An introduction of TAIL PCR Speakers Li Wing













- Slides: 36

An introduction of TAIL PCR Speakers: Li Wing Yen Francisca, Hau Pui Lei Benni, Wong Fuk Ling

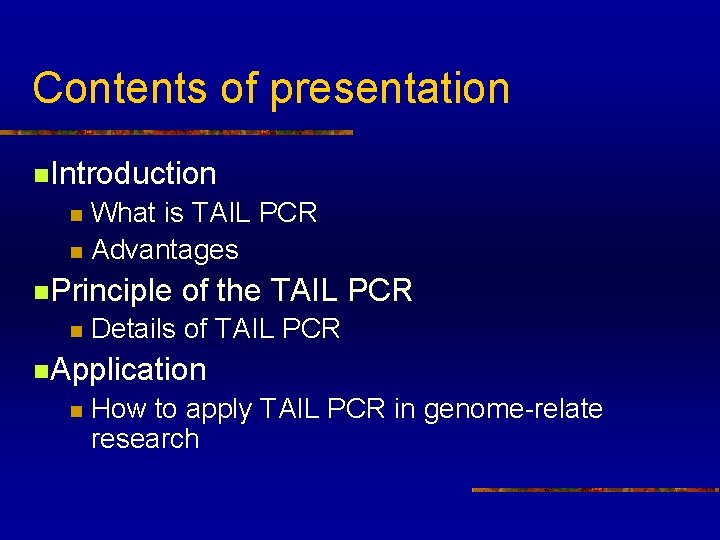
Contents of presentation n. Introduction What is TAIL PCR n Advantages n n. Principle n of the TAIL PCR Details of TAIL PCR n. Application n How to apply TAIL PCR in genome-relate research

Thermal Asymmetric Interlaced (TAIL) PCR n n A simple and powerful tool for the recovery of DNA fragments adjacent to known sequences Was developed by Liu and Whittier in 1995 Utilizes a set of nested sequence-specific primers together with a shorter arbitrary degenerate (AD) primer The relative amplification efficiencies of specific and nonspecific products can be thermally controlled

Advantages 1) 2) 3) 4) 5) 6) 7) Simplicity High specificity High efficiency Speed Less risks in chimeric artifacts Direct sequencing High sensitivity Liu & Whittier, 1995

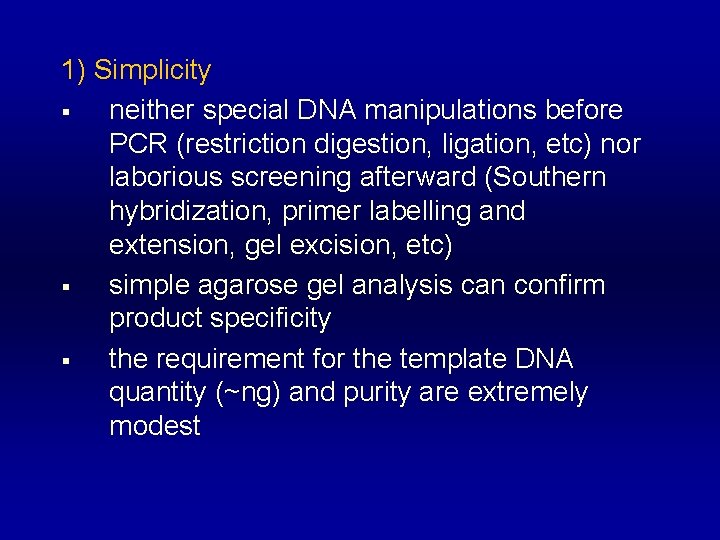
1) Simplicity § neither special DNA manipulations before PCR (restriction digestion, ligation, etc) nor laborious screening afterward (Southern hybridization, primer labelling and extension, gel excision, etc) § simple agarose gel analysis can confirm product specificity § the requirement for the template DNA quantity (~ng) and purity are extremely modest

2) High specificity § the proportion of coamplified nonspecific products is very low 3) High efficiency § 60 -80% of reactions yielded specific products with any given AD primer 4) Speed § The successive amplification reactions can all be completed in 1 day

5) Less risks in chimeric artifacts § TAIL PCR doesn't involve ligation step 6) Direct sequencing § The high specific reaction products can be added directly to the sequencing reaction , no gel excision and purification are required 7) High sensitivity § Single-copy sequences in genome can be amplified

Principle of TAIL-PCR

Important features of TAIL-PCR n n n Primer design Annealing temperature Cycling orders

Primer Design n Specific primer (SP) n n • Nested sequence specific primer complementary to vector sequence High melting temperature, Tm=58 -63 o. C Arbitrary degenerate (AD) primer – – Relatively shorter Lower melting temperature, Tm =47 -48 o. C

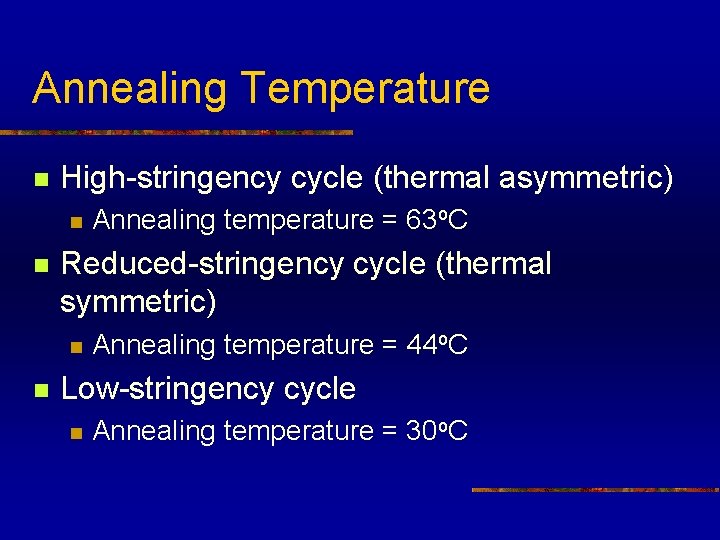
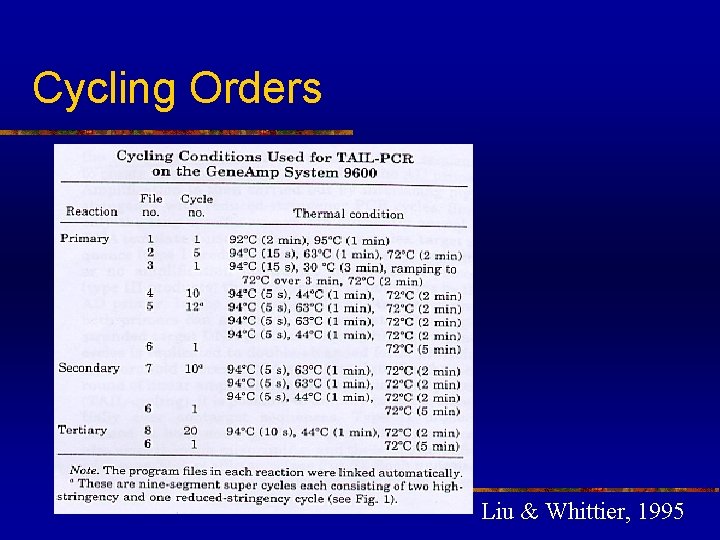
Annealing Temperature n High-stringency cycle (thermal asymmetric) n n Reduced-stringency cycle (thermal symmetric) n n Annealing temperature = 63 o. C Annealing temperature = 44 o. C Low-stringency cycle n Annealing temperature = 30 o. C

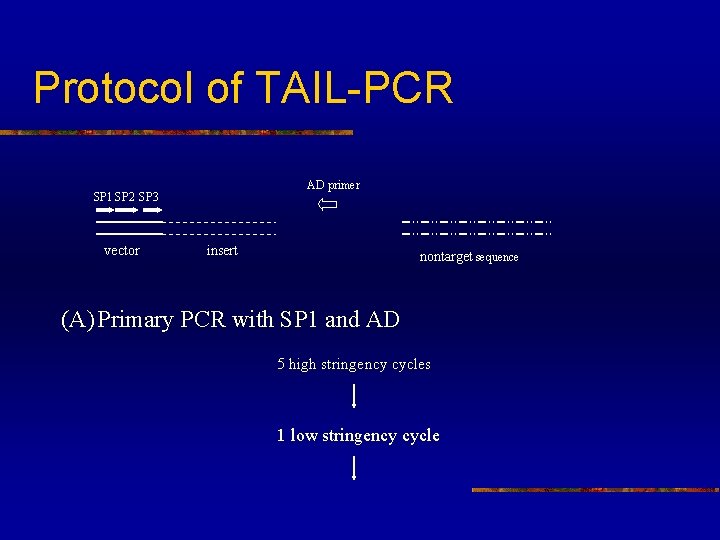
Protocol of TAIL-PCR AD primer SP 1 SP 2 SP 3 vector insert nontarget sequence (A) Primary PCR with SP 1 and AD 5 high stringency cycles 1 low stringency cycle

Protocol of TAIL-PCR 10 reduced stringency cycles 1 reduced stringency cycle (thermal symmetric) Product yield: TAIL-cycling (12 super cycles) 2 high stringency cycles (thermal asymmetric) Specific product (type I) Nonspecific product (type III) High or middle (detectable or undetectable) High (detectable) Low (undetectable)

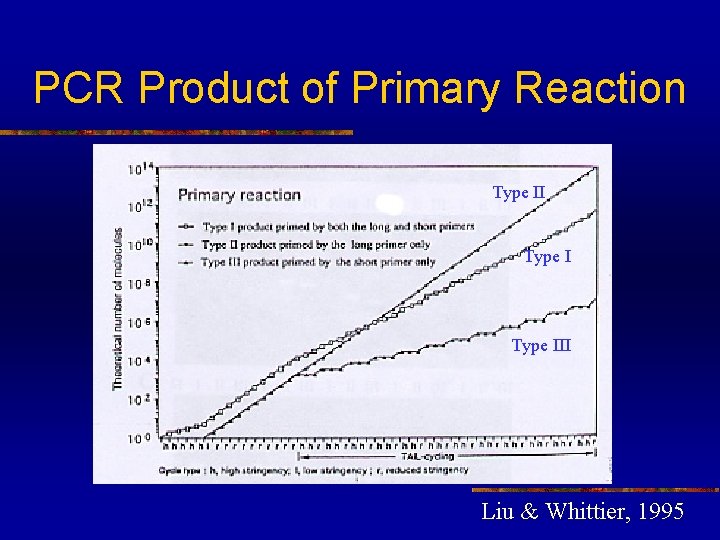
PCR Product of Primary Reaction Type III Liu & Whittier, 1995

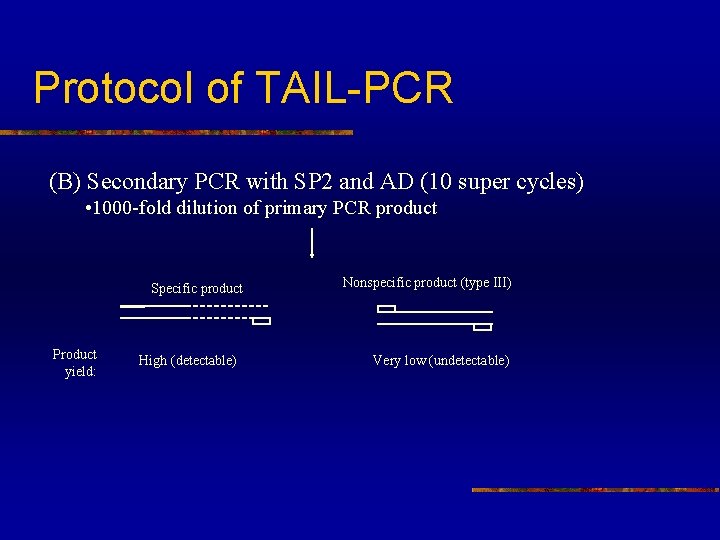
Protocol of TAIL-PCR (B) Secondary PCR with SP 2 and AD (10 super cycles) • 1000 -fold dilution of primary PCR product Specific product Product yield: High (detectable) Nonspecific product (type III) Very low (undetectable)

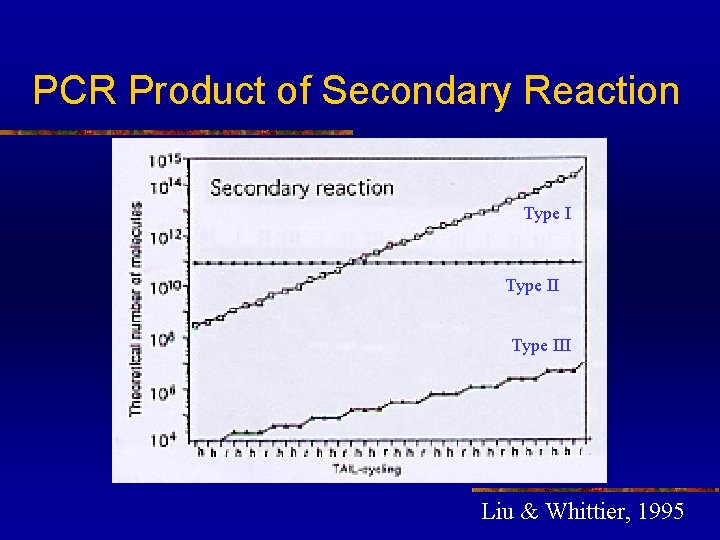
PCR Product of Secondary Reaction Type III Liu & Whittier, 1995

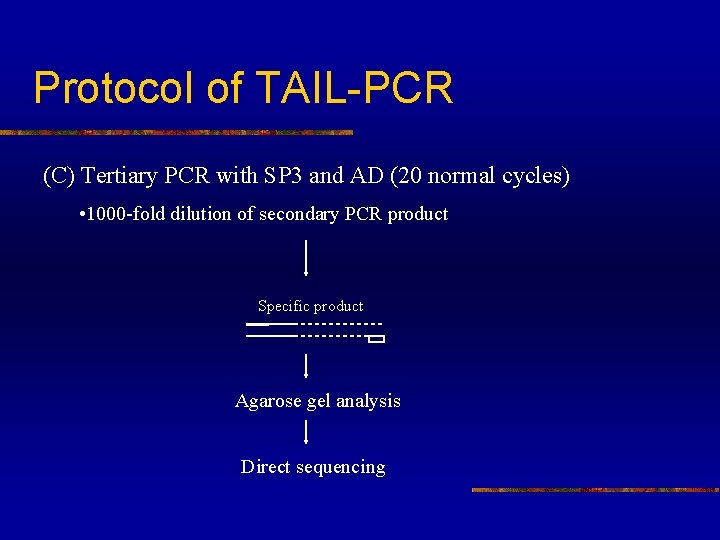
Protocol of TAIL-PCR (C) Tertiary PCR with SP 3 and AD (20 normal cycles) • 1000 -fold dilution of secondary PCR product Specific product Agarose gel analysis Direct sequencing

Cycling Orders Liu & Whittier, 1995

Application

High efficiency to amplify insert end segments from P 1, BAC and YAC clones n n n TAIL-PCR as a powerful tool for amplifying insert end segments from P 1, BAC and YAC clones The amplified products were highly specific and suitable as probes for library screening and as templates for direct sequencing The recover insert ends can also be used for chromosome walking and mapping

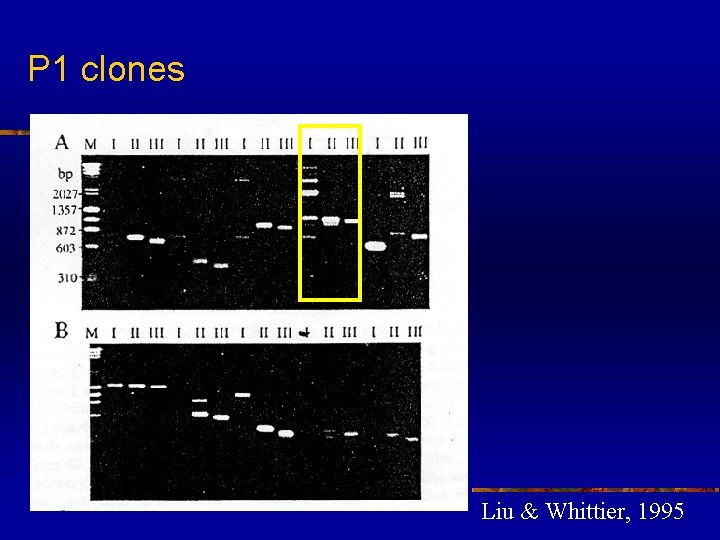
P 1 clones Liu & Whittier, 1995

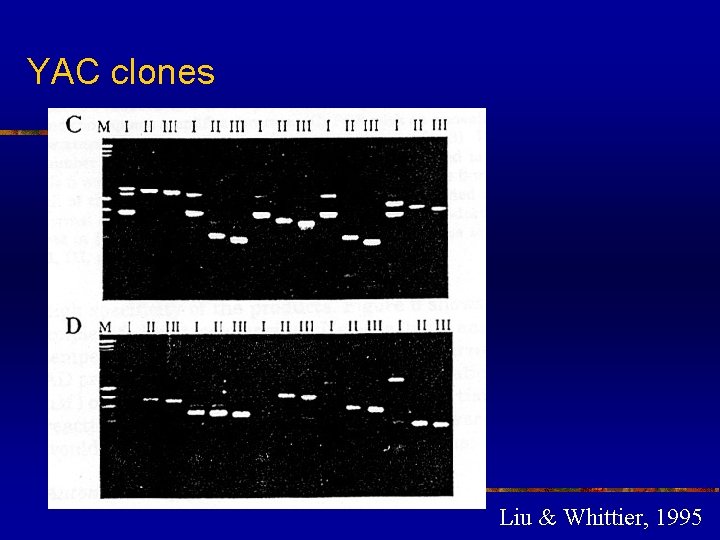
YAC clones Liu & Whittier, 1995

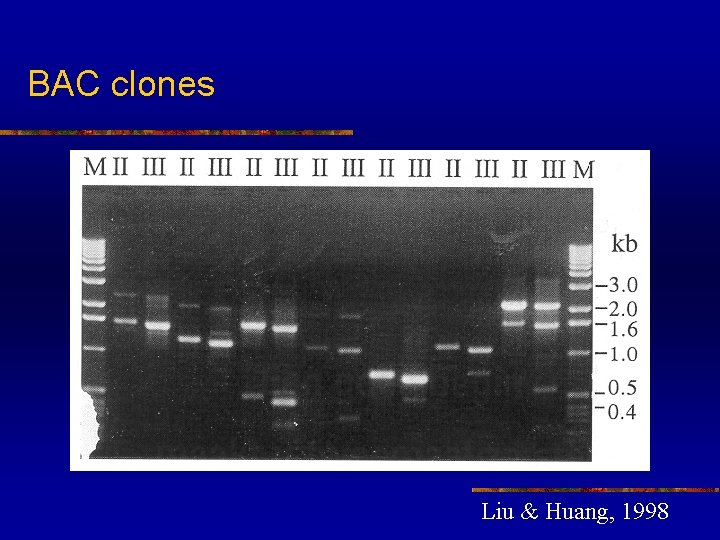
BAC clones Liu & Huang, 1998

High efficiency to amplify insert end segments from P 1, BAC and YAC clones n n Many product bands from the primary TAIL-PCR reaction disappeared after the secondary TAIL-PCR, indicating that these were non-specific type II products Specific products were not always seen in the primary reactions due to their low concentration. However, these specific products becomes visible after the subsequent secondary reaction

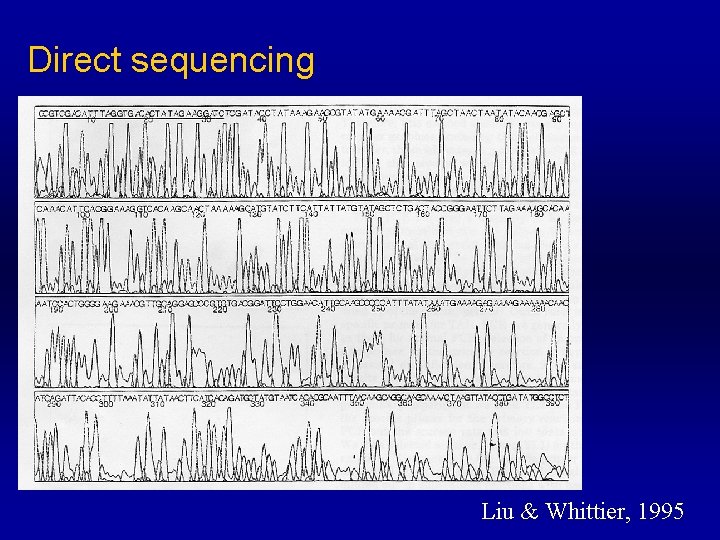
Direct Sequencing n n Because it’s high specificity, unpurified TAIL-PCR products can be directly sequenced. Unpurified products yielded clear sequencing profiles

Direct sequencing Liu & Whittier, 1995

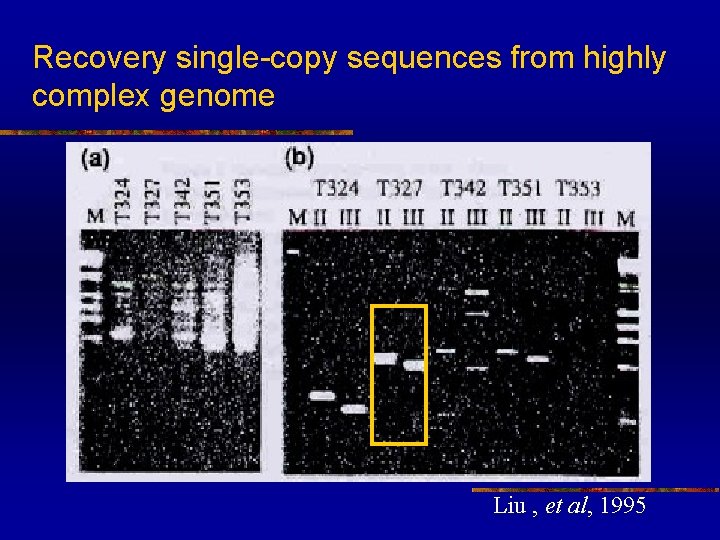
Recovery single-copy sequences from highly complex genome n n Amplification of single copy sequences was found technically more difficult in organisms with large genome. e. g. Inverse PCR is difficult to apply to genomes containing over 109 bp However, TAIL-PCR is very sensitive and can be applied to highly complex genomes

Recovery single-copy sequences from highly complex genome Liu , et al, 1995

Rapid isolation of promoter sequences n n The isolation of promoter and enhancer sequences is a crucial step in the study of the regulation of gene expression Flanking regions of genes, containing these elements, were conventionally isolated by screening genomic libraries using c. DNA as probes, which is very timeconsuming

Rapid isolation of promoter sequences n n Therefore, simpler and more reliable, and preferably PCR-based methods for promoter isolation are urgently required. Unlike Inverse PCR and ligation-mediated PCR, TAIL-PCR is a simple and efficient technique for genomic walking which does not require any restriction or ligation steps.

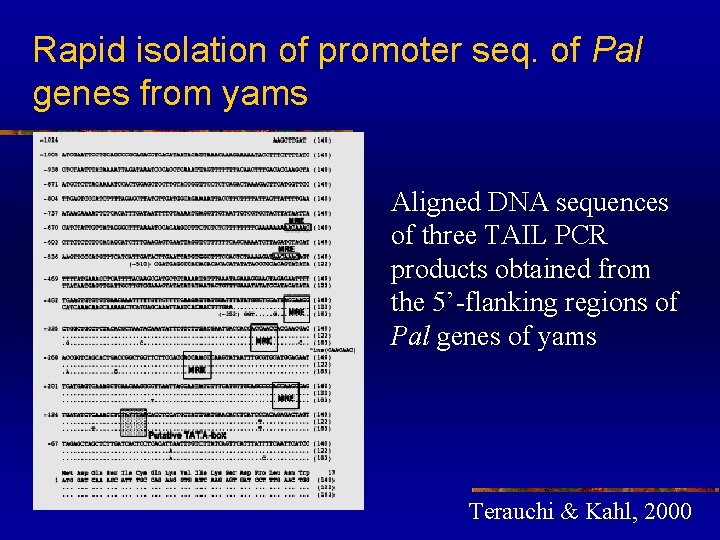
Rapid isolation of promoter seq. of Pal genes from yams Aligned DNA sequences of three TAIL PCR products obtained from the 5’-flanking regions of Pal genes of yams Terauchi & Kahl, 2000

Rapid isolation of promoter seq. of Pal from yams n DNA sequences of PCR products overlapped perfectly with the 5’-end sequence of the c. DNA. In the region isolated, a putative TATA box and several MREs could be identified.

Rapid isolation of promoter seq. of Pal genes from yams n n Isolated 5’-flanking regions of Pal and Pgi genes were fused to the GUS gene, and their activity was tested by transient transformation after delivery into tobacco BY 2 cells by particle bombardment. All the isolated 5’-flanking regions were shown to drive reporter gene expression.

Conclusion n TAIL-PCR is highly specific and efficient for amplification of DNA segments adjacent to known sequences Upon different modification, this technique could be used to handle vary tasks: Amplification of Insert Ends fragments from P 1, YAC and BAC clones for chromosome walking

Conclusion n n Isolation of 5’ flanking region of genes Isolation of promoter sequences Isolation of T-DNA insert junctions for genome physical mapping, development of sequence-tagged sites (STS), and analysis of genomic sequences flanking T-DNA, transposon or ritrovirus insertions.

 Tail pcr principle
Tail pcr principle Bat wing bird wing
Bat wing bird wing Political spectrum canada
Political spectrum canada Electromagnet in speakers
Electromagnet in speakers Agora speakers international
Agora speakers international Audio over ip speakers
Audio over ip speakers Speaker in poem
Speaker in poem Speaker's tone
Speaker's tone Teaching english to arabic speakers
Teaching english to arabic speakers Language in malay
Language in malay Nnnn speakers
Nnnn speakers Soundblock coaxial speakers creative commons pictures
Soundblock coaxial speakers creative commons pictures Banquo characteristics
Banquo characteristics Dear distinguished guests
Dear distinguished guests Agora speakers
Agora speakers Speakers
Speakers Portuguese speakers in macau
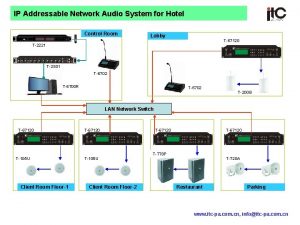
Portuguese speakers in macau Sound system for hotel
Sound system for hotel Vansina
Vansina Yeast colony pcr
Yeast colony pcr Fases pcr
Fases pcr Sispa pcr
Sispa pcr Rcp extracorpórea
Rcp extracorpórea Poonum patel
Poonum patel Pcr phases
Pcr phases Biorad pcr master mix
Biorad pcr master mix Pcr troubleshooting multiple bands
Pcr troubleshooting multiple bands Objectives of pcr
Objectives of pcr Application of pcr are
Application of pcr are Pcr en tiempo final
Pcr en tiempo final Calcul efficacité pcr quantitative
Calcul efficacité pcr quantitative Pcr abi
Pcr abi Pcr lab setup
Pcr lab setup Pcr nobel prize
Pcr nobel prize Ecr engineering change request
Ecr engineering change request Crystal digital pcr
Crystal digital pcr Missy baker
Missy baker