Light Cycler 480 RealTime PCR System Elin Shen














- Slides: 68

Light. Cycler® 480 Real-Time PCR System Elin Shen

1. LC 480 Basic training 2. LC 480 Software 2

LC 480 Basic training : 1. Real time PCR principle introduction 2. Detection formats 3. Absolute/Relative Quantification 4. Genotyping 5. Light. Cycler® 480 Instrument introduction 3

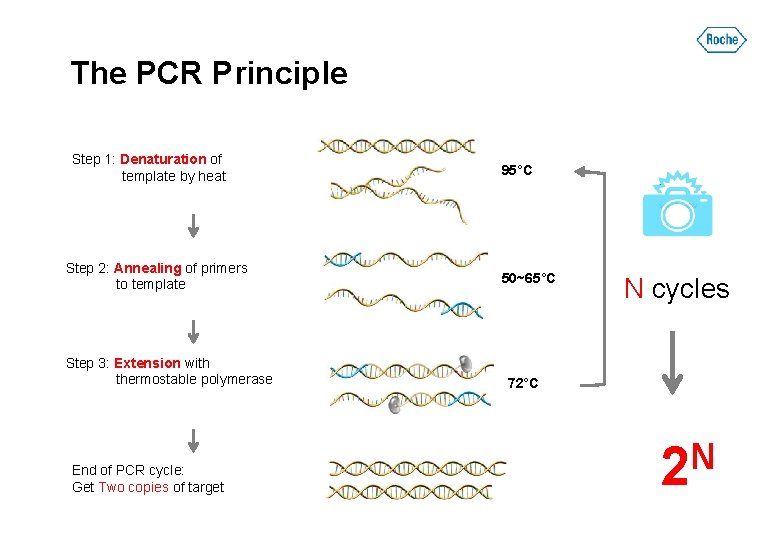
The PCR Principle Step 1: Denaturation of template by heat Step 2: Annealing of primers to template Step 3: Extension with thermostable polymerase End of PCR cycle: Get Two copies of target 95°C 50~65°C N cycles 72°C N 2

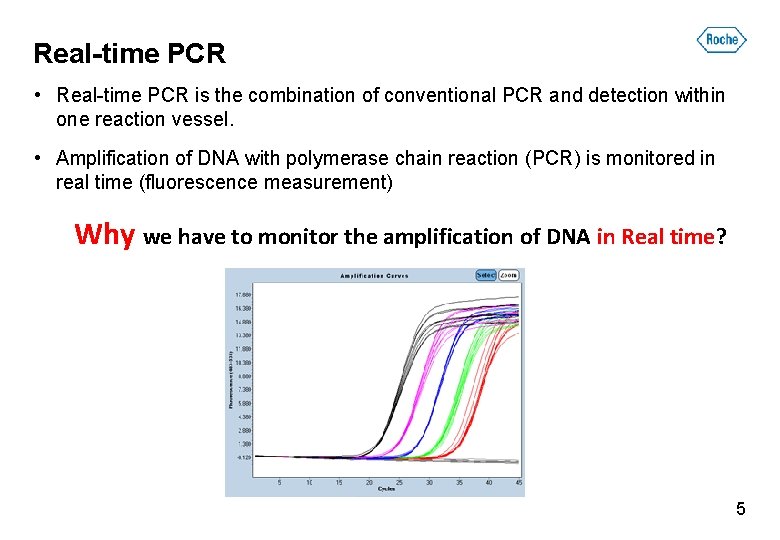
Real-time PCR • Real-time PCR is the combination of conventional PCR and detection within one reaction vessel. • Amplification of DNA with polymerase chain reaction (PCR) is monitored in real time (fluorescence measurement) Why we have to monitor the amplification of DNA in Real time? 5

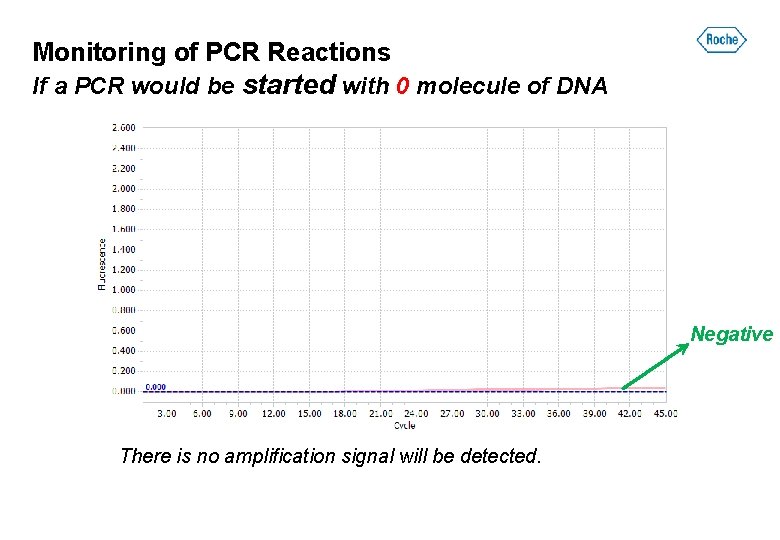
Monitoring of PCR Reactions If a PCR would be started with 0 molecule of DNA Negative There is no amplification signal will be detected.

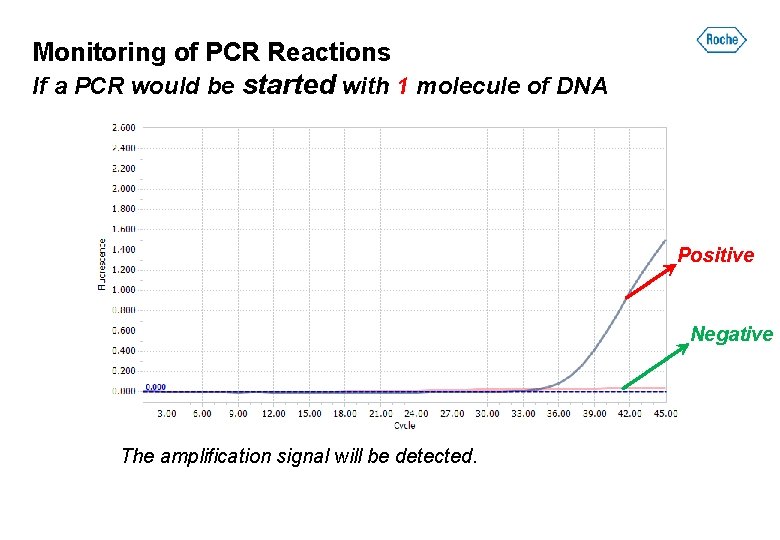
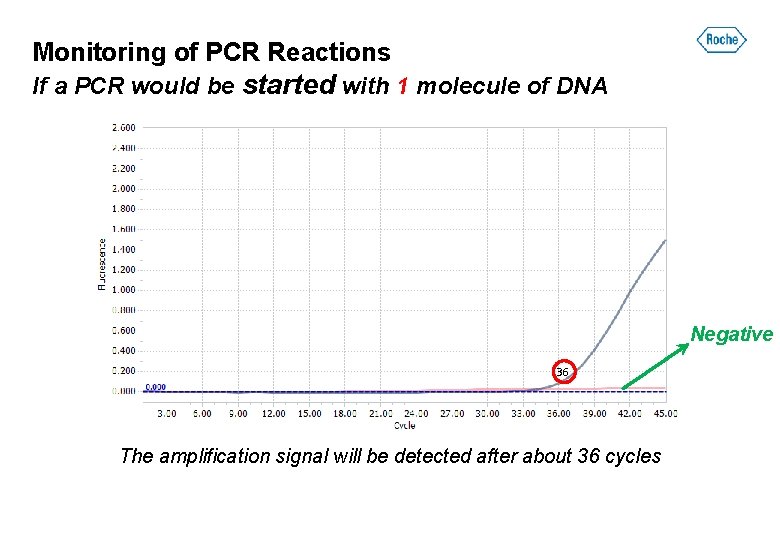
Monitoring of PCR Reactions If a PCR would be started with 1 molecule of DNA Positive Negative The amplification signal will be detected.

Monitoring of PCR Reactions If a PCR would be started with 1 molecule of DNA Negative 36 The amplification signal will be detected after about 36 cycles

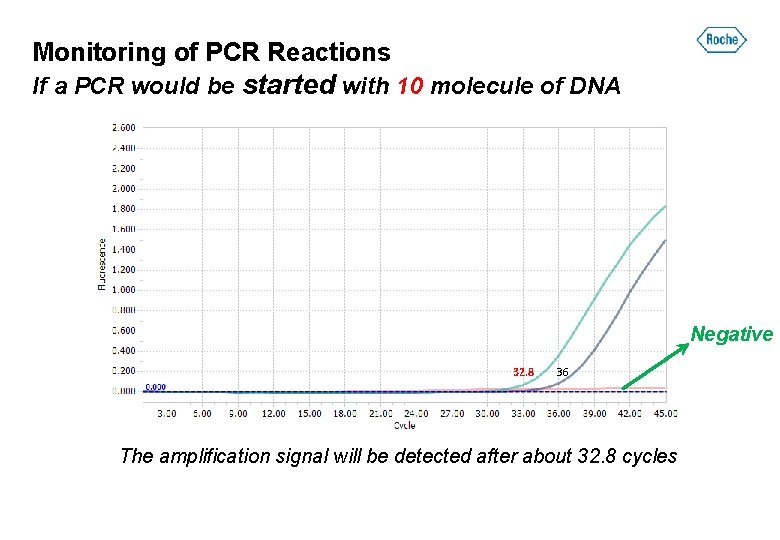
Monitoring of PCR Reactions If a PCR would be started with 10 molecule of DNA Negative 32. 8 36 The amplification signal will be detected after about 32. 8 cycles

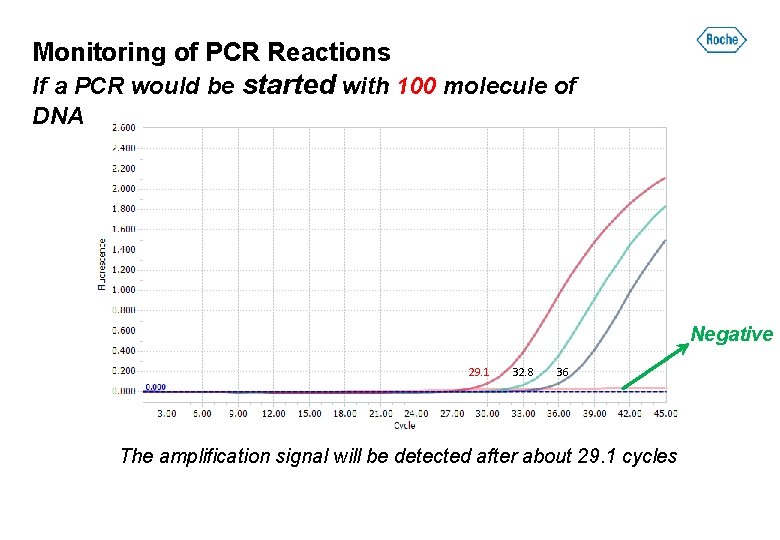
Monitoring of PCR Reactions If a PCR would be started with 100 molecule of DNA Negative 29. 1 32. 8 36 The amplification signal will be detected after about 29. 1 cycles

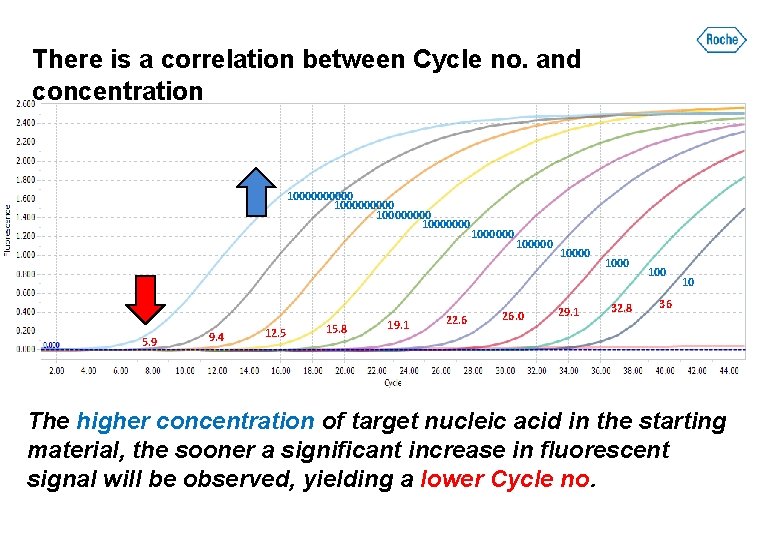
There is a correlation between Cycle no. and concentration 100000 10000000 5. 9 9. 4 12. 5 15. 8 19. 1 22. 6 1000000 100000 26. 0 10000 29. 1 1000 32. 8 100 10 36 The higher concentration of target nucleic acid in the starting material, the sooner a significant increase in fluorescent signal will be observed, yielding a lower Cycle no.

Introduction • Real-time PCR is the combination of conventional PCR and detection within one reaction vessel. • Amplification of DNA with polymerase chain reaction (PCR) is monitored in real time (fluorescence measurement) How to monitor the amplification of DNA in Real time? 12

LC 480 Basic training : 1. Real time PCR principle introduction 2. Detection formats 3. Absolute/Relative Quantification 4. Genotyping 5. Light. Cycler® 480 Instrument introduction 13

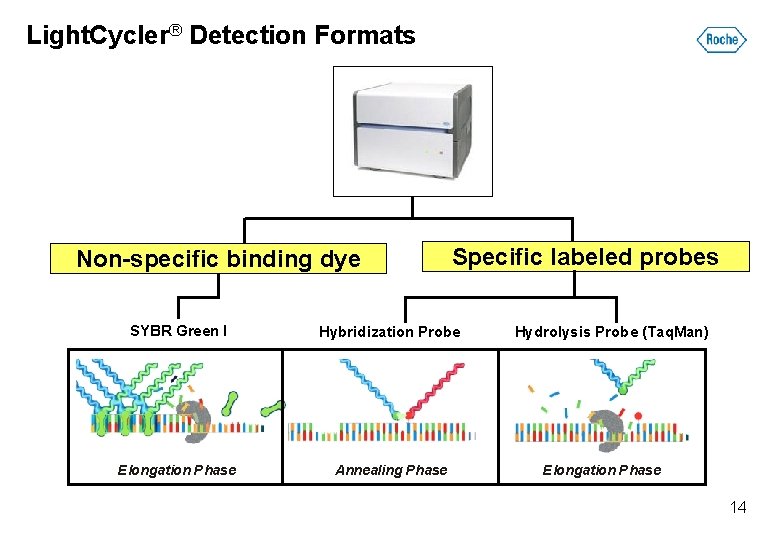
Light. Cycler® Detection Formats Non-specific binding dye Specific labeled probes SYBR Green I Hybridization Probe Elongation Phase Annealing Phase Hydrolysis Probe (Taq. Man) Elongation Phase 14

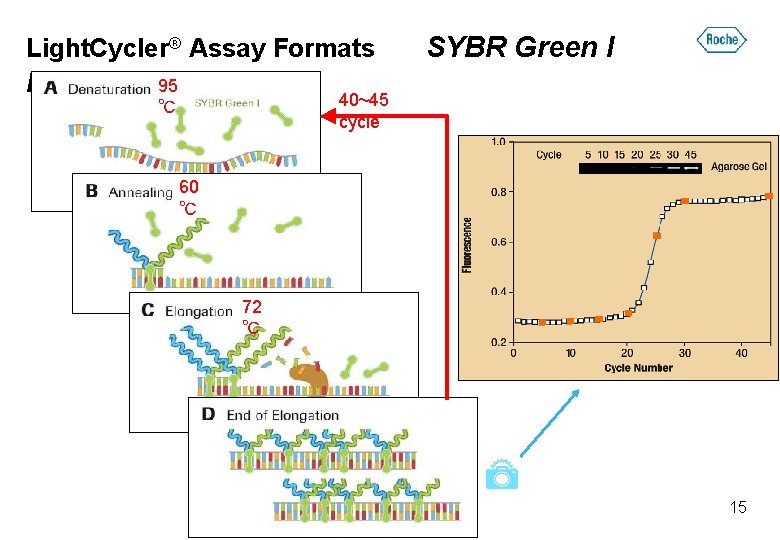
Light. Cycler® Assay Formats Format 95 ℃ SYBR Green I 40~45 cycle 60 ℃ 72 ℃ 15

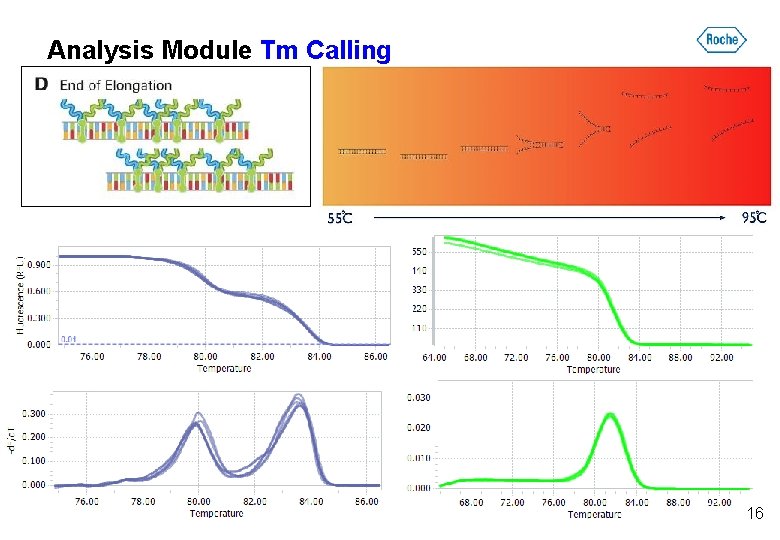
Analysis Module Tm Calling 16

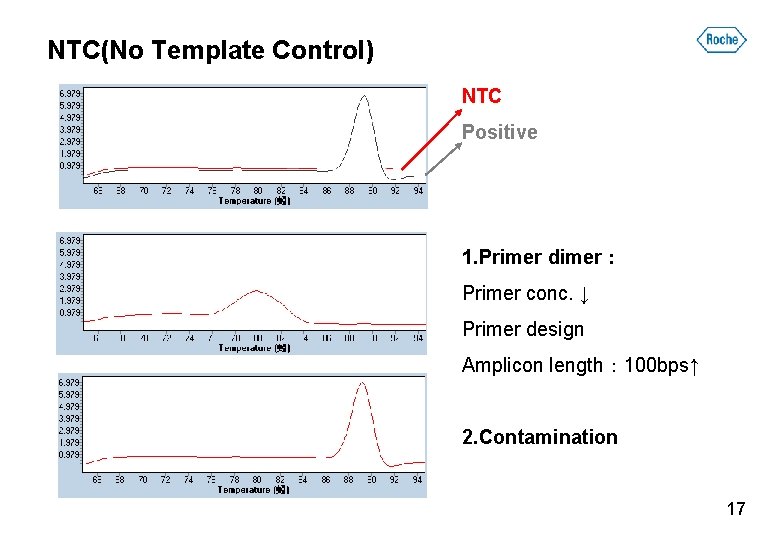
NTC(No Template Control) NTC Positive 1. Primer dimer: Primer conc. ↓ Primer design Amplicon length: 100 bps↑ 2. Contamination 17

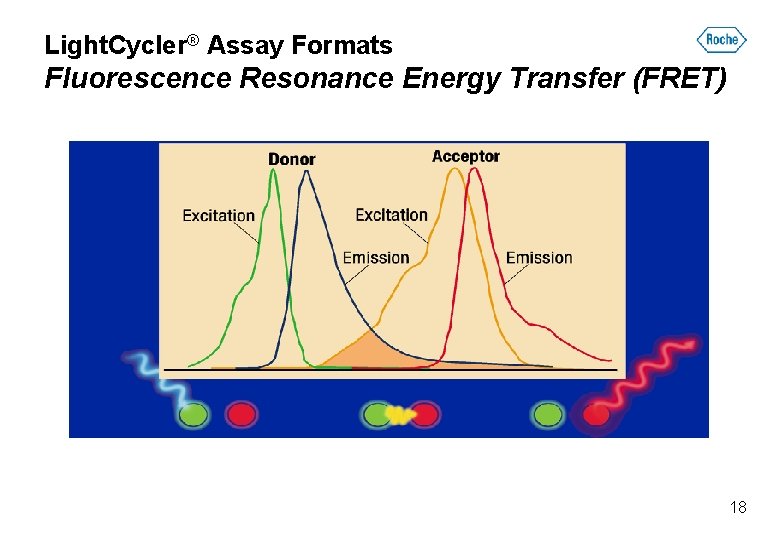
Light. Cycler® Assay Formats Fluorescence Resonance Energy Transfer (FRET) 18

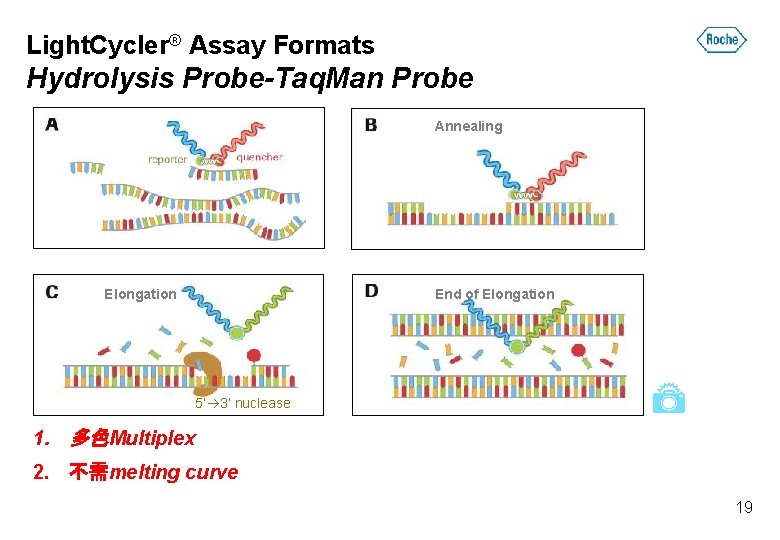
Light. Cycler® Assay Formats Hydrolysis Probe-Taq. Man Probe Annealing End of Elongation 5’ 3’ nuclease 1. 多色Multiplex 2. 不需melting curve 19

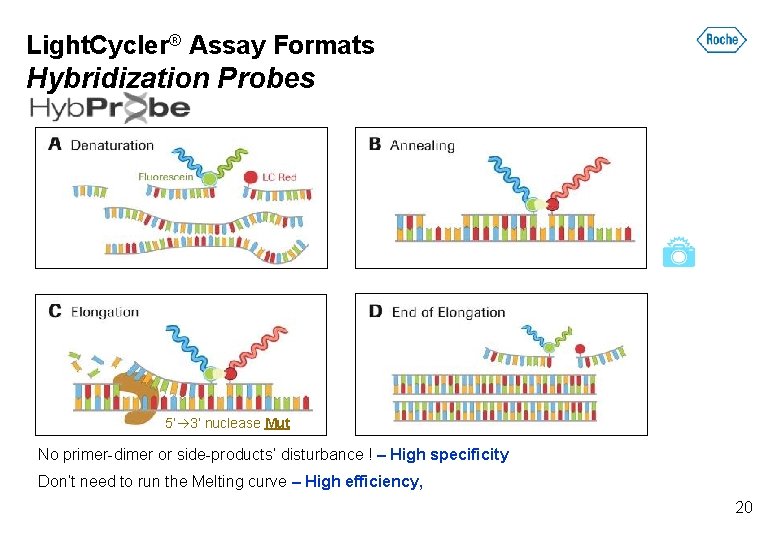
Light. Cycler® Assay Formats Hybridization Probes 5’ 3’ nuclease Mut No primer-dimer or side-products’ disturbance ! – High specificity Don’t need to run the Melting curve – High efficiency, 20

LC 480 Basic training : 1. Real time PCR principle introduction 2. Detection formats 3. Absolute/Relative Quantification 4. Genotyping 5. Light. Cycler® 480 Instrument introduction 21

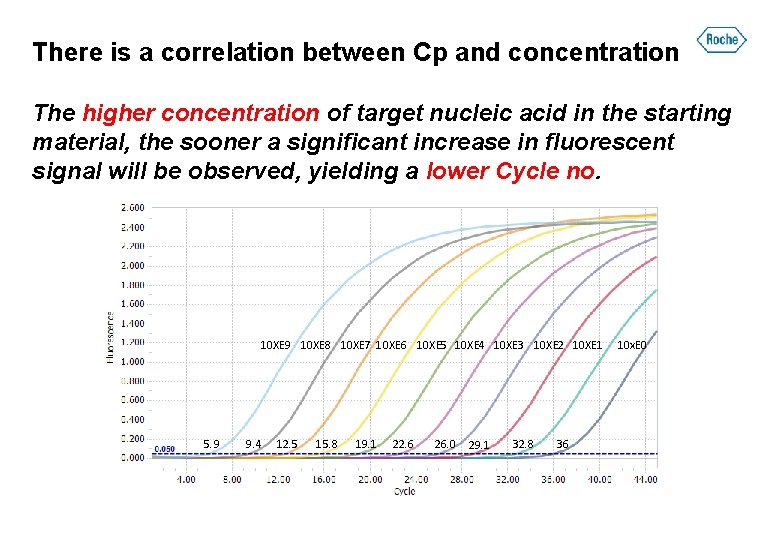
There is a correlation between Cp and concentration The higher concentration of target nucleic acid in the starting material, the sooner a significant increase in fluorescent signal will be observed, yielding a lower Cycle no. 10 XE 9 10 XE 8 10 XE 7 10 XE 6 10 XE 5 10 XE 4 10 XE 3 10 XE 2 10 XE 1 5. 9 9. 4 12. 5 15. 8 19. 1 22. 6 26. 0 29. 1 32. 8 36 10 x. E 0

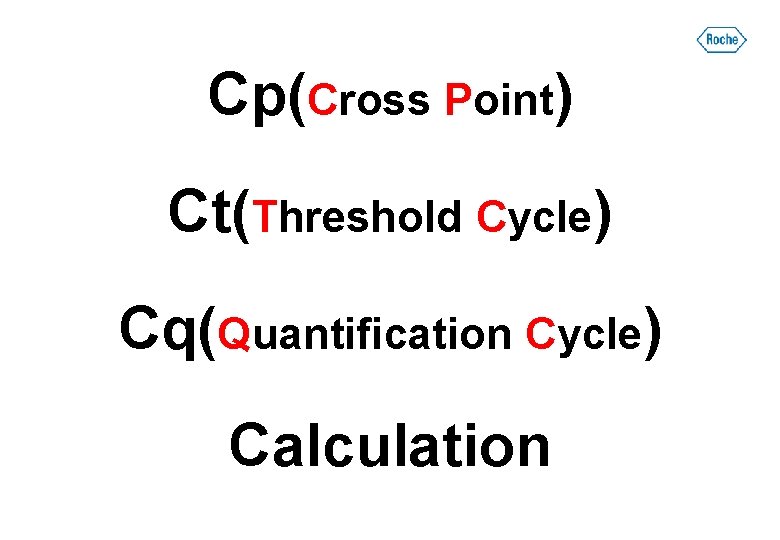
Cp(Cross Point) Ct(Threshold Cycle) Cq(Quantification Cycle) Calculation

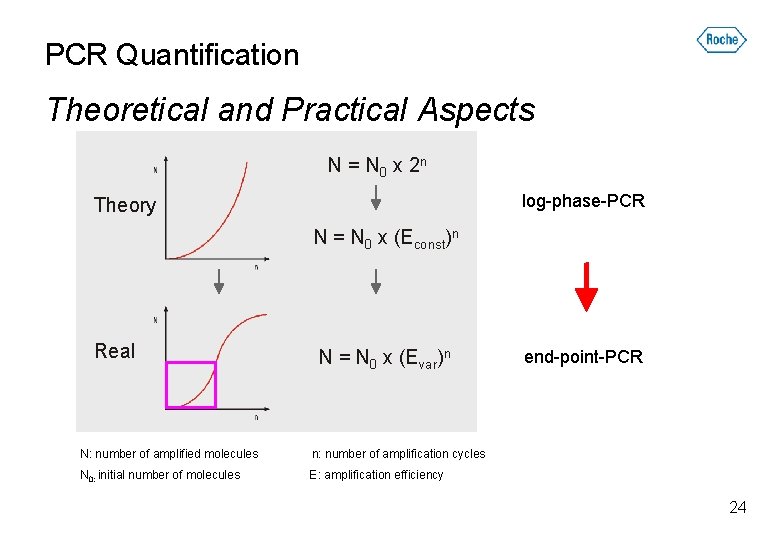
PCR Quantification Theoretical and Practical Aspects N = N 0 x 2 n log-phase-PCR Theory N = N 0 x (Econst)n Real N = N 0 x (Evar)n end-point-PCR N: number of amplified molecules n: number of amplification cycles N 0: initial number of molecules E: amplification efficiency 24

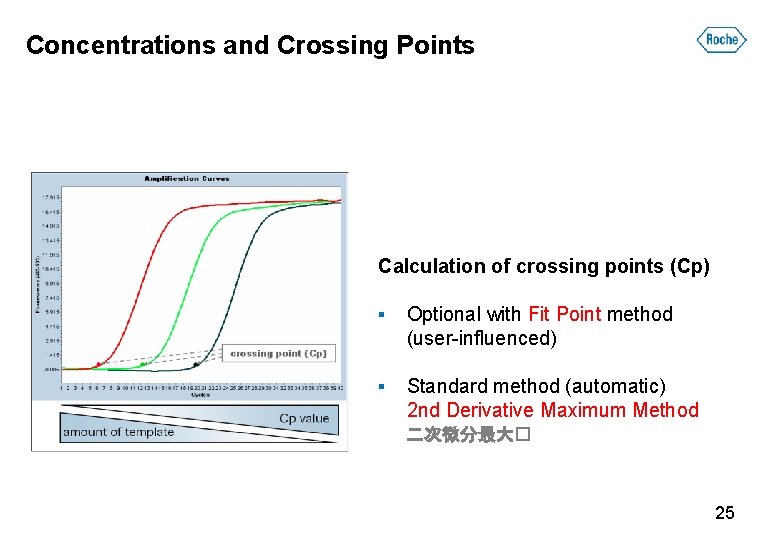
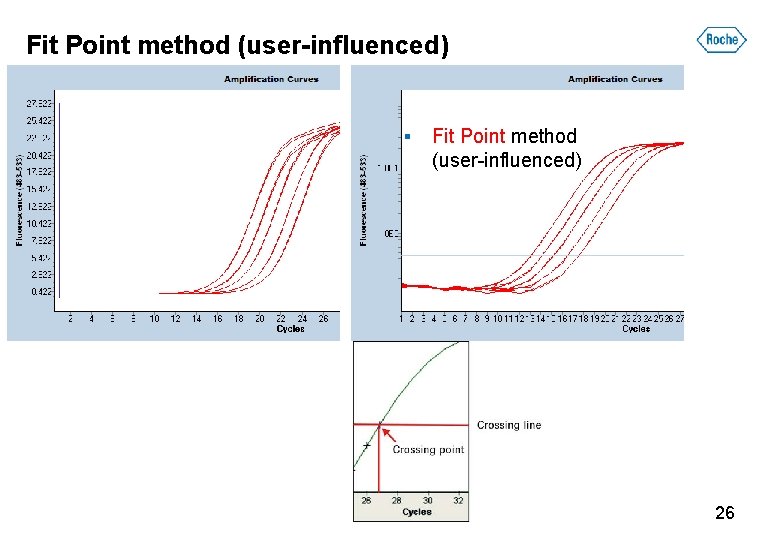
Concentrations and Crossing Points Calculation of crossing points (Cp) ▪ Optional with Fit Point method (user-influenced) ▪ Standard method (automatic) 2 nd Derivative Maximum Method 二次微分最大� 25

Fit Point method (user-influenced) ▪ Fit Point method (user-influenced) 26

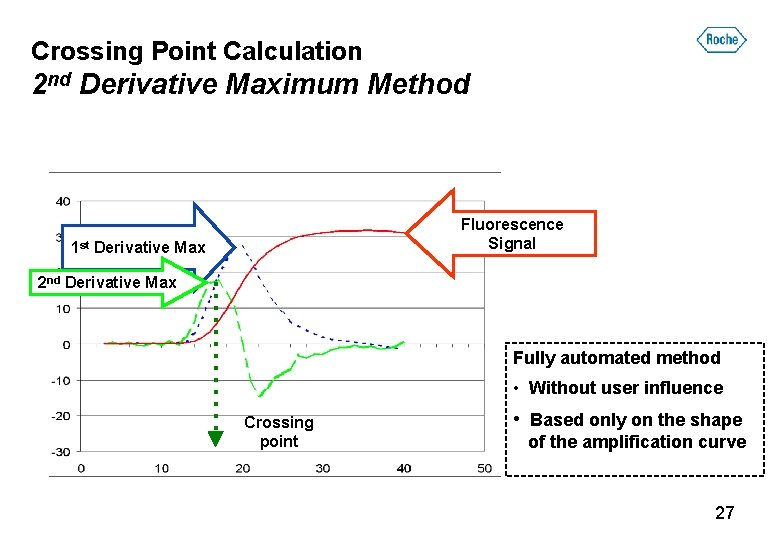
Crossing Point Calculation 2 nd Derivative Maximum Method Fluorescence Signal 1 st Derivative Max 2 nd Derivative Max Fully automated method • Without user influence Crossing point • Based only on the shape of the amplification curve 27

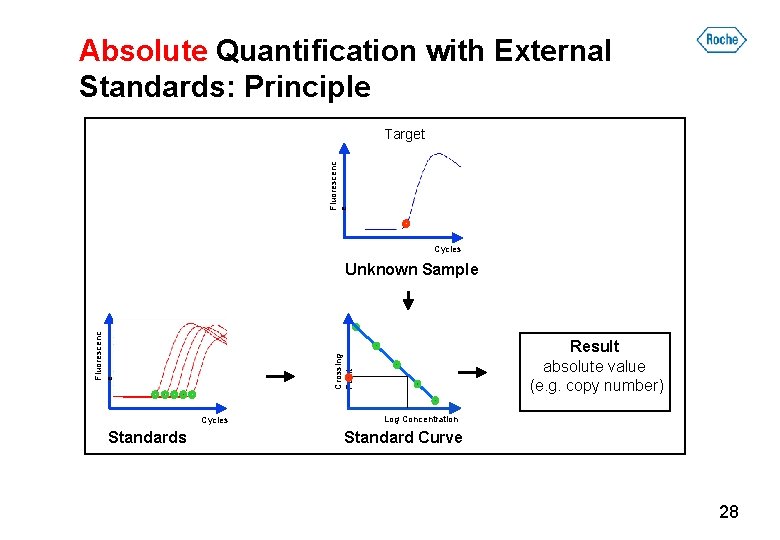
Absolute Quantification with External Standards: Principle Fluorescenc e Target Cycles Standards Result absolute value (e. g. copy number) Crossing Point Fluorescenc e Unknown Sample Log Concentration Standard Curve 28

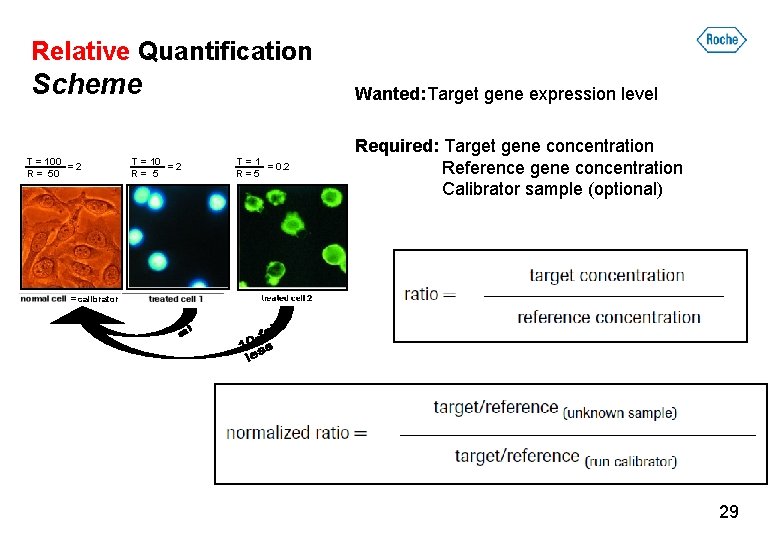
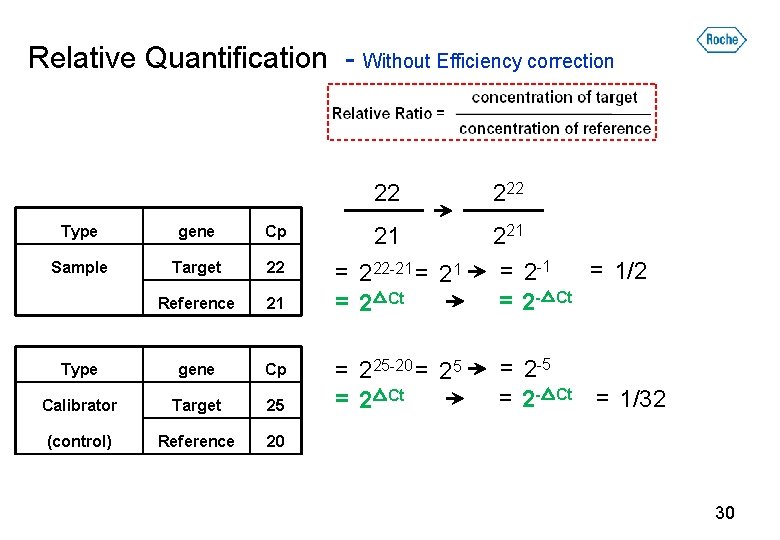
Relative Quantification Scheme T = 100 =2 R = 50 T = 10 =2 R= 5 Wanted: Target gene expression level T=1 = 0. 2 R=5 Required: Target gene concentration Reference gene concentration Calibrator sample (optional) = calibrator 29

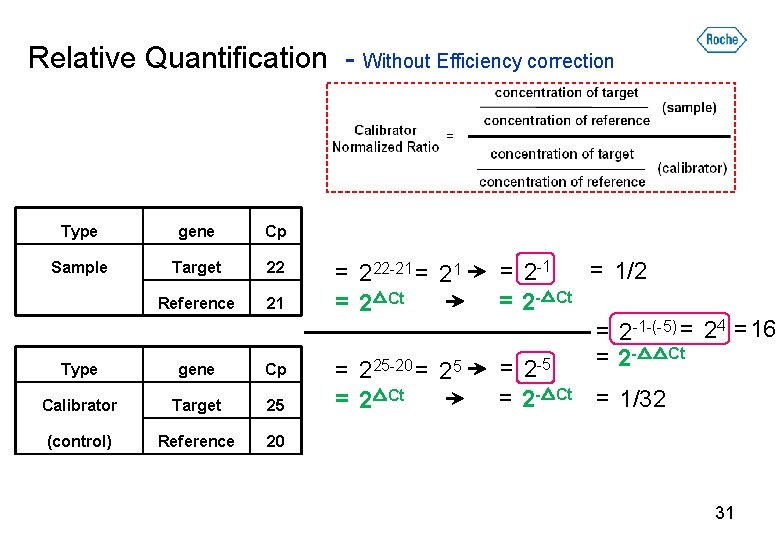
Relative Quantification - Without Efficiency correction Type gene Cp Sample Target 22 Reference 21 Type gene Cp Calibrator Target 25 (control) Reference 20 22 21 = 222 -21 = 2△Ct 221 = 1/2 = 2 -1 = 2 -△Ct = 225 -20 = 25 = 2△Ct = 2 -5 = 2 -△Ct = 1/32 30

Relative Quantification - Without Efficiency correction Type gene Cp Sample Target 22 Reference 21 Type gene Cp Calibrator Target 25 (control) Reference 20 = 222 -21 = 2△Ct = 225 -20 = 25 = 2△Ct = 1/2 = 2 -1 = 2 -△Ct = 2 -1 -(-5) = 24 = 16 -△△Ct = 2 -5 = 2 -△Ct = 1/32 31

LC 480 Basic training : 1. Real time PCR principle introduction 2. Detection formats 3. Absolute/Relative Quantification 4. Genotyping 5. Light. Cycler® 480 Instrument introduction 32

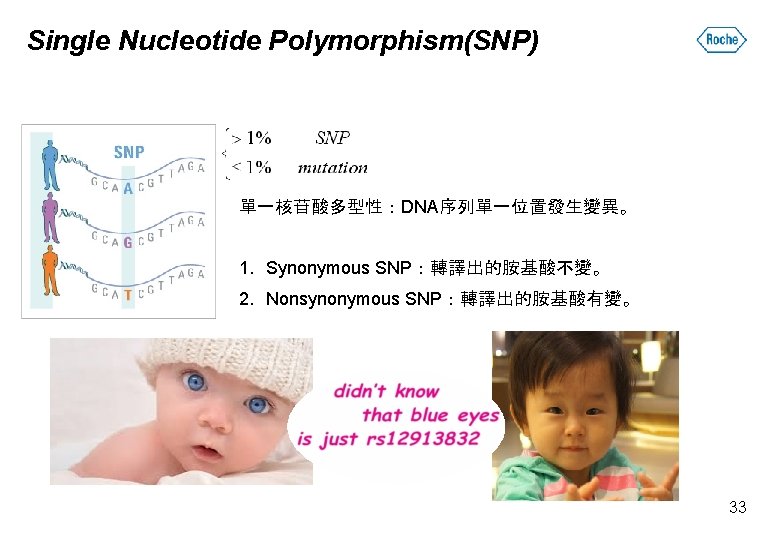
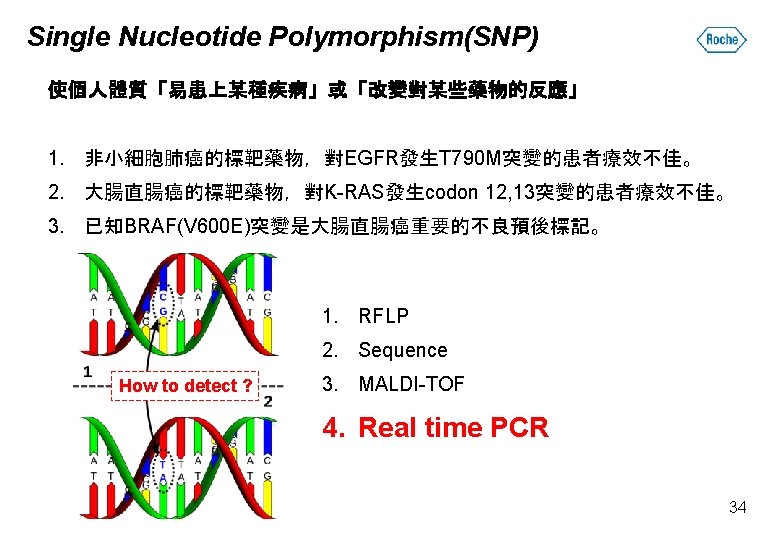
Single Nucleotide Polymorphism(SNP) 單一核苷酸多型性:DNA序列單一位置發生變異。 1. Synonymous SNP:轉譯出的胺基酸不變。 2. Nonsynonymous SNP:轉譯出的胺基酸有變。 33


Endpoint Genotyping (allelic discrimination) Principle 35

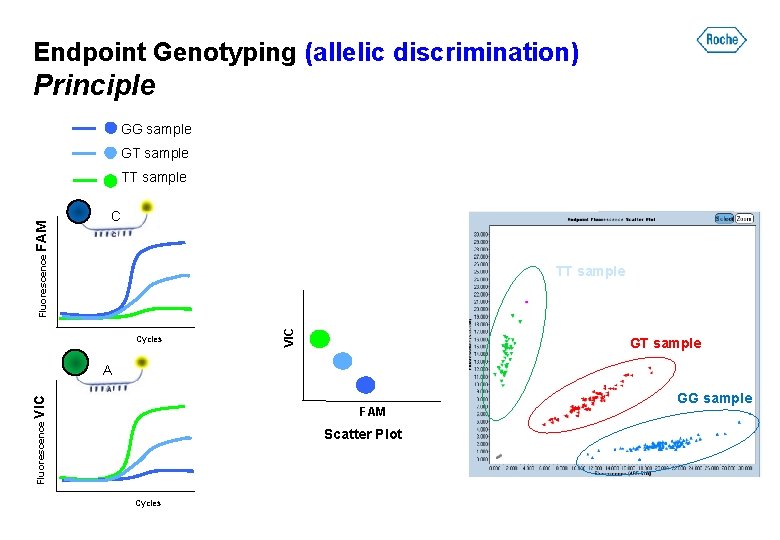
Endpoint Genotyping (allelic discrimination) Principle GG sample GT sample C TT sample Cycles VIC Fluorescence FAM TT sample GT sample Fluorescence VIC A FAM Scatter Plot Cycles GG sample

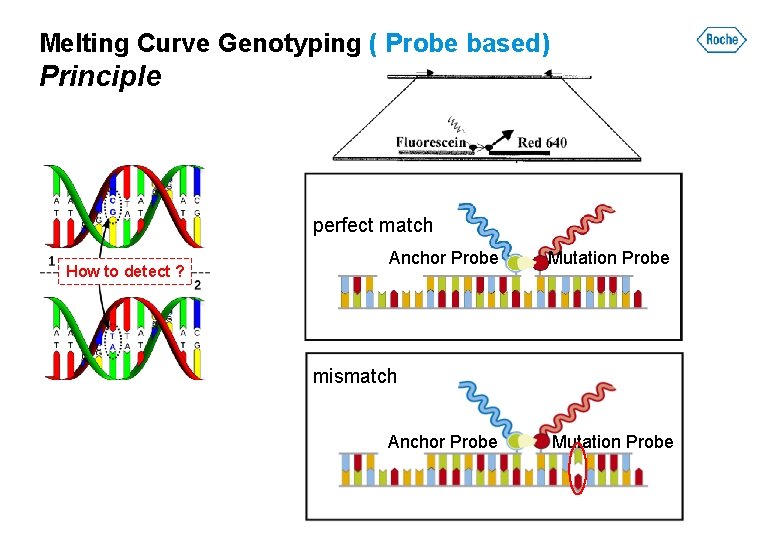
Melting Curve Genotyping ( Probe based) Principle perfect match How to detect ? Anchor Probe Mutation Probe mismatch Anchor Probe Mutation Probe

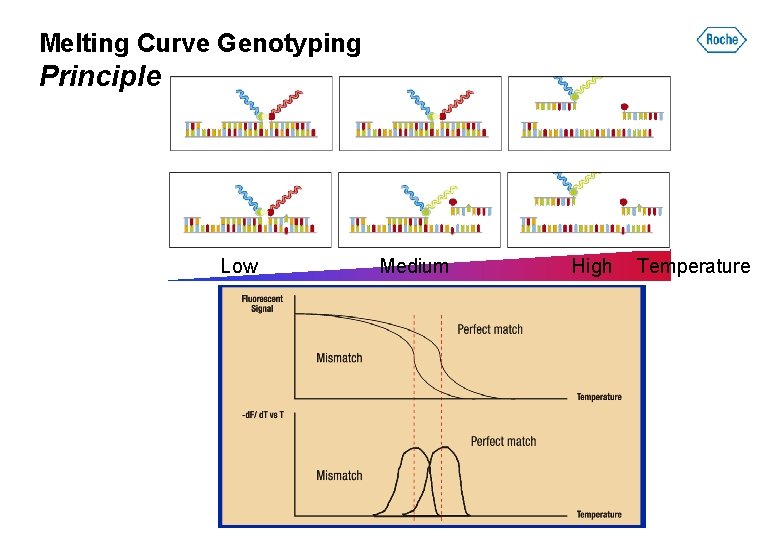
Melting Curve Genotyping Principle Low Medium High Temperature

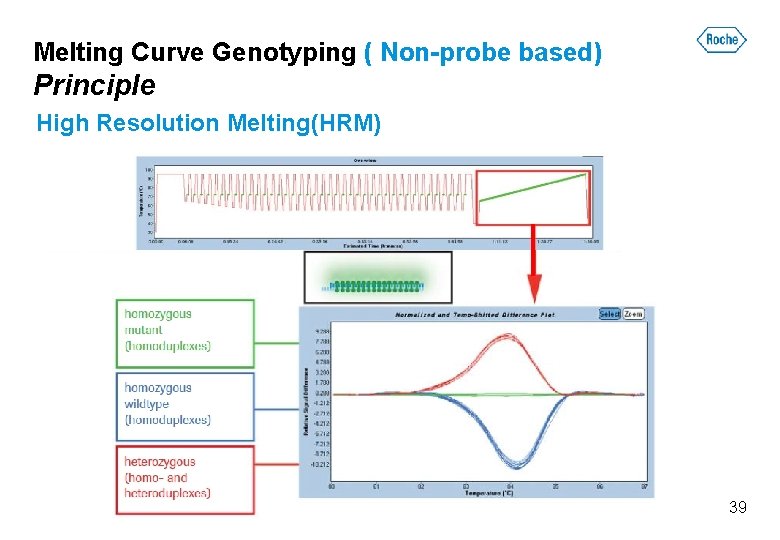
Melting Curve Genotyping ( Non-probe based) Principle High Resolution Melting(HRM) 39

LC 480 Basic training : 1. Real time PCR principle introduction 2. Detection formats 3. Absolute/Relative Quantification 4. Genotyping 5. Light. Cycler® 480 Instrument introduction 40

Light. Cycler® 480 Real-Time PCR System 41

Light. Cycler® 480 System Components 42

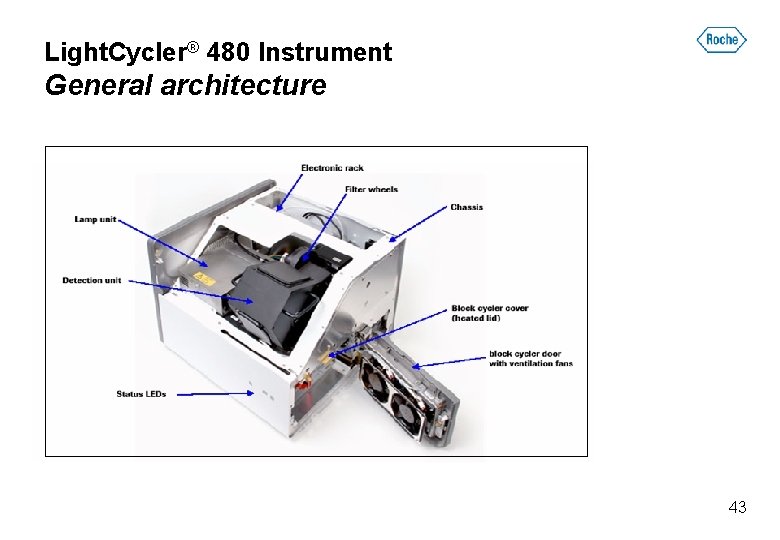
Light. Cycler® 480 Instrument General architecture 43

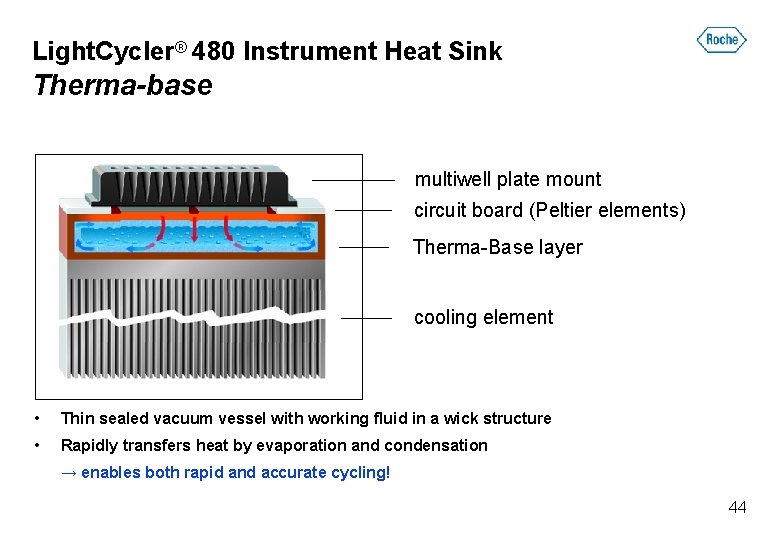
Light. Cycler® 480 Instrument Heat Sink Therma-base multiwell plate mount circuit board (Peltier elements) Therma-Base layer cooling element • Thin sealed vacuum vessel with working fluid in a wick structure • Rapidly transfers heat by evaporation and condensation → enables both rapid and accurate cycling! 44

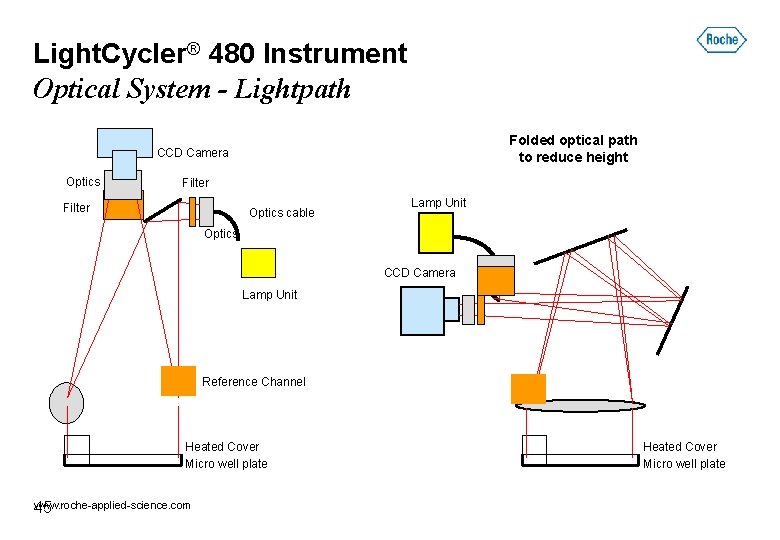
Light. Cycler® 480 Instrument Optical System - Lightpath Folded optical path to reduce height CCD Camera Optics Filter Optics cable Lamp Unit Optics CCD Camera Lamp Unit Reference Channel Heated Cover Micro well plate www. roche-applied-science. com 45 Heated Cover Micro well plate

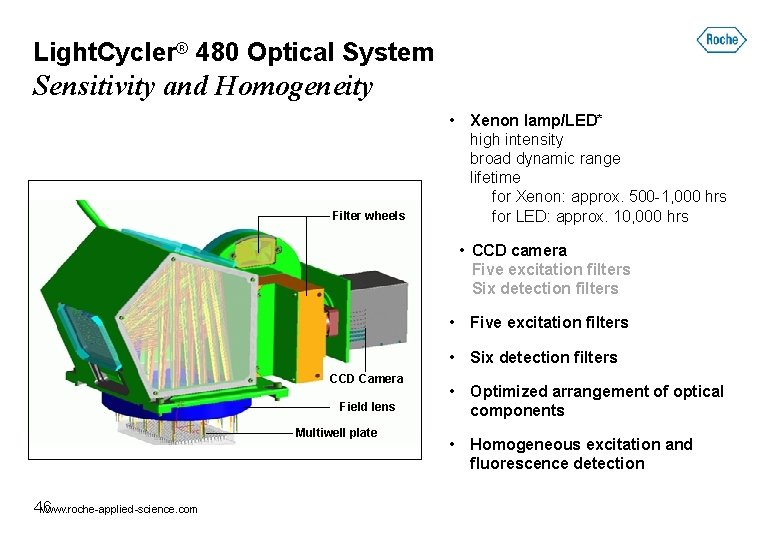
Light. Cycler® 480 Optical System Sensitivity and Homogeneity Filter wheels • Xenon lamp/LED* high intensity broad dynamic range lifetime for Xenon: approx. 500 -1, 000 hrs for LED: approx. 10, 000 hrs • CCD camera Five excitation filters Six detection filters • Five excitation filters • Six detection filters CCD Camera Field lens Multiwell plate www. roche-applied-science. com 46 • Optimized arrangement of optical components • Homogeneous excitation and fluorescence detection

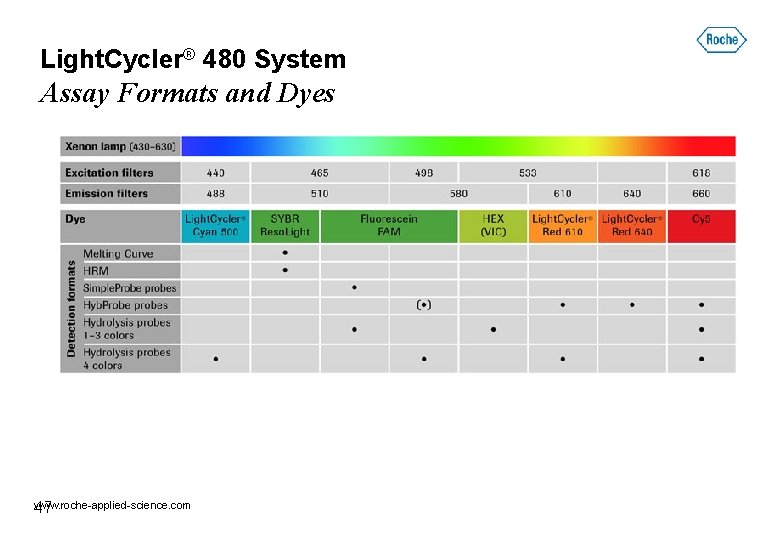
Light. Cycler® 480 System Assay Formats and Dyes www. roche-applied-science. com 47

Q&A

Light. Cycler 480 Software: 1. New experiment from template 2. Subset & sample set up 3. Data analysis & report 4. Color Compensation Calibration 49

System Start-Up 50

System Start-Up 51

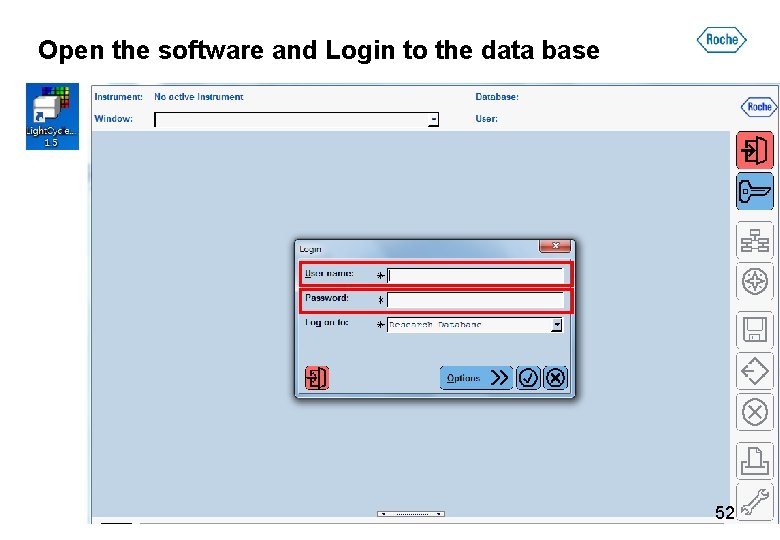
Open the software and Login to the data base 52

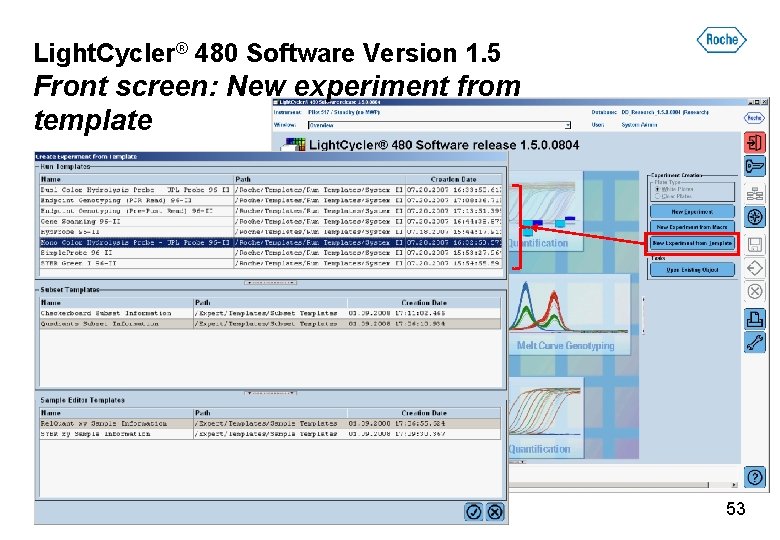
Light. Cycler® 480 Software Version 1. 5 Front screen: New experiment from template 53

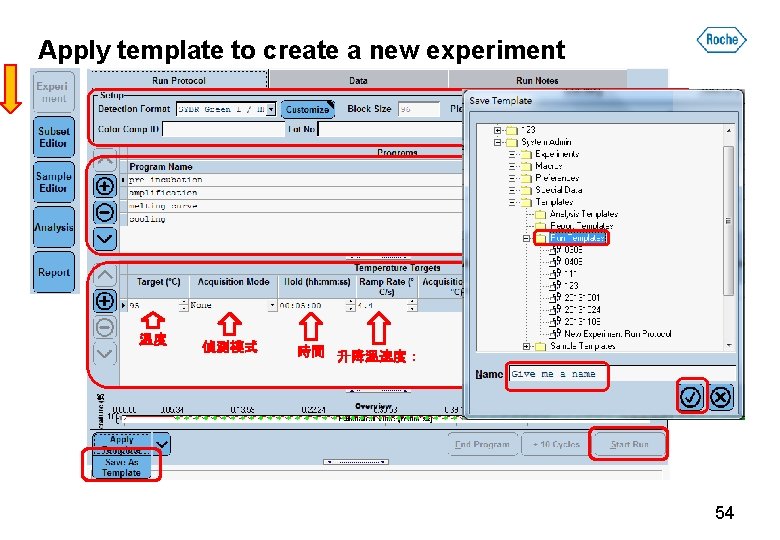
Apply template to create a new experiment 1 2 溫度 偵測模式 3 時間 升降溫速度: 54

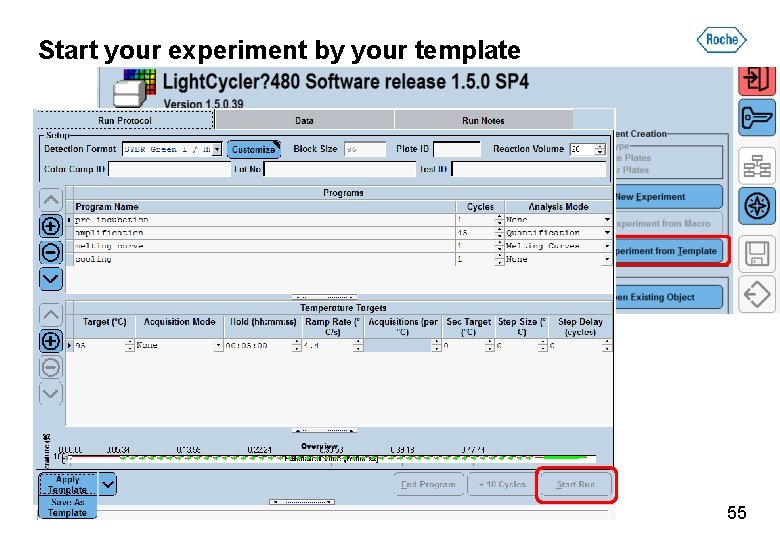
Start your experiment by your template 55

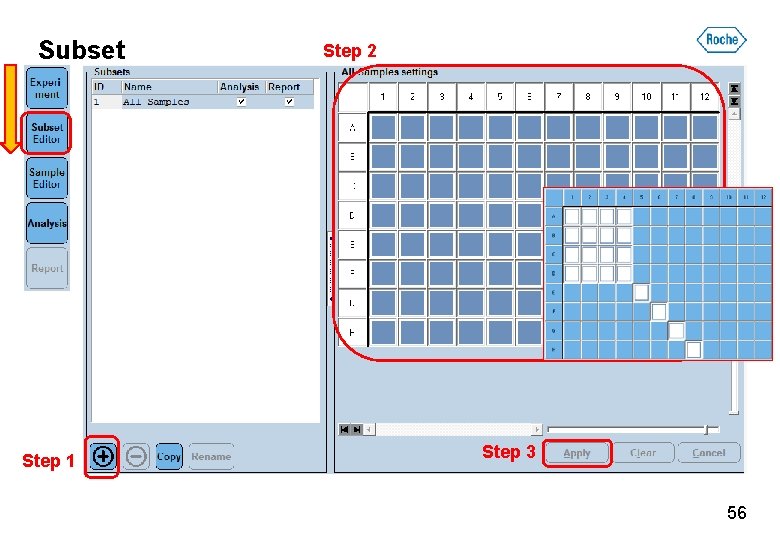
Subset Step 1 Step 2 Step 3 56

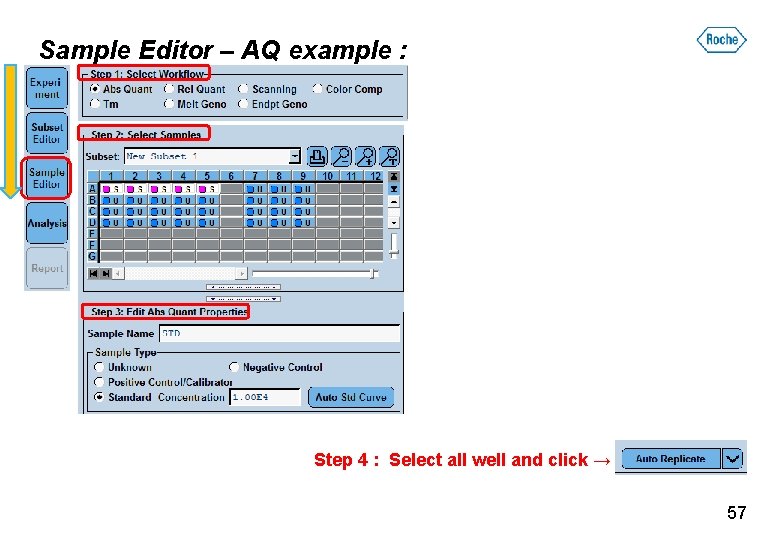
Sample Editor – AQ example : Step 4 : Select all well and click → 57

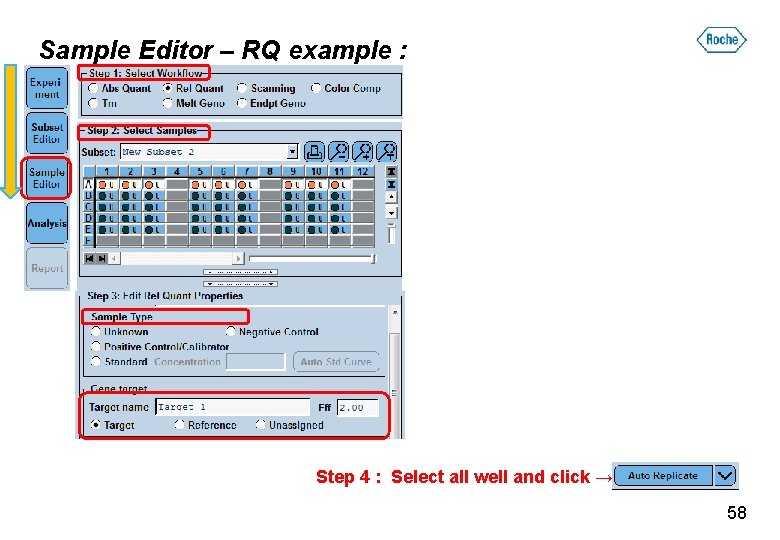
Sample Editor – RQ example : Step 4 : Select all well and click → 58

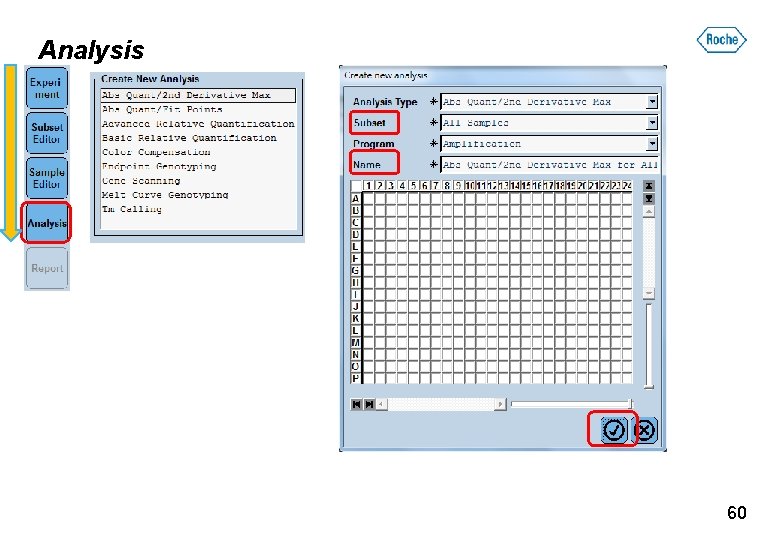
Analysis 分析模式簡要說明: 分析模式 說明 Abs Quant / 2 nd Derivative Max �對定量 (2 nd. Max) Abs Quant / Fit Points �對定量 (Fit Point) Advanced Relative Quantification 進階相對定量(2 nd. Max or Fit Point) Basic Relative Quantification 基礎相對定量 (Fit Point only) Endpoint Genotyping 基因分型(適用於Taq. Man probe) Melt Curve Genotyping 基因分型(適用於Hyprobe) Tm Calling Tm分析 59

Analysis 60

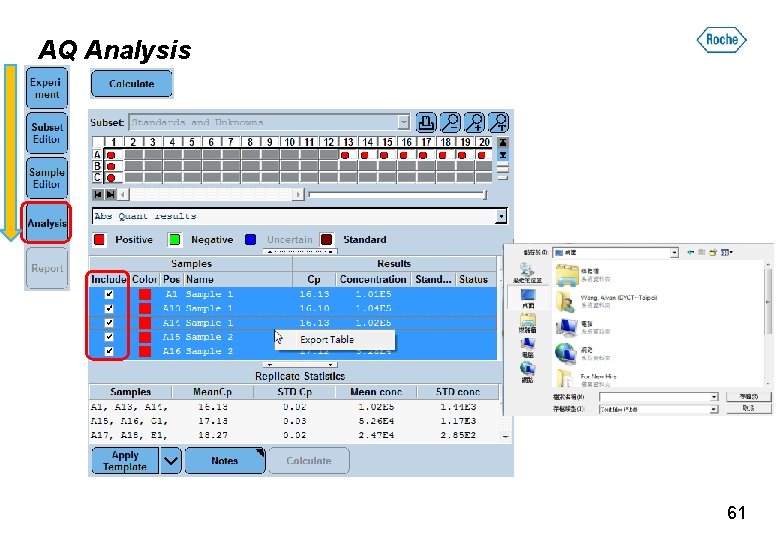
AQ Analysis 61

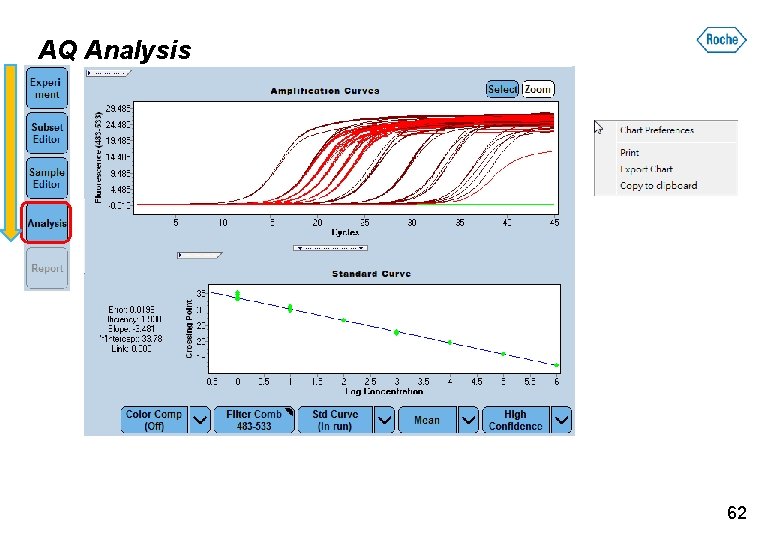
AQ Analysis 62

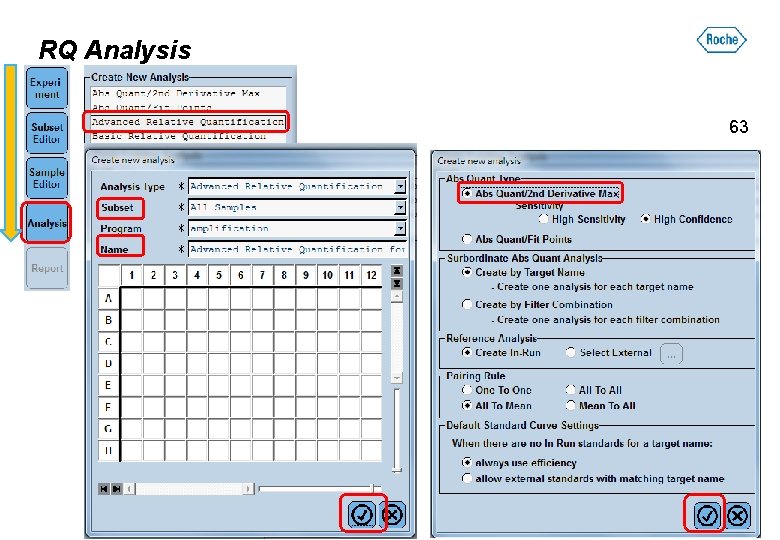
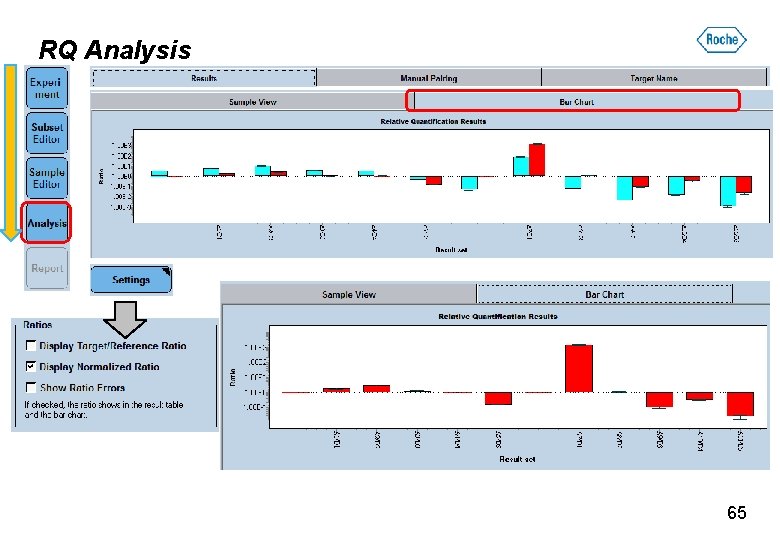
RQ Analysis 63

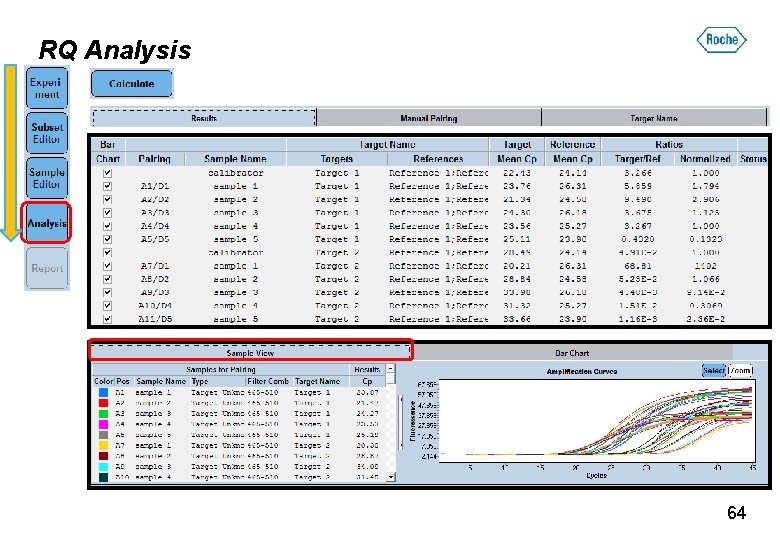
RQ Analysis 64

RQ Analysis 65

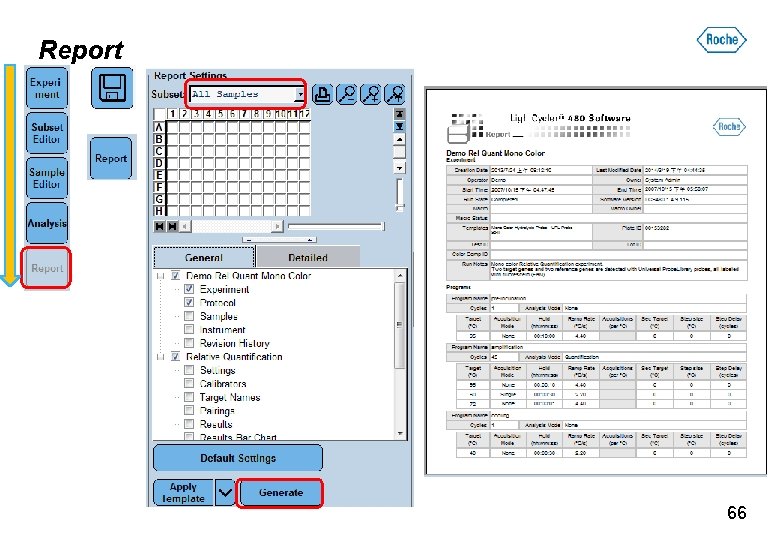
Report 66

Q&A

Doing now what patients need next