The polymerase chain reaction PCR Experiment Goals Understand












- Slides: 31

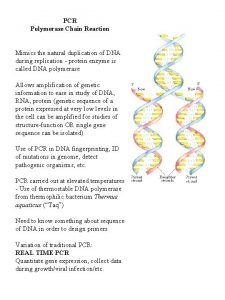
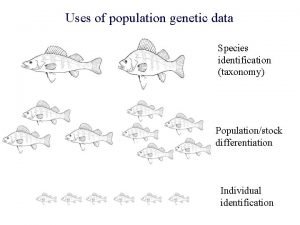
The polymerase chain reaction (PCR)

Experiment Goals • Understand how PCR technique works • Perform PCR experiment • Analyze PCR products

What is PCR? • Definition § The polymerase chain reaction (PCR) is a technique to amplify a piece of DNA very rapidly outside a living cell

Development of PCR § 1971: Khorana described basic principle of DNA replication using DNA primers § 1983: Dr. Karry Mullis developed PCR technique, for which he received the Nobel Prize in Chemistry in 1993.

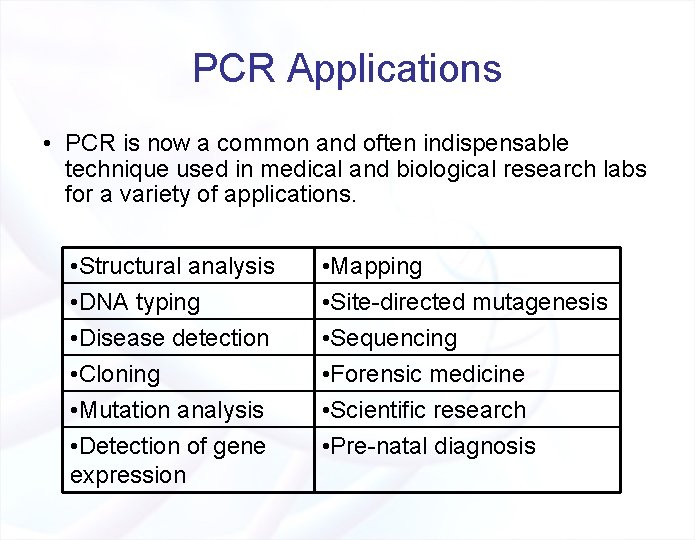
PCR Applications • PCR is now a common and often indispensable technique used in medical and biological research labs for a variety of applications. • Structural analysis • DNA typing • Disease detection • Cloning • Mutation analysis • Detection of gene expression • Mapping • Site-directed mutagenesis • Sequencing • Forensic medicine • Scientific research • Pre-natal diagnosis

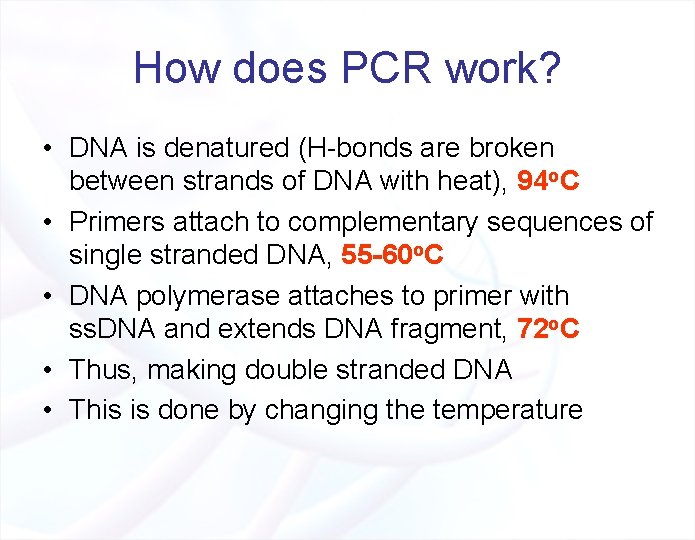
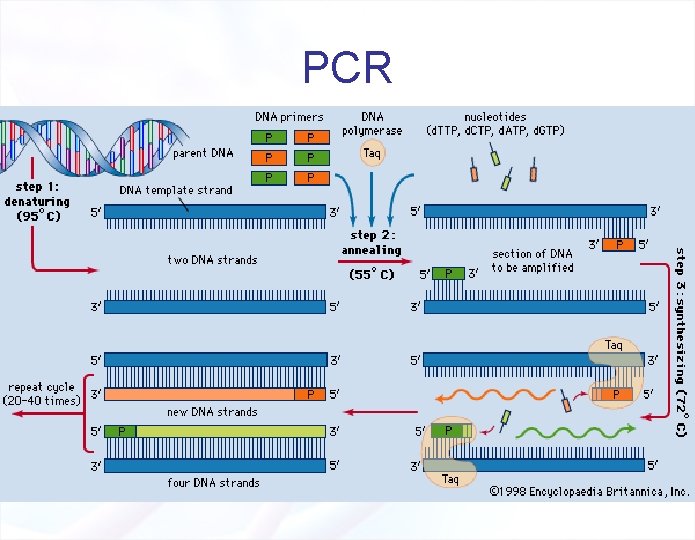
How does PCR work? • DNA is denatured (H-bonds are broken between strands of DNA with heat), 94 o. C • Primers attach to complementary sequences of single stranded DNA, 55 -60 o. C • DNA polymerase attaches to primer with ss. DNA and extends DNA fragment, 72 o. C • Thus, making double stranded DNA • This is done by changing the temperature

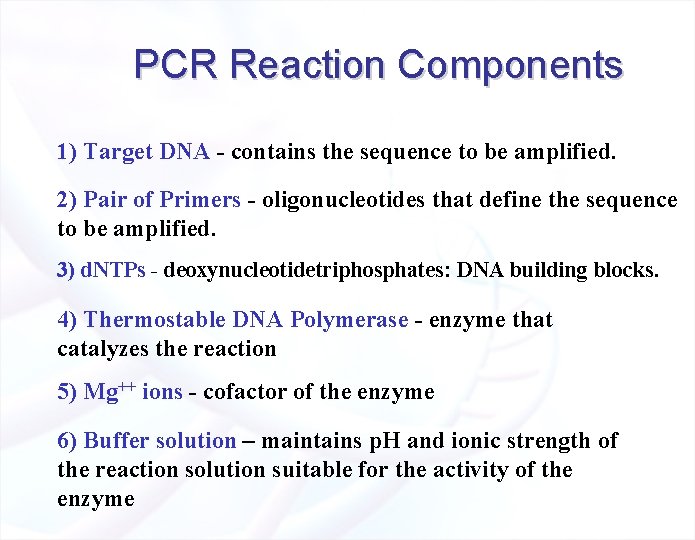
PCR Reaction Components 1) Target DNA - contains the sequence to be amplified. 2) Pair of Primers - oligonucleotides that define the sequence to be amplified. 3) d. NTPs - deoxynucleotidetriphosphates: DNA building blocks. 4) Thermostable DNA Polymerase - enzyme that catalyzes the reaction 5) Mg++ ions - cofactor of the enzyme 6) Buffer solution – maintains p. H and ionic strength of the reaction solution suitable for the activity of the enzyme

1) Target DNA § Target of DNA can be Ø a single gene Ø part of a gene Ø or a non-coding sequence

DNA Quality • DNA should be intact and free of contaminants that inhibit amplification. – Contaminants can be purified from the original DNA source. • Heme from blood, and melanin from hair – Contaminants can be introduced during the purification process. • Phenol, ethanol, sodium dodecyl sulfate (SDS) and other detergents, and salts.

DNA quantity • More template is not necessarily better. – Too much template can cause nonspecific amplification. – Too little template will result in little or no PCR product.

How Big A Target is? • Amplification products are typically in the size range 100 -1500 bp. • Longer targets are amplifiable >25 kb. • Requires modified reaction buffer, cocktails of polymerases, and longer extension times.

2) Pair of Primers • Primers define the DNA sequence to be amplified—they give the PCR specificity. • Primers bind (anneal) to the DNA template and act as starting points for the DNA polymerase, – DNA polymerases cannot initiate DNA synthesis without a primer. • The distance between the two primers determines the length of the newly synthesized DNA molecules.

3) d. NTPs (deoxynucleotidetriphosphates) – The building blocks for the newly synthesized DNA strands. – d. ATP, d. GTP, d. CTP or d. TTP

4) Thermostable DNA Polymerase • DNA Polymerase is the enzyme responsible for copying the sequence starting at the primer from the single DNA strand • Commonly use Taq, an enzyme from the hyperthermophilic organisms Thermus aquaticus, isolated first at a thermal spring • This enzyme is heat-tolerant – it is thermally tolerant (survives the melting T of DNA denaturation) – which also means the process is more specific, higher temps result in less mismatch – more specific replication

Running PCR • The PCR is commonly carried out in a reaction volume of 15 -100 μl in small reaction tubes (0. 2 -0. 5 ml volumes) in a thermal cycler. • The thermal cycler allows heating and cooling of the reaction tubes to control the temperature required at each reaction step. • Thin-walled reaction tubes permit favorable thermal conductivity to allow for rapid thermal equilibration. • Most thermal cyclers have heated lids to prevent condensation at the top of the reaction tube. • Older thermocyclers lacking a heated lid require a layer of oil on top of the reaction mixture or a ball of wax inside the tube.

Initialization step • Prior to the first cycle, there is an initialization step – the PCR reaction is often heated to a temperature of 94 -96°C, and this temperature is then held for 1 -9 minutes – This first hold is employed to ensure that most of the DNA template and primers are denatured, – Also, some PCR polymerases require this step for activation

PCR Reaction Cycles • One PCR cycle consists of a DNA denaturation step, a primer annealing step and a primer extension step. § § § DNA Denaturation: Expose the DNA template to high temperatures to separate the two DNA strands and allow access by DNA polymerase and PCR primers. Primer Annealing: Lower the temperature to allow primers to anneal to their complementary sequence. Primer Extension: Adjust the temperature for optimal thermostable DNA polymerase activity to extend primers.

PCR

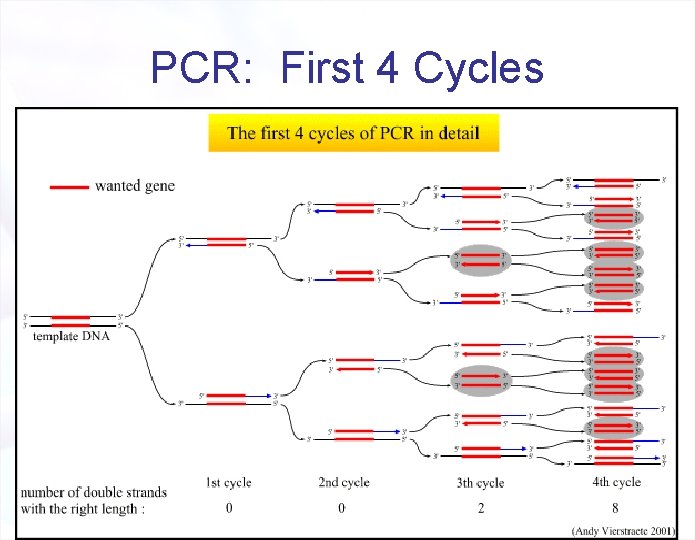
PCR: First 4 Cycles

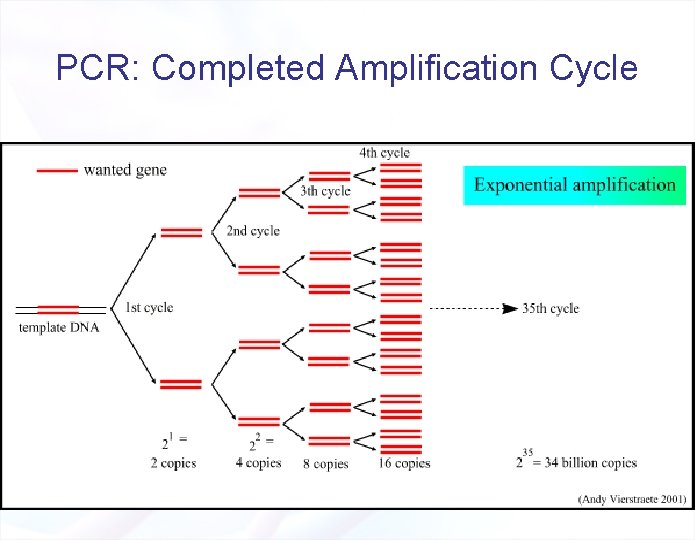
PCR: Completed Amplification Cycle

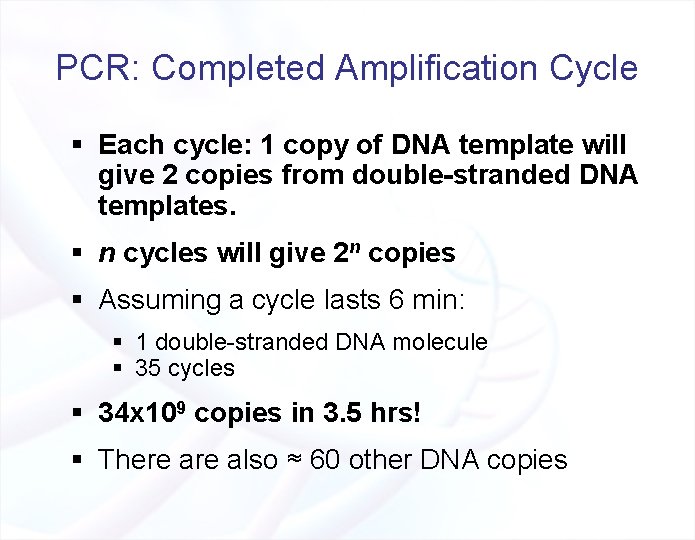
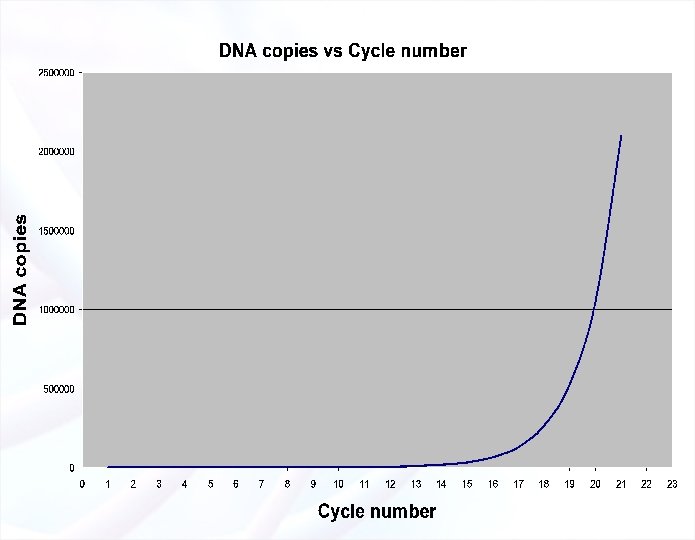
PCR: Completed Amplification Cycle § Each cycle: 1 copy of DNA template will give 2 copies from double-stranded DNA templates. § n cycles will give 2 n copies § Assuming a cycle lasts 6 min: § 1 double-stranded DNA molecule § 35 cycles § 34 x 109 copies in 3. 5 hrs! § There also ≈ 60 other DNA copies


Analyze PCR products • Check a sample by gel electrophoresis. • Is the product the size that you expected? • Is there more than one band? • Is any band the correct size? • May need to optimize the reaction conditions.

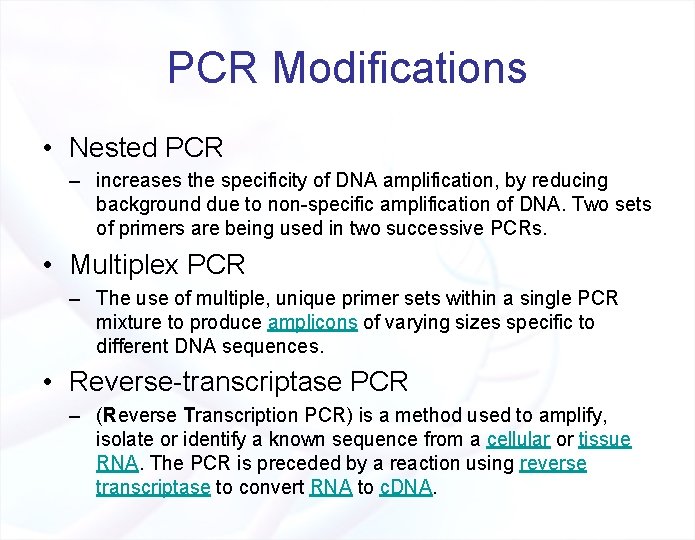
PCR Modifications • Nested PCR – increases the specificity of DNA amplification, by reducing background due to non-specific amplification of DNA. Two sets of primers are being used in two successive PCRs. • Multiplex PCR – The use of multiple, unique primer sets within a single PCR mixture to produce amplicons of varying sizes specific to different DNA sequences. • Reverse-transcriptase PCR – (Reverse Transcription PCR) is a method used to amplify, isolate or identify a known sequence from a cellular or tissue RNA. The PCR is preceded by a reaction using reverse transcriptase to convert RNA to c. DNA.

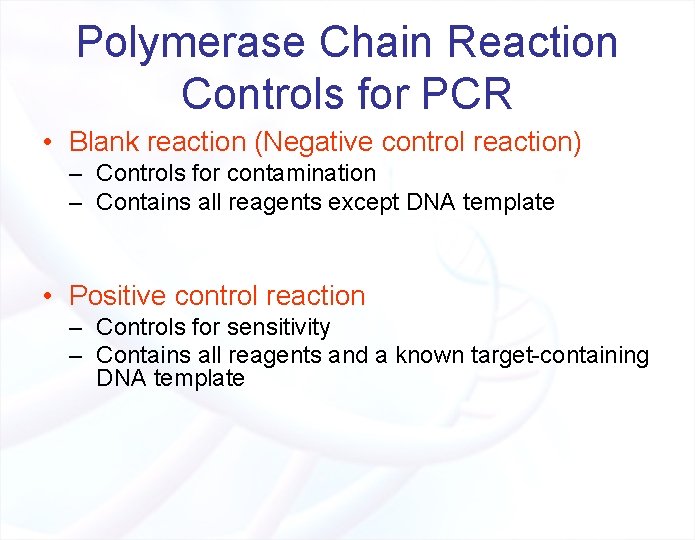
Polymerase Chain Reaction Controls for PCR • Blank reaction (Negative control reaction) – Controls for contamination – Contains all reagents except DNA template • Positive control reaction – Controls for sensitivity – Contains all reagents and a known target-containing DNA template

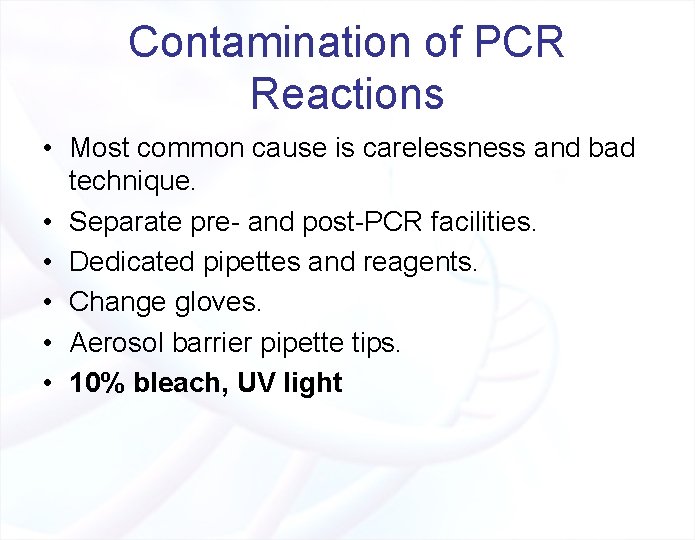
Contamination of PCR Reactions • Most common cause is carelessness and bad technique. • Separate pre- and post-PCR facilities. • Dedicated pipettes and reagents. • Change gloves. • Aerosol barrier pipette tips. • 10% bleach, UV light

Procedure 1 - Prepare master Mix 2 - Program thermocycler 3 - Run the samples on thermocycler 4 - Analysis of PCR products

Target DNA • Amplification of part of the Human growth hormone gene • Specific primers used • Forward primer: 5’TCCCTTCCCAACCATTCCCTTA-3’ • Reverse primer: 5’CCACTCACGGATTTCTGTTGTGTTTC 3’

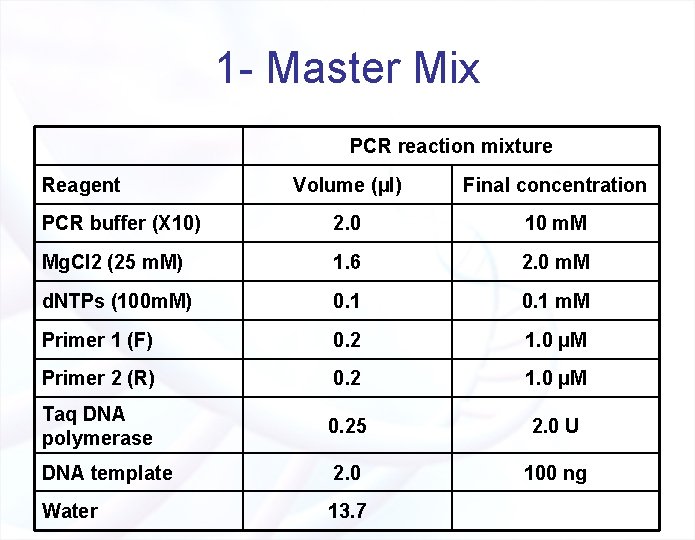
1 - Master Mix PCR reaction mixture Reagent Volume (µl) Final concentration PCR buffer (X 10) 2. 0 10 m. M Mg. Cl 2 (25 m. M) 1. 6 2. 0 m. M d. NTPs (100 m. M) 0. 1 m. M Primer 1 (F) 0. 2 1. 0 µM Primer 2 (R) 0. 2 1. 0 µM Taq DNA polymerase 0. 25 2. 0 U DNA template 2. 0 100 ng Water 13. 7

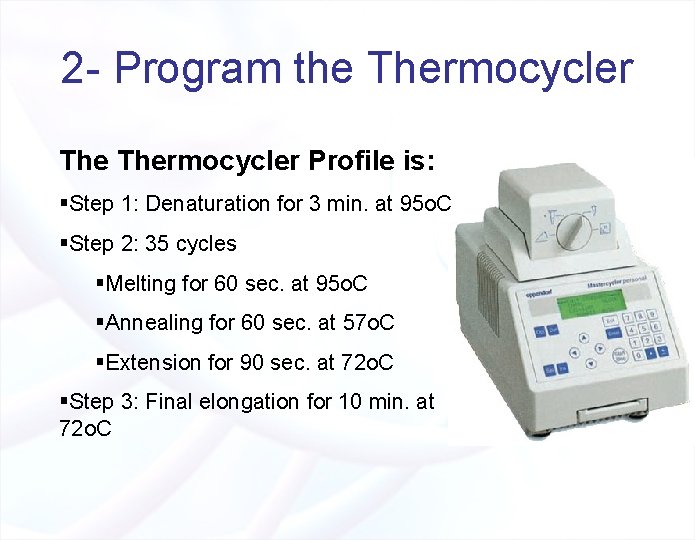
2 - Program the Thermocycler Profile is: §Step 1: Denaturation for 3 min. at 95 o. C §Step 2: 35 cycles §Melting for 60 sec. at 95 o. C §Annealing for 60 sec. at 57 o. C §Extension for 90 sec. at 72 o. C §Step 3: Final elongation for 10 min. at 72 o. C

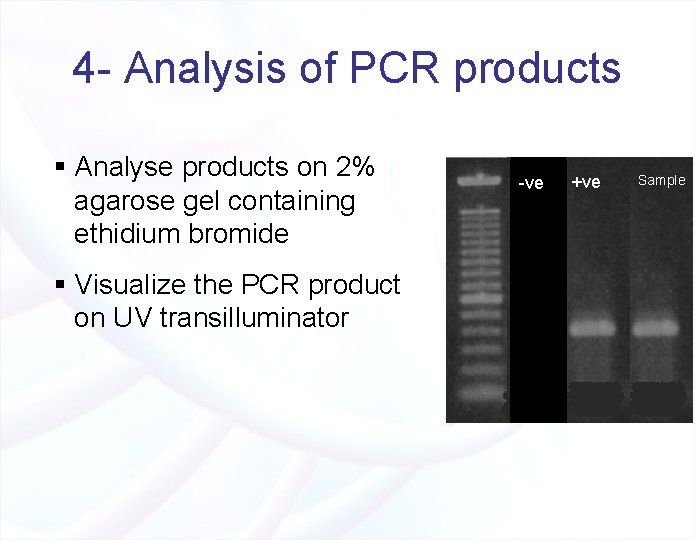
4 - Analysis of PCR products § Analyse products on 2% agarose gel containing ethidium bromide § Visualize the PCR product on UV transilluminator -ve +ve Sample
 Pcr application
Pcr application Polymerase chain reaction
Polymerase chain reaction Polymerase chain reaction
Polymerase chain reaction Site:slidetodoc.com
Site:slidetodoc.com Polymerase chain reaction uses
Polymerase chain reaction uses Polymerase chain reaction
Polymerase chain reaction The three steps of polymerase chain reaction
The three steps of polymerase chain reaction Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals To understand recursion you must understand recursion
To understand recursion you must understand recursion Food chain food chain food chain
Food chain food chain food chain General goals and specific goals
General goals and specific goals Examples of generic goals and product-specific goals
Examples of generic goals and product-specific goals Rna polymerase 1 2 3
Rna polymerase 1 2 3 Whats primase
Whats primase Dna polymerase iii
Dna polymerase iii Replication
Replication Dna polymerase proofreading
Dna polymerase proofreading How many dna polymerase in eukaryotes
How many dna polymerase in eukaryotes Types of dna polymerase in eukaryotes
Types of dna polymerase in eukaryotes Dna polymerase
Dna polymerase Transcription initiation in eukaryotes
Transcription initiation in eukaryotes Dna polymerase
Dna polymerase Rt pcr primer efficiency
Rt pcr primer efficiency Rna polymerase
Rna polymerase Clonaggio
Clonaggio Adn polymérase
Adn polymérase Taq polymerase
Taq polymerase Dna polymerase
Dna polymerase The principal enzyme involved in dna replication is
The principal enzyme involved in dna replication is Template strand, new strand, base pair, and dna polymerase.
Template strand, new strand, base pair, and dna polymerase. Blue bottle experiment
Blue bottle experiment