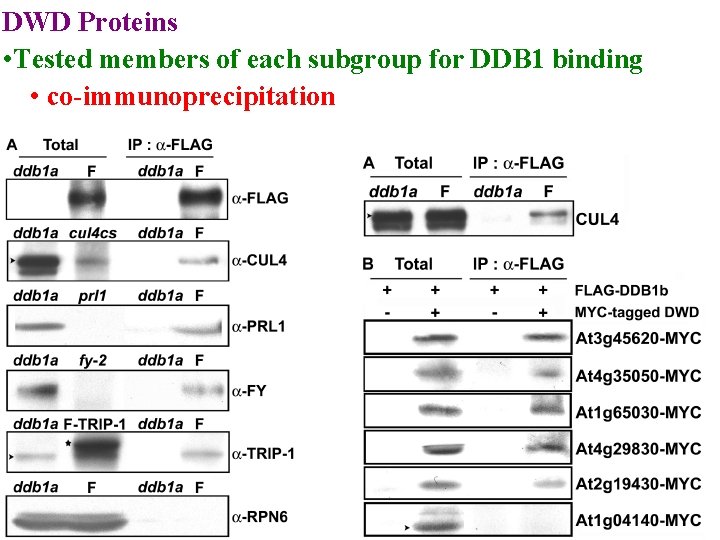
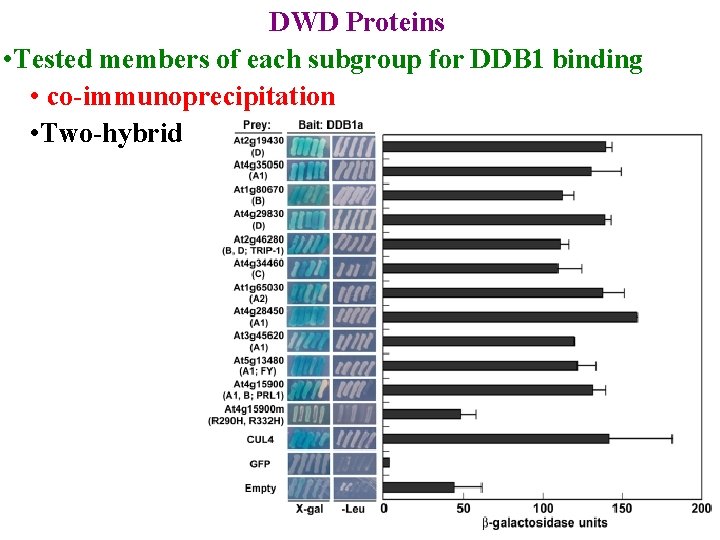
DWD Proteins • used bioinformatics to find all Arabidopsis & rice DWDs • Placed in subgroups based on DWD sequence • Tested members of each subgroup for DDB 1 binding

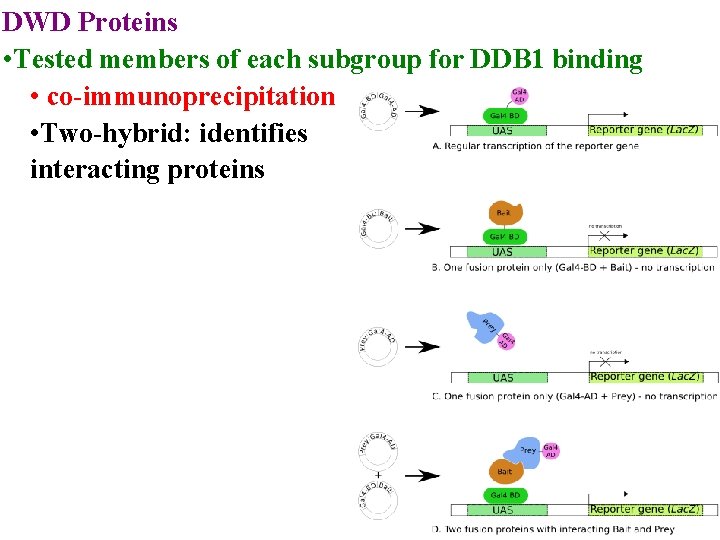
DWD Proteins • Tested members of each subgroup for DDB 1 binding • co-immunoprecipitation

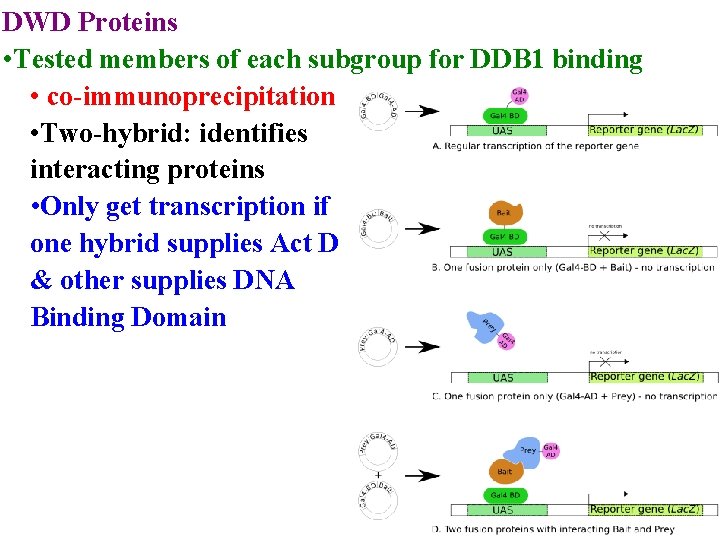
DWD Proteins • Tested members of each subgroup for DDB 1 binding • co-immunoprecipitation • Two-hybrid: identifies interacting proteins

DWD Proteins • Tested members of each subgroup for DDB 1 binding • co-immunoprecipitation • Two-hybrid: identifies interacting proteins • Only get transcription if one hybrid supplies Act D & other supplies DNA Binding Domain

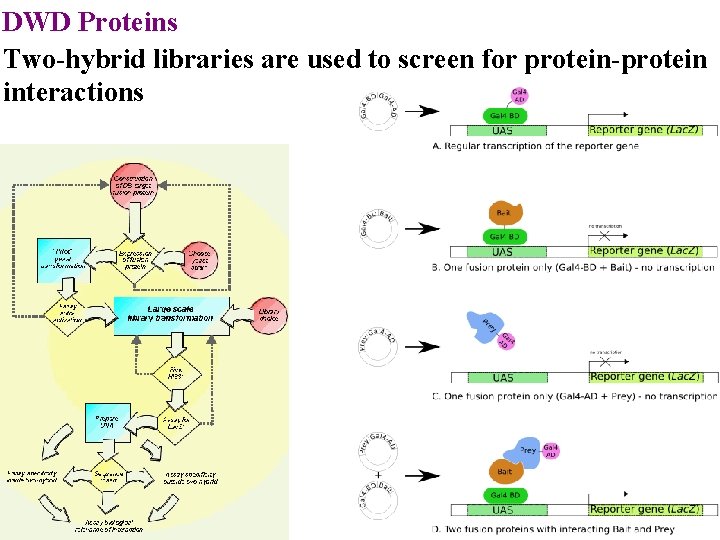
DWD Proteins Two-hybrid libraries are used to screen for protein-protein interactions

DWD Proteins • Tested members of each subgroup for DDB 1 binding • co-immunoprecipitation • Two-hybrid

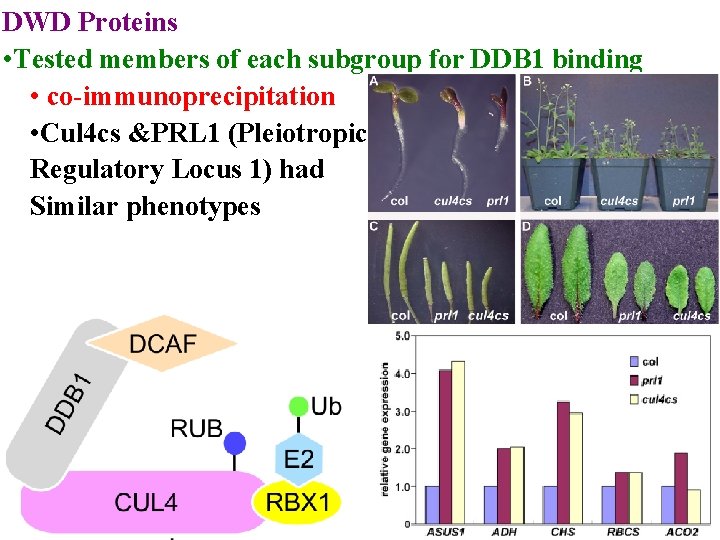
DWD Proteins • Tested members of each subgroup for DDB 1 binding • co-immunoprecipitation • Cul 4 cs &PRL 1 (Pleiotropic Regulatory Locus 1) had Similar phenotypes

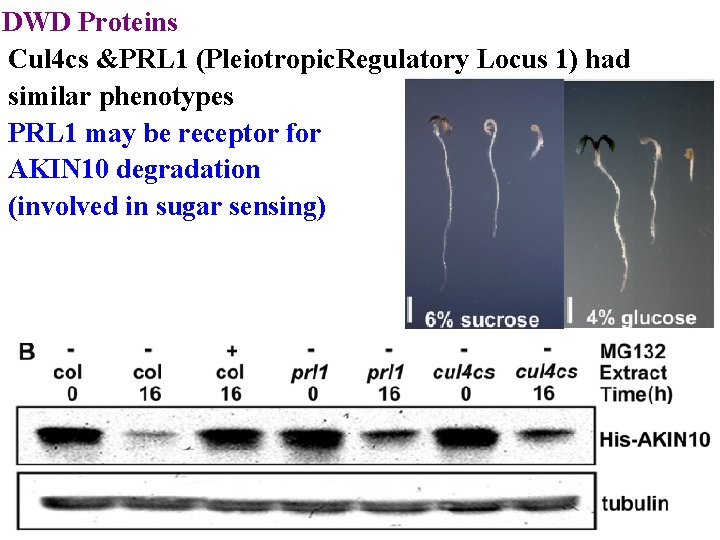
DWD Proteins Cul 4 cs &PRL 1 (Pleiotropic. Regulatory Locus 1) had similar phenotypes PRL 1 may be receptor for AKIN 10 degradation (involved in sugar sensing)

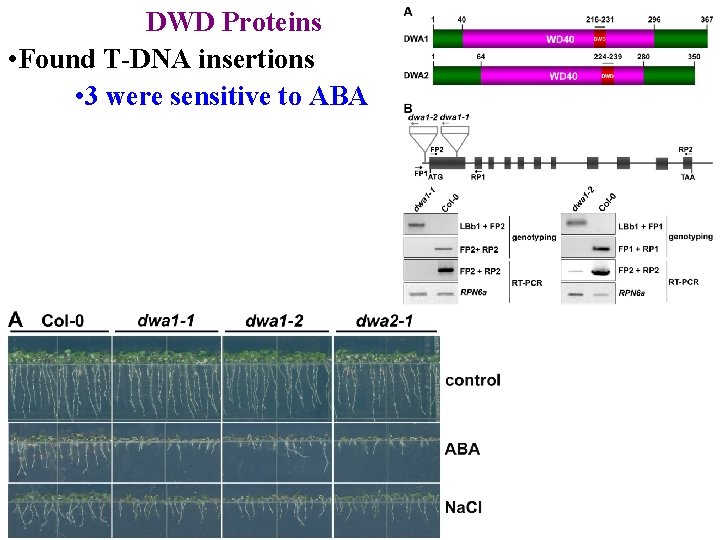
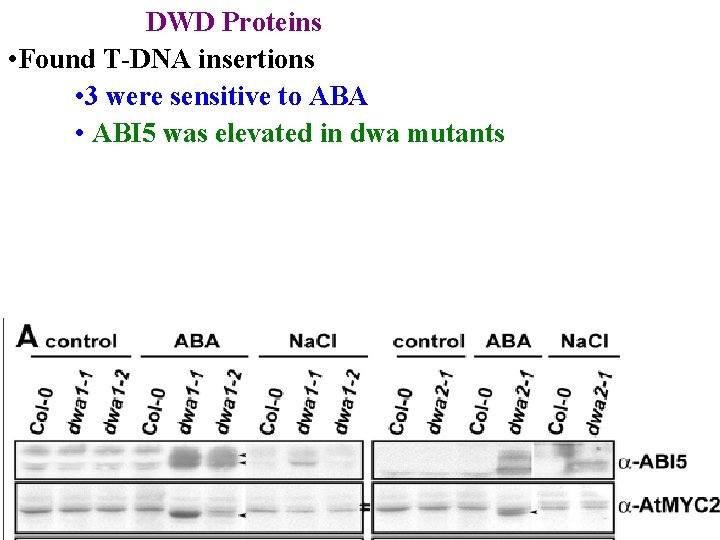
DWD Proteins • Found T-DNA insertions • 3 were sensitive to ABA

DWD Proteins • Found T-DNA insertions • 3 were sensitive to ABA • ABI 5 was elevated in dwa mutants

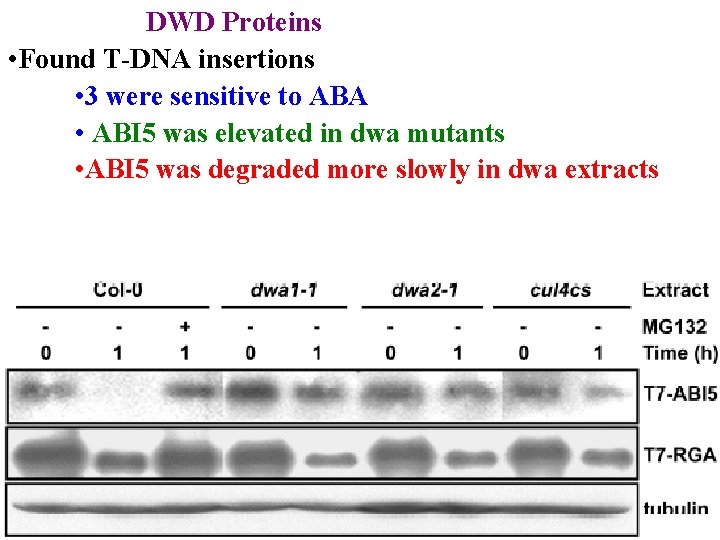
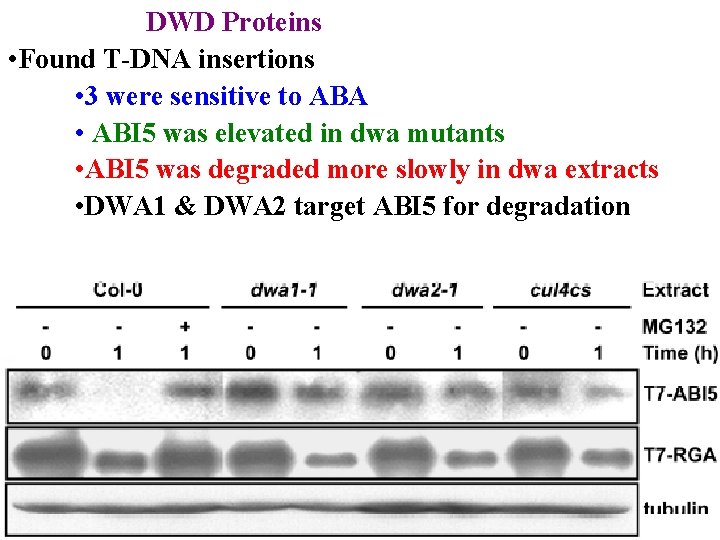
DWD Proteins • Found T-DNA insertions • 3 were sensitive to ABA • ABI 5 was elevated in dwa mutants • ABI 5 was degraded more slowly in dwa extracts

DWD Proteins • Found T-DNA insertions • 3 were sensitive to ABA • ABI 5 was elevated in dwa mutants • ABI 5 was degraded more slowly in dwa extracts • DWA 1 & DWA 2 target ABI 5 for degradation

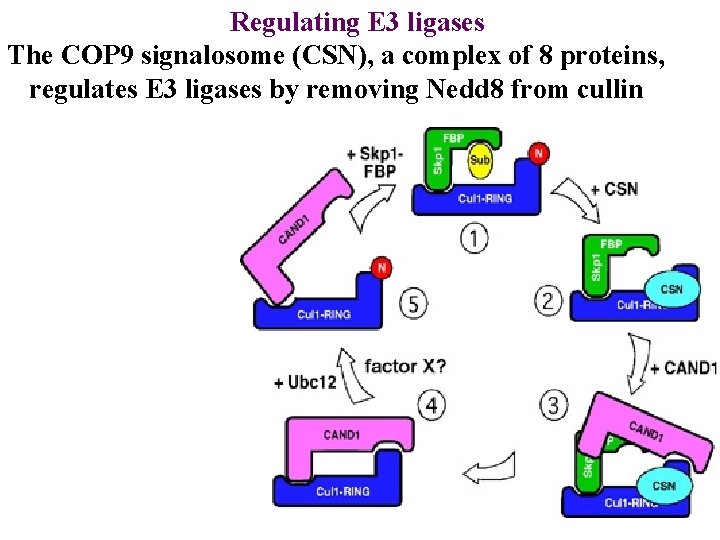
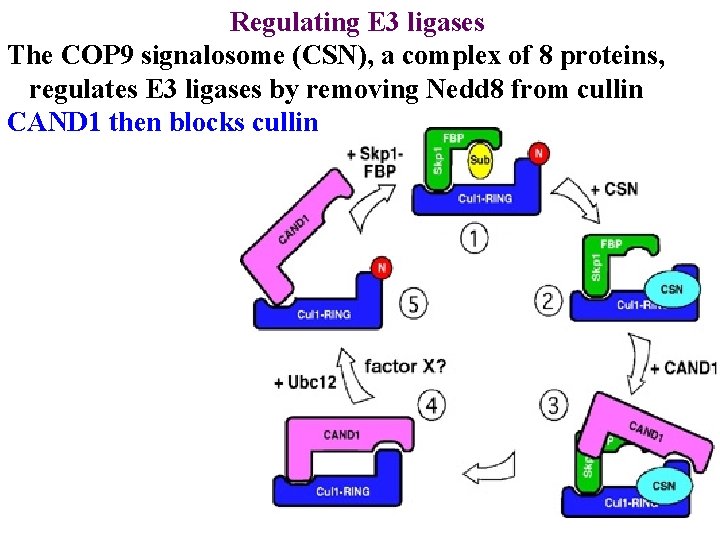
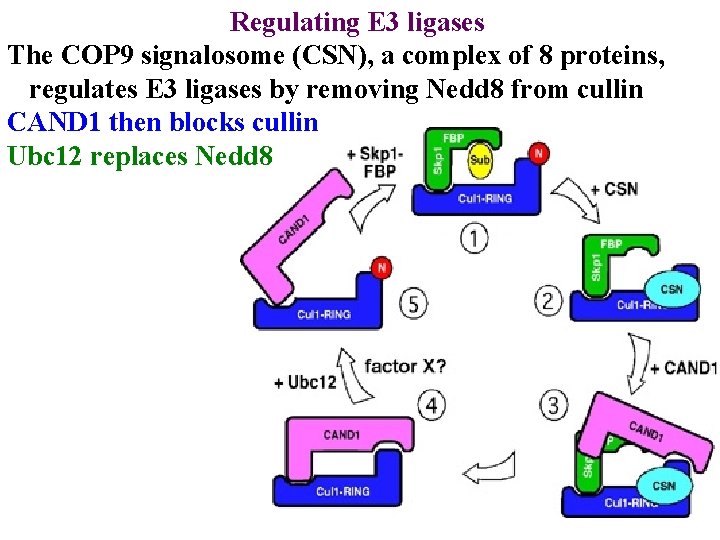
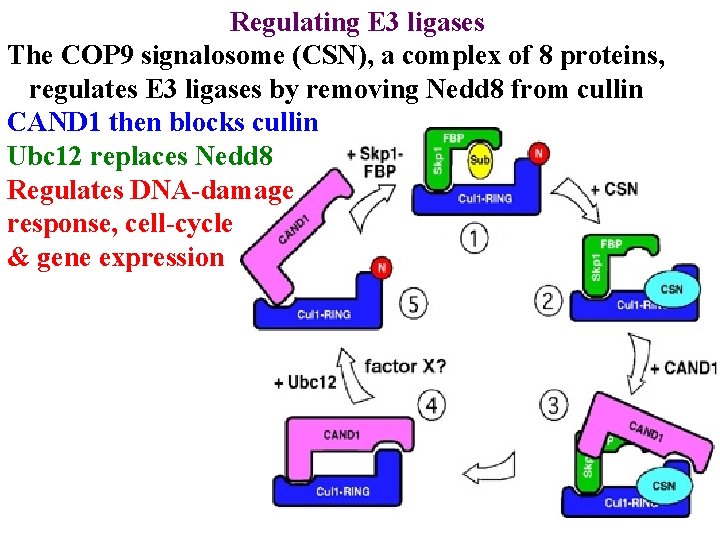
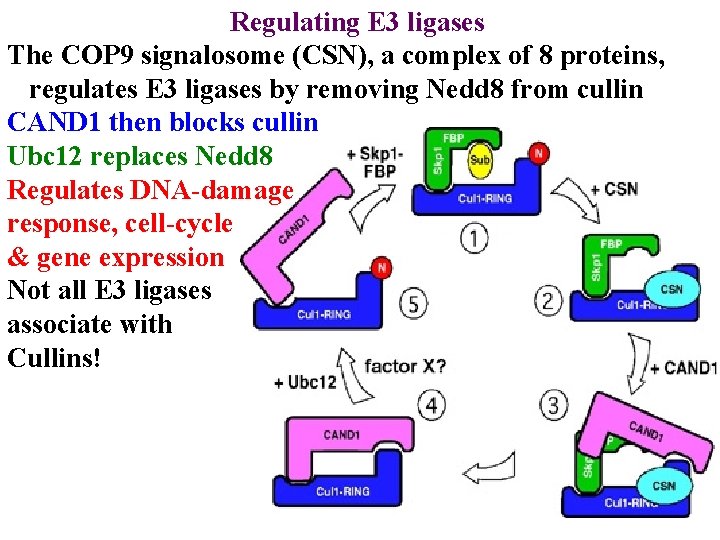
Regulating E 3 ligases The COP 9 signalosome (CSN), a complex of 8 proteins, regulates E 3 ligases by removing Nedd 8 from cullin

Regulating E 3 ligases The COP 9 signalosome (CSN), a complex of 8 proteins, regulates E 3 ligases by removing Nedd 8 from cullin CAND 1 then blocks cullin

Regulating E 3 ligases The COP 9 signalosome (CSN), a complex of 8 proteins, regulates E 3 ligases by removing Nedd 8 from cullin CAND 1 then blocks cullin Ubc 12 replaces Nedd 8

Regulating E 3 ligases The COP 9 signalosome (CSN), a complex of 8 proteins, regulates E 3 ligases by removing Nedd 8 from cullin CAND 1 then blocks cullin Ubc 12 replaces Nedd 8 Regulates DNA-damage response, cell-cycle & gene expression

Regulating E 3 ligases The COP 9 signalosome (CSN), a complex of 8 proteins, regulates E 3 ligases by removing Nedd 8 from cullin CAND 1 then blocks cullin Ubc 12 replaces Nedd 8 Regulates DNA-damage response, cell-cycle & gene expression Not all E 3 ligases associate with Cullins!

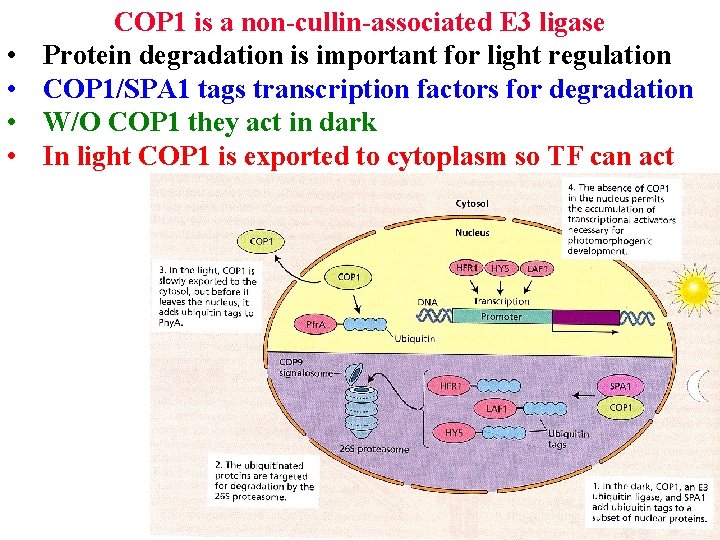
• • COP 1 is a non-cullin-associated E 3 ligase Protein degradation is important for light regulation COP 1/SPA 1 tags transcription factors for degradation W/O COP 1 they act in dark In light COP 1 is exported to cytoplasm so TF can act

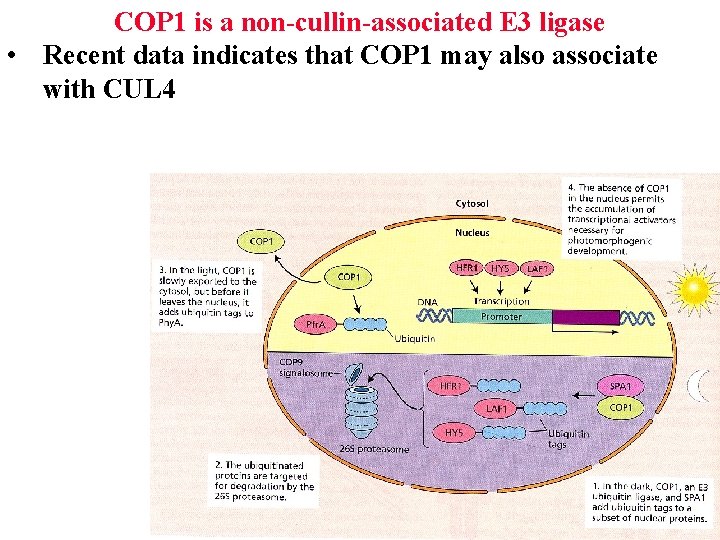
COP 1 is a non-cullin-associated E 3 ligase • Recent data indicates that COP 1 may also associate with CUL 4

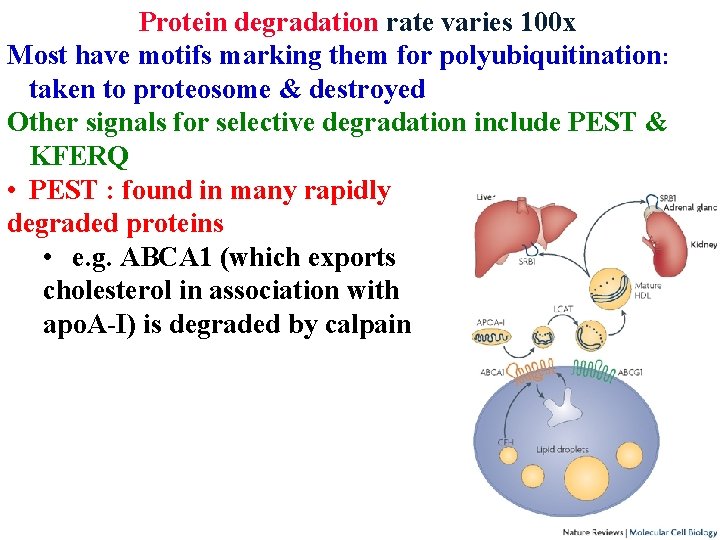
Protein degradation rate varies 100 x Most have motifs marking them for polyubiquitination: taken to proteosome & destroyed Other signals for selective degradation include PEST & KFERQ • PEST : found in many rapidly degraded proteins • e. g. ABCA 1 (which exports cholesterol in association with apo. A-I) is degraded by calpain

Protein degradation rate varies 100 x Other signals for selective degradation include PEST & KFERQ • PEST : found in many rapidly degraded proteins • e. g. ABCA 1 (which exports cholesterol in association with apo. A-I) is degraded by calpain • Deletion increases t 1/2 10 x, adding PEST drops t 1/2 10 x

Protein degradation rate varies 100 x Other signals for selective degradation include PEST & KFERQ • PEST : found in many rapidly degraded proteins • e. g. ABCA 1 (which exports cholesterol in association with apo. A-I) is degraded by calpain • Deletion increases t 1/2 10 x, adding PEST drops t 1/2 10 x • Sometimes targets poly-Ub

Protein degradation rate varies 100 x Other signals for selective degradation include PEST & KFERQ • PEST : found in many rapidly degraded proteins • e. g. ABCA 1 (which exports cholesterol in association with apo. A-I) is degraded by calpain • Deletion increases t 1/2 10 x, adding PEST drops t 1/2 10 x • Sometimes targets poly-Ub • Recent yeast study doesn’t support general role

Protein degradation rate varies 100 x Other signals for selective degradation include PEST & KFERQ • PEST : found in many rapidly degraded proteins • e. g. ABCA 1 (which exports cholesterol in association with apo. A-I) is degraded by calpain • Deletion increases t 1/2 10 x, adding PEST drops t 1/2 10 x • Sometimes targets poly-Ub • Recent yeast study doesn’t support general role • KFERQ: cytosolic proteins with KFERQ are selectively taken up by lysosomes in chaperone-mediated autophagy under conditions of nutritional or oxidative stress.