Transcriptome PROMOTERS AND EXPRESSION The transcriptome transcriptome is













- Slides: 45

Transcriptome PROMOTERS AND EXPRESSION

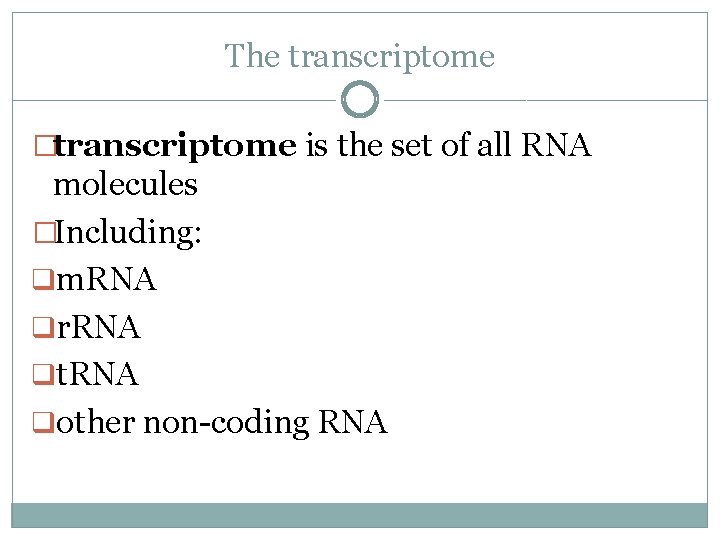
The transcriptome �transcriptome is the set of all RNA molecules �Including: qm. RNA qr. RNA qt. RNA qother non-coding RNA

Outline �Measuring gene expression levels �New understandings of promoters directionality

Motivation �Question: How can we measure protein levels in the body? �The current way to measure was to look at genes expression levels But… �New understandings show that we should measure each of the genes isoforms separately

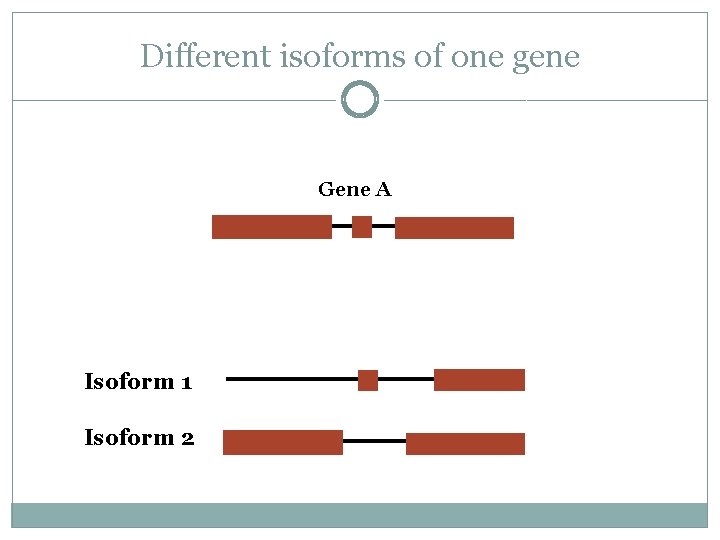
We would like to… 1. Compare between expression levels of different genes 2. Compare between expression levels of different isoforms of a single gene

Different isoforms of one gene Gene A Isoform 1 Isoform 2

RNA-seq: reminder CCGAT T G AG

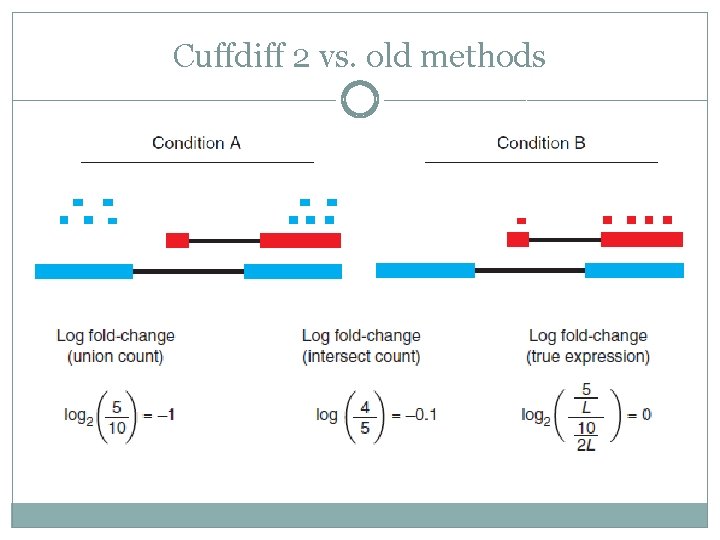
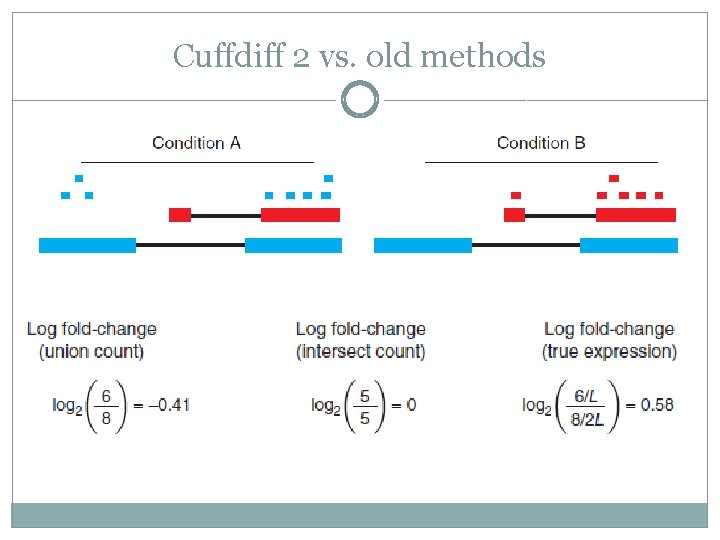
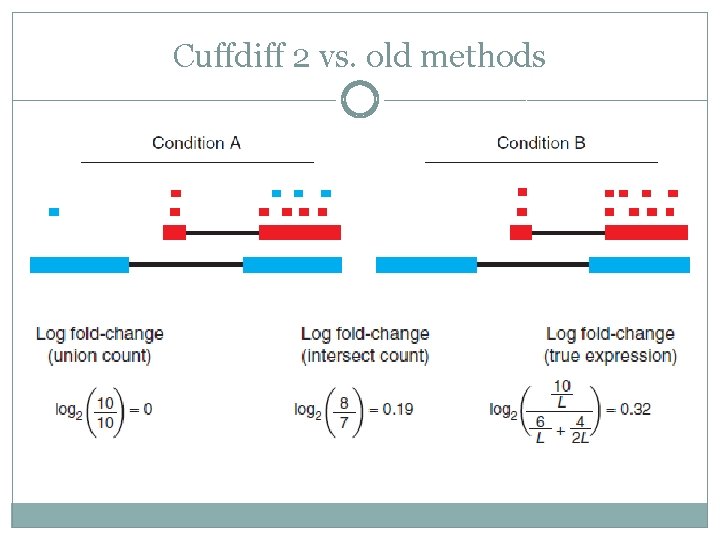
What’s after RNA-seq? �We will describe two different methods to approximate the gene expression Union count Intersect count

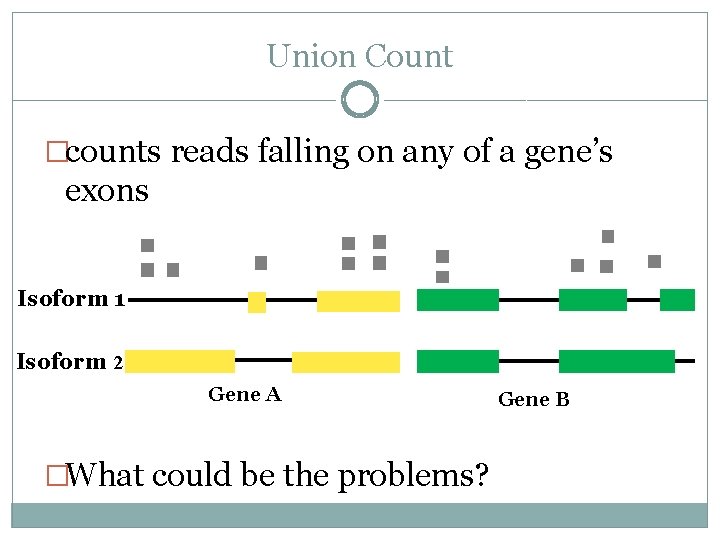
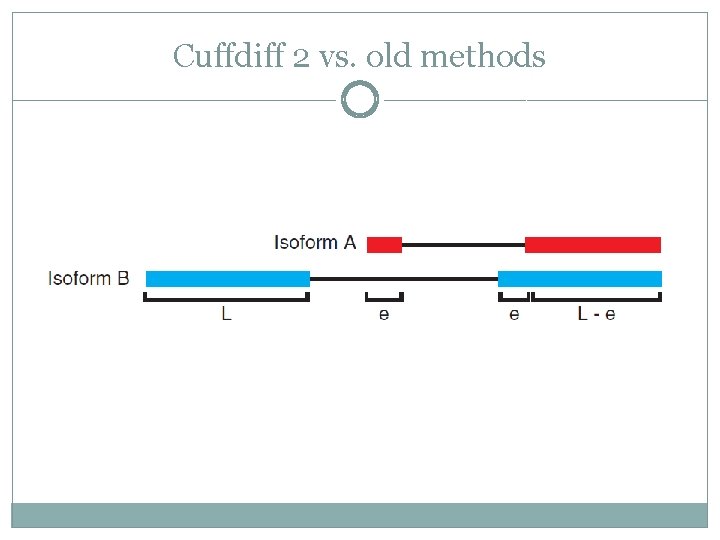
Union Count �counts reads falling on any of a gene’s exons Isoform 1 Isoform 2 Gene A �What could be the problems? Gene B

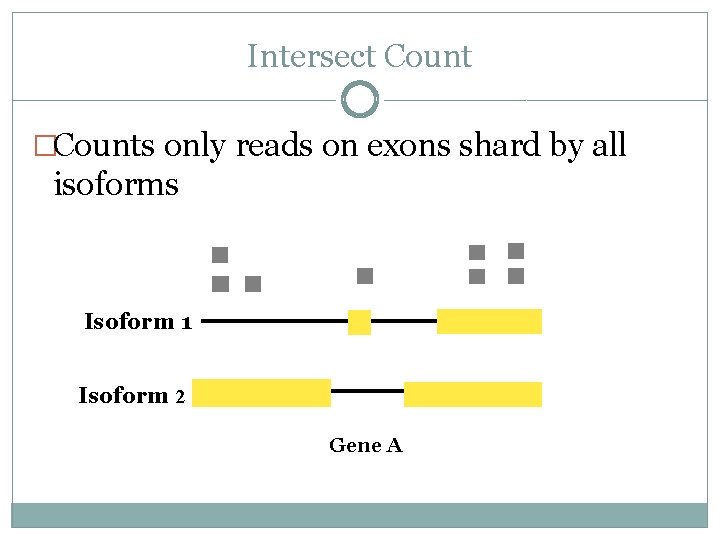
Intersect Count �Counts only reads on exons shard by all isoforms Isoform 1 Isoform 2 Gene A

Two main problems �How can we compare between isoforms of the same gene? �What about the length of the gene?

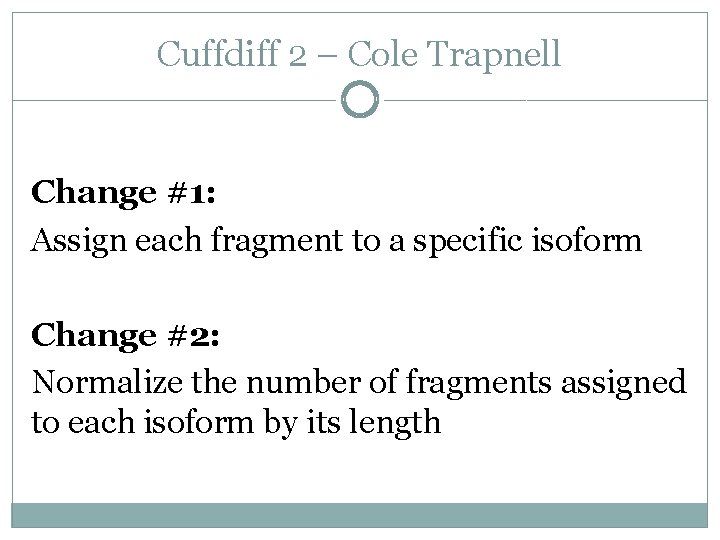
Cuffdiff 2 – Cole Trapnell Change #1: Assign each fragment to a specific isoform Change #2: Normalize the number of fragments assigned to each isoform by its length

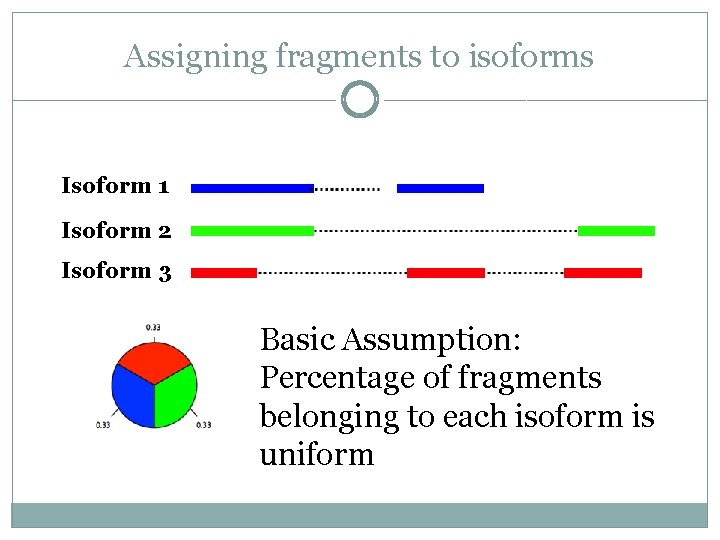
Assigning fragments to isoforms Isoform 1 Isoform 2 Isoform 3 Basic Assumption: Percentage of fragments belonging to each isoform is uniform

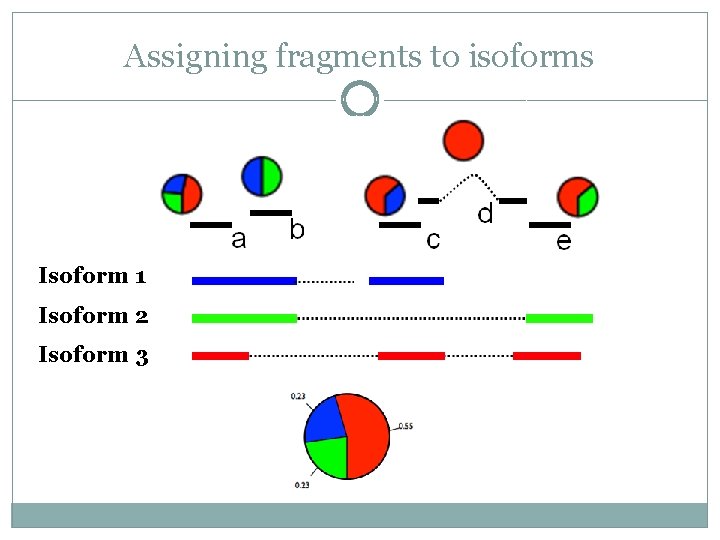
Assigning fragments to isoforms Isoform 1 Isoform 2 Isoform 3

Cuffdiff 2 vs. old methods

Cuffdiff 2 vs. old methods

Cuffdiff 2 vs. old methods

Cuffdiff 2 vs. old methods

Outline �Measuring gene expression levels �New understandings of promoters directionality

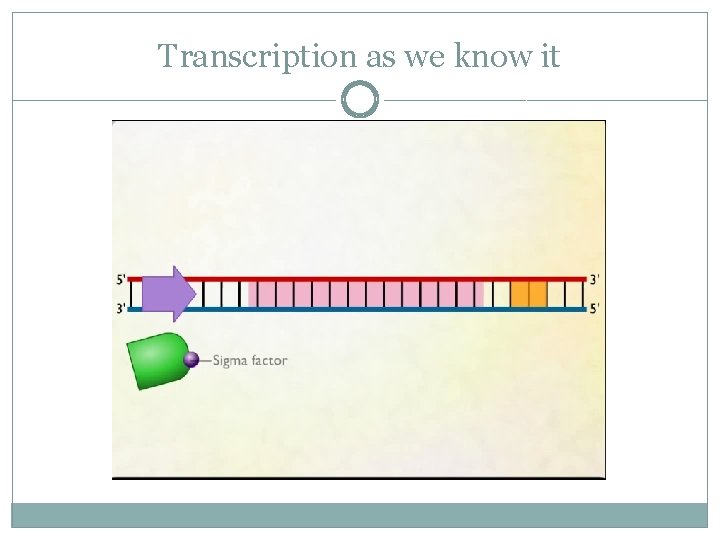
Transcription as we know it

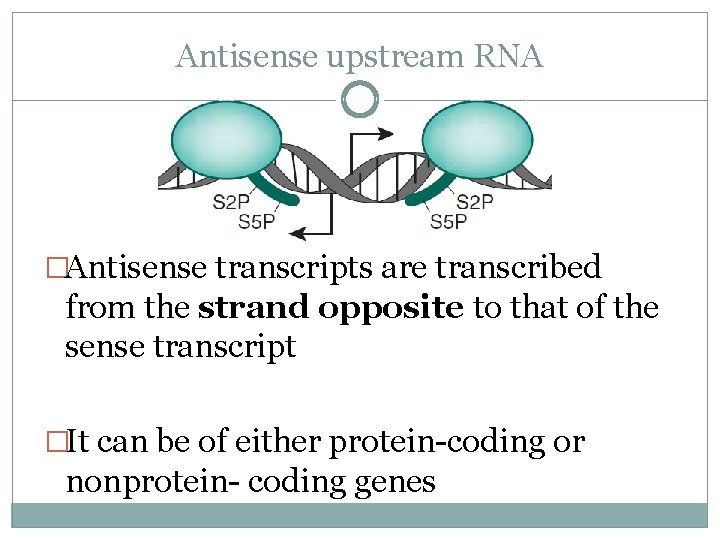
Antisense upstream RNA �Antisense transcripts are transcribed from the strand opposite to that of the sense transcript �It can be of either protein-coding or nonprotein- coding genes

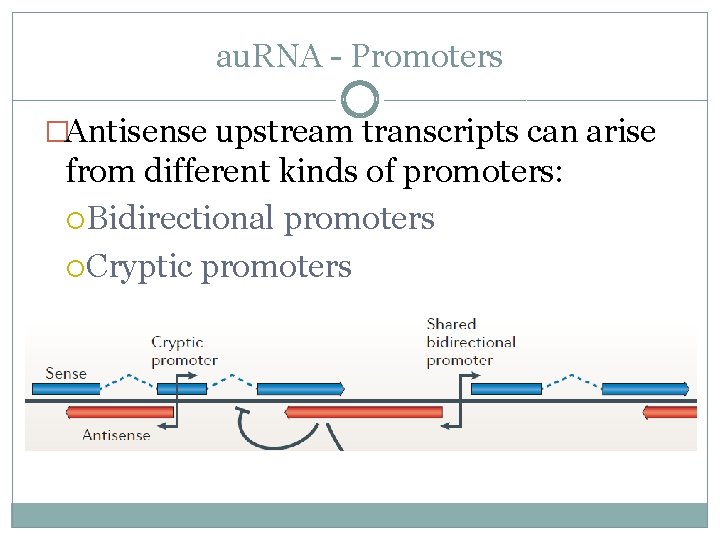
au. RNA - Promoters �Antisense upstream transcripts can arise from different kinds of promoters: Bidirectional promoters Cryptic promoters

au. RNA - Characteristics • • Aside from their antisense orientation, they do not have unique features They lack of protein - coding potential Can contain specific domains that interact with DNA, RNA or proteins (like many nc. RNAs) Many of them carry out specific functions

So which strand is the downstream direction? And where are those antisense transcripts?

A new approach Christopher B. Burge Phillip A. Sharp MIT

ua. RNAs do not elongate efficiently �Two potential mechanisms: Inefficient release of paused RNAP 2 Early termination of transcription

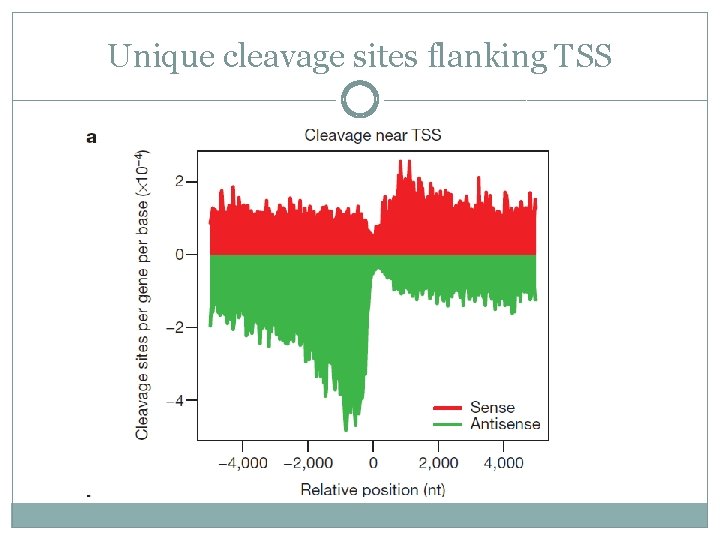
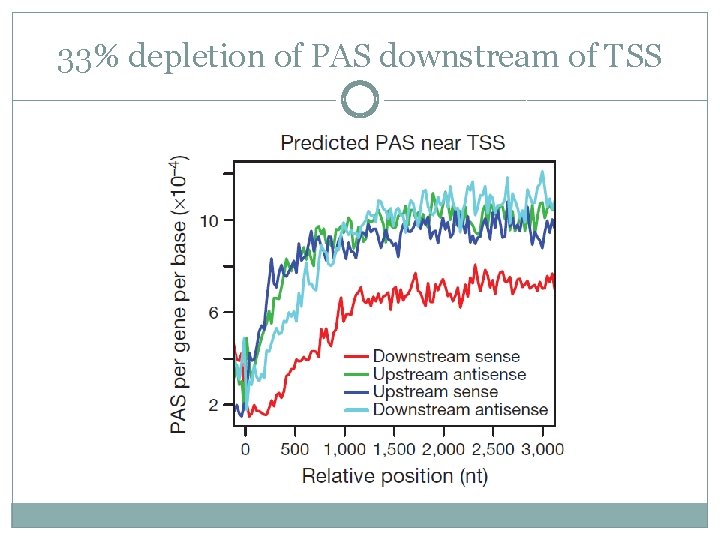
Early termination? �It was found that upstream antisense region has a higher number of cleavage sites compared to the downstream sense sites

Unique cleavage sites flanking TSS

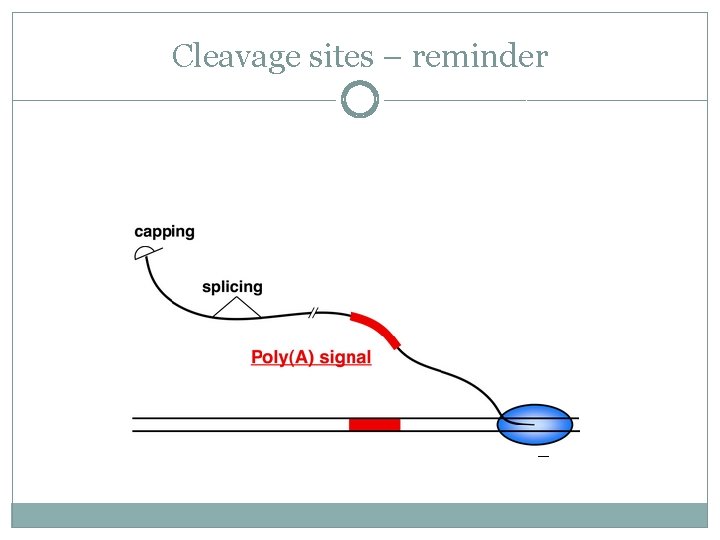
Cleavage sites – reminder

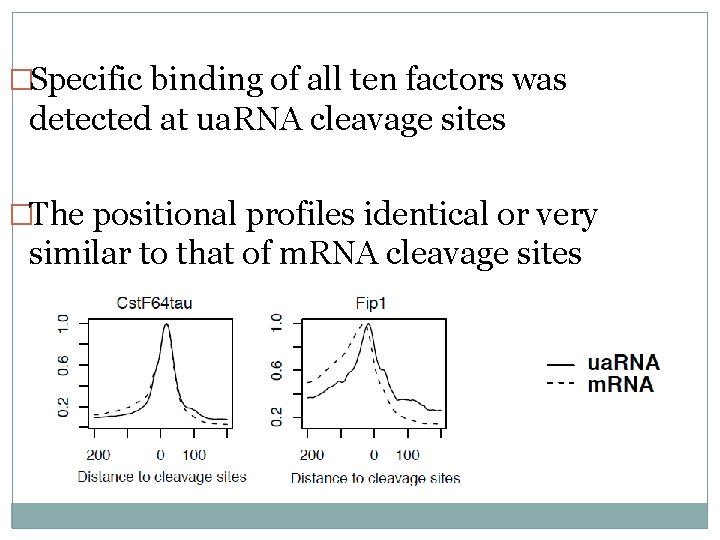
The first step �Determine whether members of the canonical cleavage machinery bind to ua. RNAs �Ten 3’ end processing factors were analyzed to find protein – RNA interactions (CLIP method)

�Specific binding of all ten factors was detected at ua. RNA cleavage sites �The positional profiles identical or very similar to that of m. RNA cleavage sites

Understanding the molecular mechanism So, how come there are 2 times the cleavage areas in the upstream direction?

Understanding the molecular mechanism Examine PAS frequencies!

33% depletion of PAS downstream of TSS

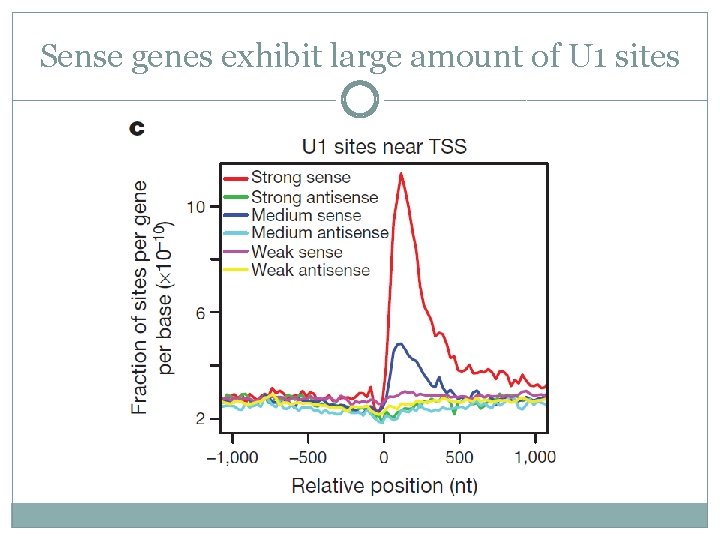
Sense genes exhibit large amount of U 1 sites

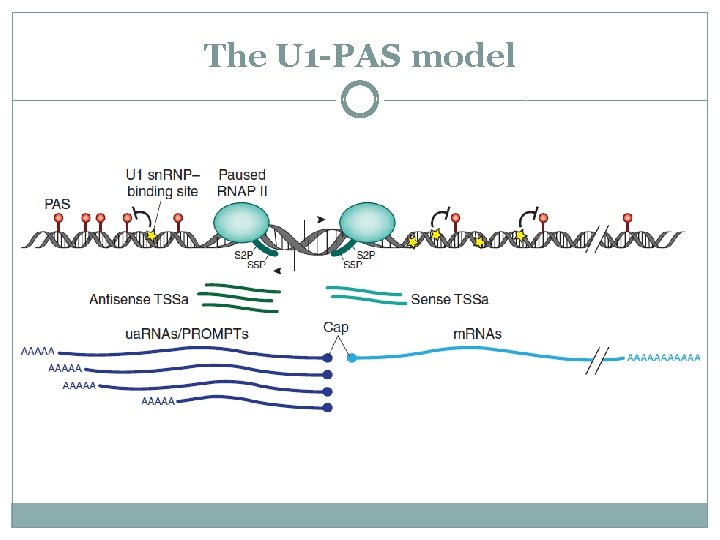
The U 1 -PAS model U 1 sn. RNP complex suppresses cleavage and polyadenylation near a U 1 site

The U 1 -PAS model


au. RNA – Mechanisms of gene regulation �We will discuss the different steps: Effects on transcription initiation Effects on transcription elongation Post transcriptional effects

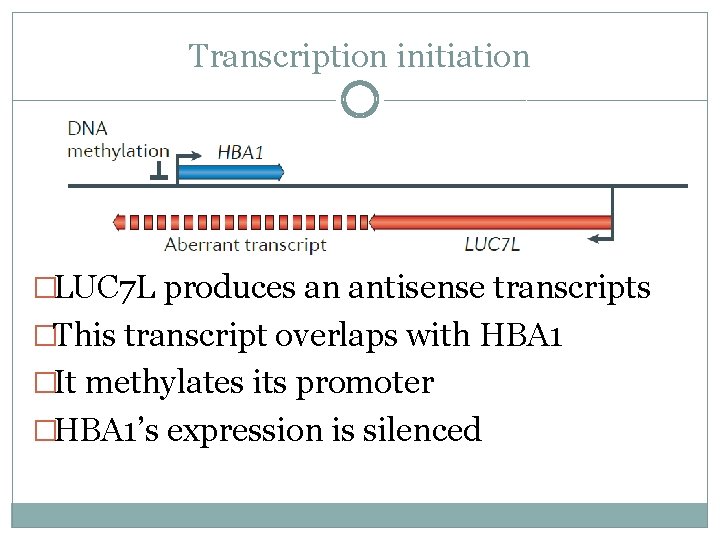
Transcription initiation �LUC 7 L produces an antisense transcripts �This transcript overlaps with HBA 1 �It methylates its promoter �HBA 1’s expression is silenced

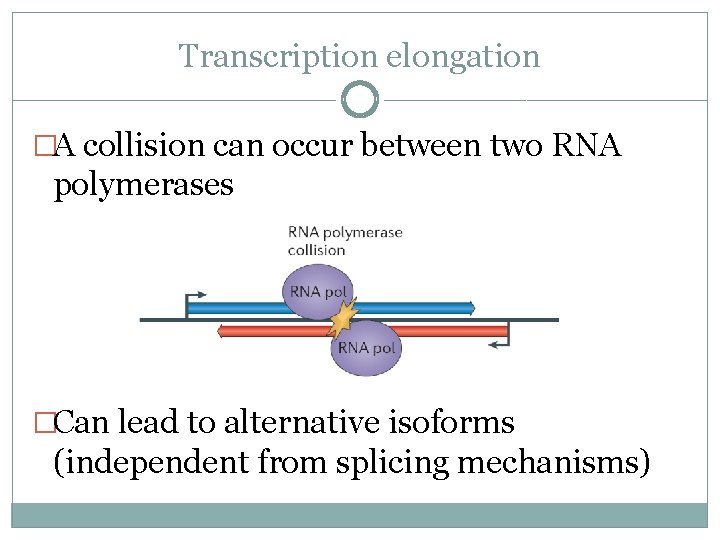
Transcription elongation �A collision can occur between two RNA polymerases �Can lead to alternative isoforms (independent from splicing mechanisms)

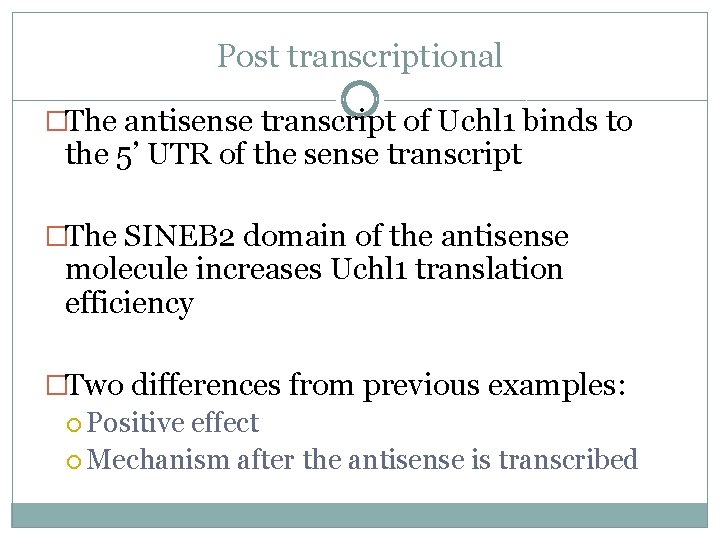
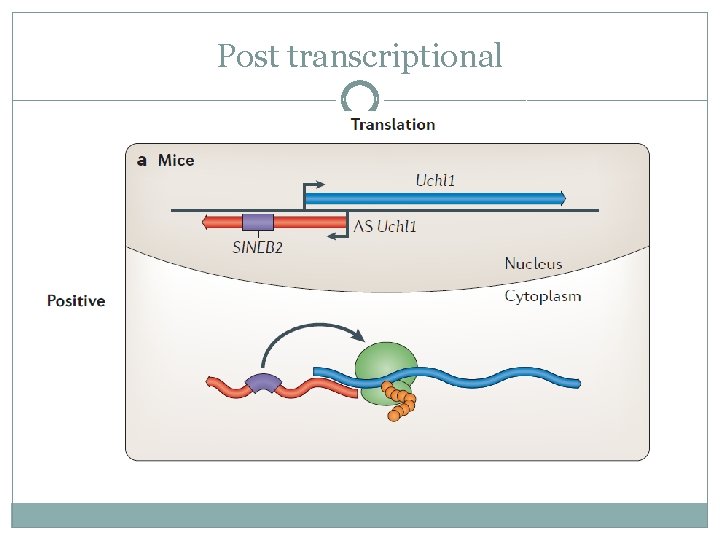
Post transcriptional �The antisense transcript of Uchl 1 binds to the 5’ UTR of the sense transcript �The SINEB 2 domain of the antisense molecule increases Uchl 1 translation efficiency �Two differences from previous examples: Positive effect Mechanism after the antisense is transcribed

Post transcriptional

Summary �New technologies enable us to further understand genes expression levels �Transcription process is not as “simple” as we thought �Every transcript has its role

Questions ?