Crystal digital PCR for detection and quantification of



- Slides: 13

Crystal digital PCR for detection and quantification of circulating EGFR mutations in advanced nonsmall cell lung cancer Cécile Jovelet Postdocoral fellow Translational research lab Gustave Roussy Institute, France

Introduction l Lung cancer: third most frequent cancer > Non-Small Cell Lung Cancer (NSCLC): 80% cases of lung cancer l Sensitivity to EGFR-TKIs for patients harboring EGFR activating mutations > 11% of NSCLC > Deletion exon 19, L 858 R, L 861 Q l Development of secondary resistance to EGFR-TKIs > 50% of resistance: mutation T 790 M in exon 20 of the EGFR gene l Third-generation EGFR-TKIs: active on tumor cells harboring also EGFR T 790 M resistant mutations. Their prescription: requirement of detection of resistance mutation in a new tissue biopsy. 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 2

l Limitations for NSCLC rebiopsying: > lack of available tissue for molecular profile assessment, > the location or size of the tumor, > the insufficient quality or quantity of genetic material, > and the risk of complications l Plasma circulating free DNA (cf. DNA) analysis: minimally-invasive alternative to tissue biopsy l Potential applications of cf. DNA based analysis: > molecular abnormalities characterization at diagnosis, > patient selection for targeted therapies, > monitoring of treatment response, > and screening for acquired mutations at the time of resistance 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 3

l Main objectives: > Detect EGFR activating and resistance mutations in cf. DNA of NSCLC patients by digital PCR. > Compare the d. PCR results with NGS analysis > Monitor the cf. DNA in longitudinal samples during follow-up. l Methods: > Extraction: • Blood sample: 10 m. L, EDTA tubes • Double centrifugation, < 4 h after sampling • Extraction: 3 m. L Plasma, QIAamp circulating nucleic acid Kit (Qiagen) > Detection of mutation: NGS (PGM, Ion Torrent), Crystal digital PCR (Stilla) 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 4

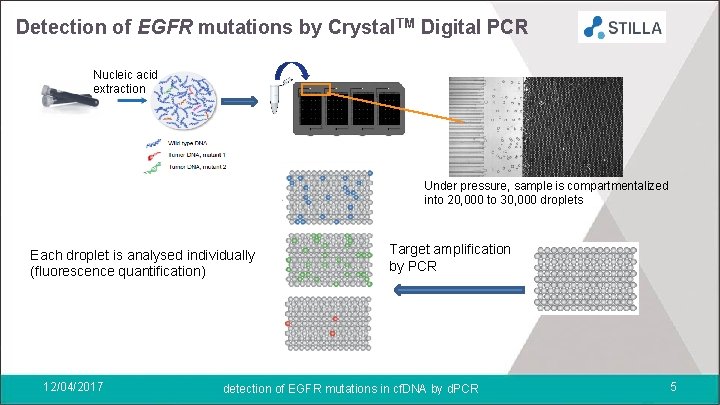
Detection of EGFR mutations by Crystal. TM Digital PCR Nucleic acid extraction Under pressure, sample is compartmentalized into 20, 000 to 30, 000 droplets Each droplet is analysed individually (fluorescence quantification) 12/04/2017 Target amplification by PCR detection of EGFR mutations in cf. DNA by d. PCR 5

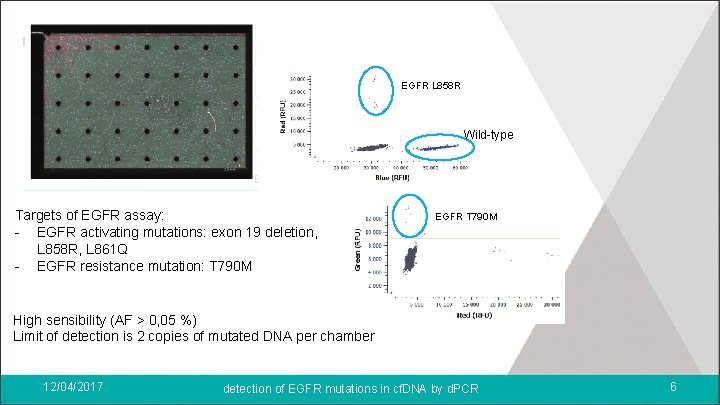
EGFR L 858 R Wild-type Targets of EGFR assay: - EGFR activating mutations: exon 19 deletion, L 858 R, L 861 Q - EGFR resistance mutation: T 790 M EGFR T 790 M High sensibility (AF > 0, 05 %) Limit of detection is 2 copies of mutated DNA per chamber 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 6

Detection of EGFR mutations by NGS l l l Plasma samples were analyzed by PGM (Ion Torrent) 10 ng of cf. DNA Cancer Hotspot Panel v 2 (CHP 2) targeting 50 cancer genes (EGFR) Patients Investigation of the EGFR mutational status in cf. DNA of 61 patients with advanced stage IV NSCLC and for whom re-biopsy was not feasible. > 11 patients: no tumor EGFR mutation in non-synchronous tumor tissue (by NGS) > 50 patients: EGFR mutations detected in non-synchronous tumor tissue (by NGS), treated by EGFR TKI therapies l Among the 50 patients with EGFR mutation in tumor tissue, > 14 patients had at least one follow-up sample > 7 patients had at least 3 samples. in order to monitor mutations detected at baseline or to detect new mutations l 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 7

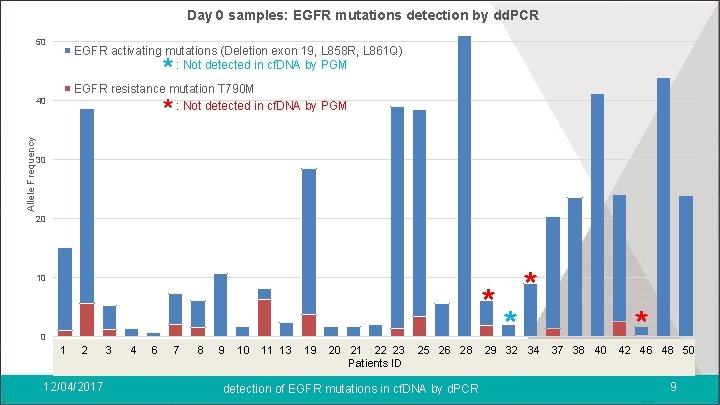
Results l 11 patients with no EGFR mutation detected in tumor DNA > No EGFR mutation detected in cf. DNA (d. PCR and NGS) l 50 patients harboring EGFR activating mutations in tumor DNA (nonsynchronous) In Day 0 samples: > 29 EGFR activating mutations > 14 EGFR resistance mutations T 790 M 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 8

Day 0 samples: EGFR mutations detection by dd. PCR 50 EGFR activating mutations (Deletion exon 19, L 858 R, L 861 Q) : Not detected in cf. DNA by PGM * EGFR resistance mutation T 790 M * : Not detected in cf. DNA by PGM Allele Frequency 40 30 20 10 0 11 22 3 3 44 56 6 7 78 89 9 10 10 11 11 13 12 19 13 20 14 21 1522 16 23 * * 17 25 18 26 19 28 20 29 21 32 22 34 23 37 24 38 25 40 2642 27 46 28 48 29 50 Patients ID 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 9

12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 10

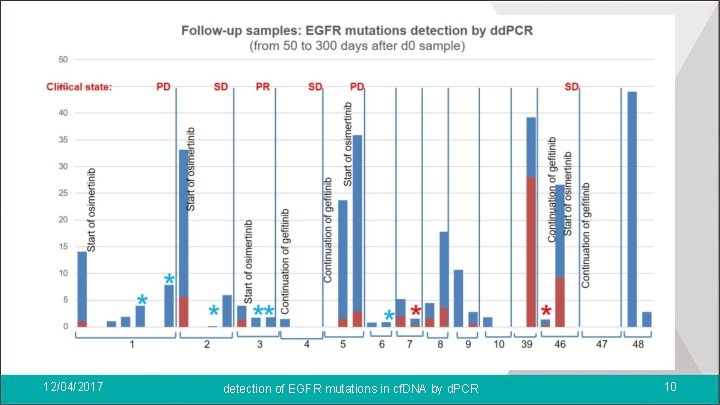
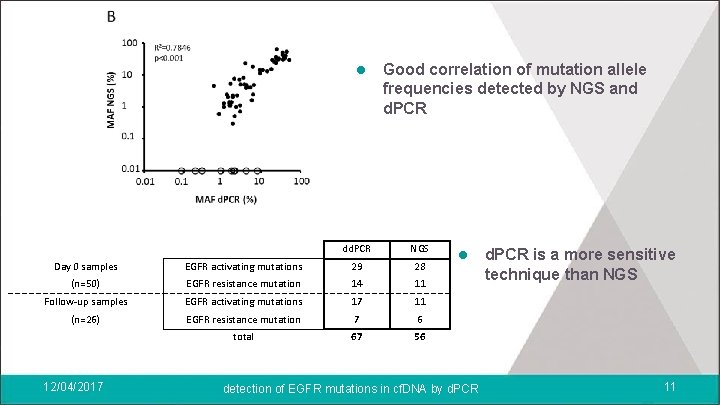
l Good correlation of mutation allele frequencies detected by NGS and d. PCR dd. PCR NGS Day 0 samples EGFR activating mutations 29 28 (n=50) EGFR resistance mutation 14 11 Follow-up samples EGFR activating mutations 17 11 (n=26) EGFR resistance mutation 7 6 total 67 56 12/04/2017 l detection of EGFR mutations in cf. DNA by d. PCR is a more sensitive technique than NGS 11

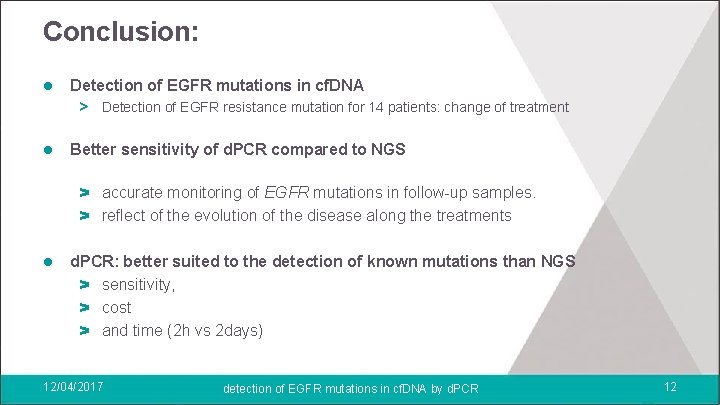
Conclusion: l Detection of EGFR mutations in cf. DNA > Detection of EGFR resistance mutation for 14 patients: change of treatment l Better sensitivity of d. PCR compared to NGS > accurate monitoring of EGFR mutations in follow-up samples. > reflect of the evolution of the disease along the treatments l d. PCR: better suited to the detection of known mutations than NGS > sensitivity, > cost > and time (2 h vs 2 days) 12/04/2017 detection of EGFR mutations in cf. DNA by d. PCR 12

Thank you for your attention Thanks to Dr. Ludovic Lacroix l Dr. Benjamin Besse l Translationnal research Lab Team l Patients l 12/04/2017 Dr. Magali Droniou l Dr. Jordan Madic l ? ? l 13