The Transcriptome Gene Discovery Quantitation of Gene Expression














- Slides: 35

The Transcriptome Gene Discovery Quantitation of Gene Expression Reading: Ch 15. 1 BIO 520 Bioinformatics Jim Lund

WHY? • The genes (proteins) expressed determine the state of the cell. – – Signaling. Metabolic capabilities. Differentiation state (cell type). Response to changes in environment. • Verifies gene predictions. • Transcriptional regulation – Normal vs. abnormal – Conditional expression

Transcriptome Analysis • Gene (transcript) discovery – transcripts – alternative splicing/processing • • Transcript assays Promoter analysis Transcription Factors Cellular control networks

Gene Discovery • Inference from genomic DNA – Prokaryotes & fungi OK • c. DNA characterization – EST – SAGE

EST (Expressed Sequence Tag) • Sequence c. DNA libraries – proportional libraries – subtracted or normalized libraries • Which end? – 5’ or 3’ or Whole

Library Type • “regular” or proportional • Subtracted – Miss alternate transcripts • normalized • Tissue • Primer – d. T vs random

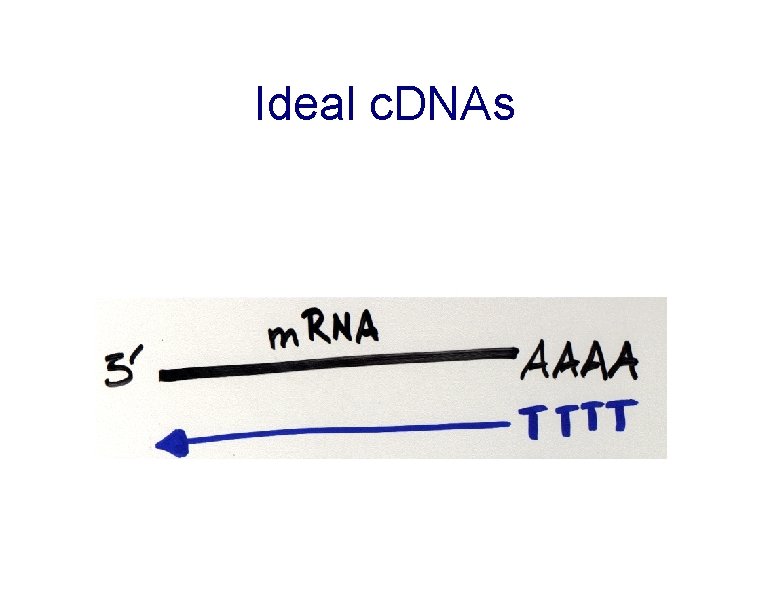
Ideal c. DNAs

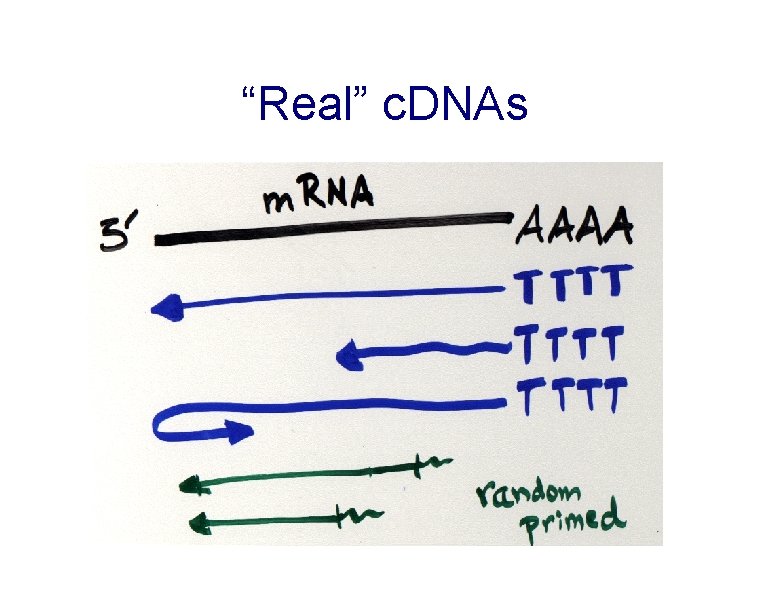
“Real” c. DNAs

Which end? • Whole c. DNA – BEST & HARDEST (Long) • 3’-end – Consistent technically, limited information • 5’end – Coding “identity” highest • 5’ AND 3’ – Good, but technical & informatic challenge


EST Data Analyses • Clustering Analysis – – Assemble ESTs into genes. Alternative splicing forms Find coding SNPs. Truncated, unspliced, and junk ESTs can be misleading – Project: Unigene – Program: stack. PACK • Frequency analysis – Digital Differential Display • DDD is a computational method for comparing sequence-based gene representation profiles among individual c. DNA libraries or pools of libraries.

EST Results (old) • Known genes (30%) • Similarities to other ORFs, ESTs (30%) – Infer Function? • Novel Class (30%, w/ time)

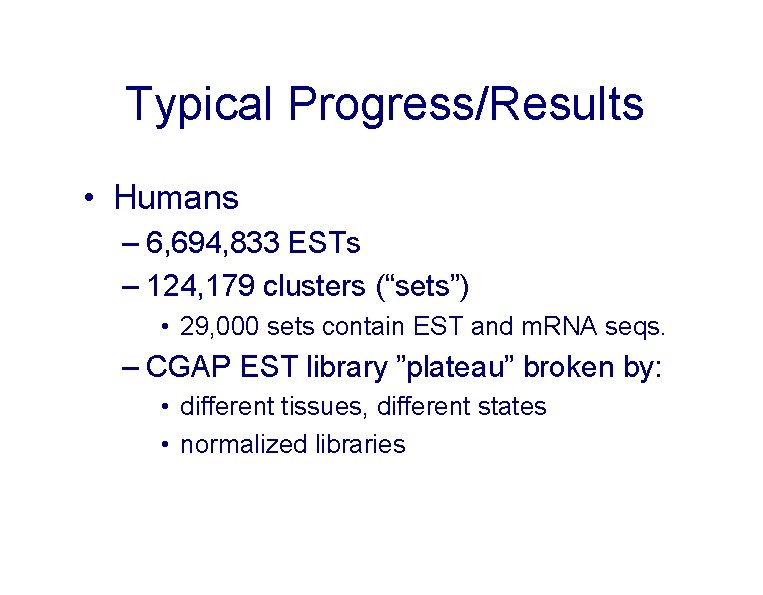
Typical Progress/Results • Humans – 6, 694, 833 ESTs – 124, 179 clusters (“sets”) • 29, 000 sets contain EST and m. RNA seqs. – CGAP EST library ”plateau” broken by: • different tissues, different states • normalized libraries

Data Quality Considerations • 99% correct data (1% errors!). • Frameshifts-effects depend on tools – BLASTX tool to “find” frameshifts • How sensitive? – TBLASTX, TBLASTN to “use” in other projects • How sensitive?

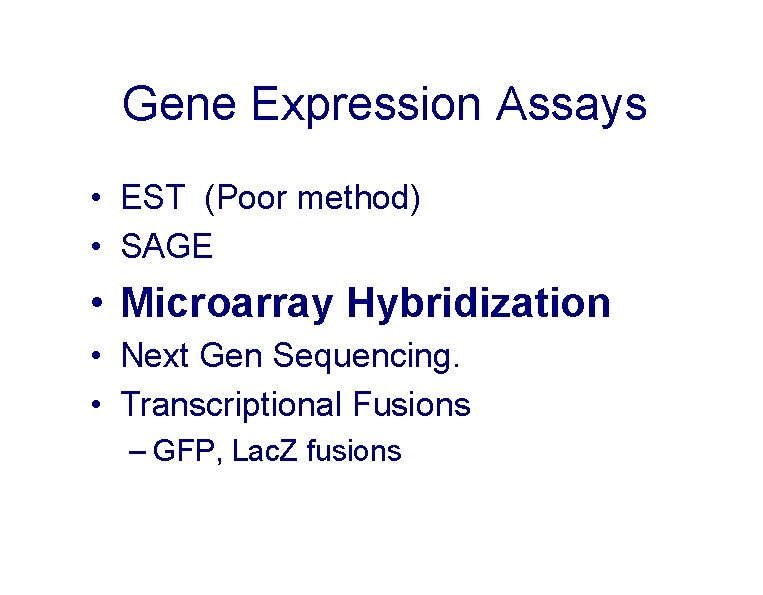
Gene Expression Assays • EST (Poor method) • SAGE • Microarray Hybridization • Next Gen Sequencing. • Transcriptional Fusions – GFP, Lac. Z fusions

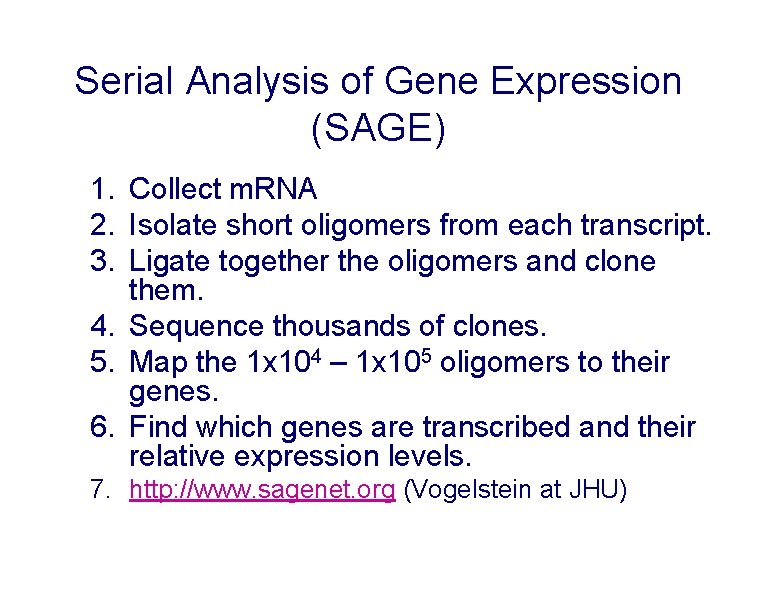
Serial Analysis of Gene Expression (SAGE) 1. Collect m. RNA 2. Isolate short oligomers from each transcript. 3. Ligate together the oligomers and clone them. 4. Sequence thousands of clones. 5. Map the 1 x 104 – 1 x 105 oligomers to their genes. 6. Find which genes are transcribed and their relative expression levels. 7. http: //www. sagenet. org (Vogelstein at JHU)

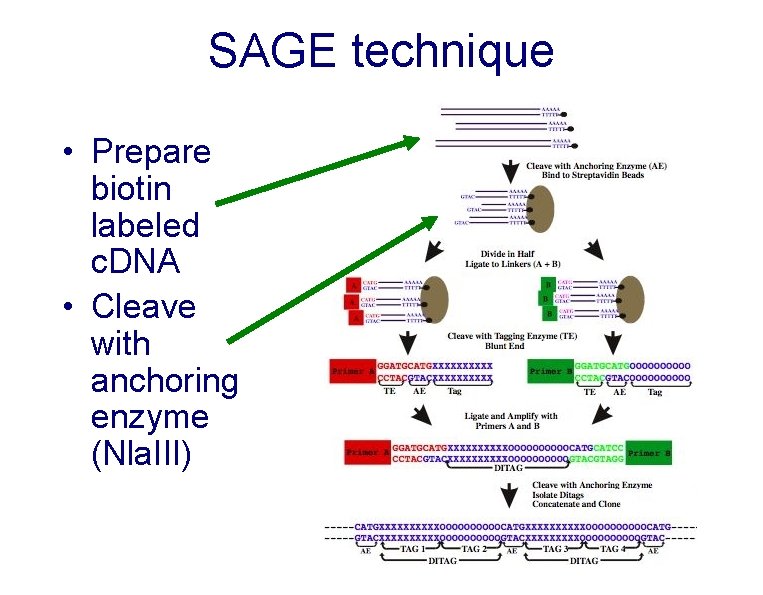
SAGE technique • Prepare biotin labeled c. DNA • Cleave with anchoring enzyme (Nla. III)

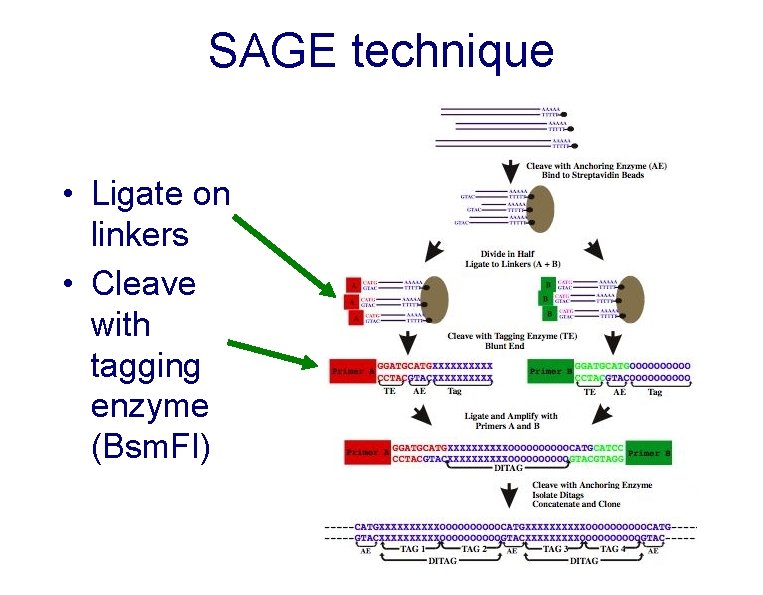
SAGE technique • Ligate on linkers • Cleave with tagging enzyme (Bsm. FI)

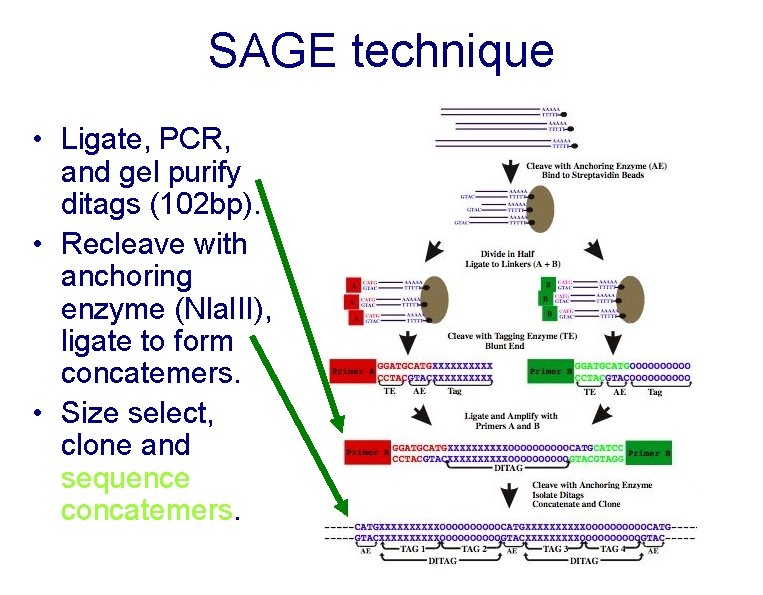
SAGE technique • Ligate, PCR, and gel purify ditags (102 bp). • Recleave with anchoring enzyme (Nla. III), ligate to form concatemers. • Size select, clone and sequence concatemers.

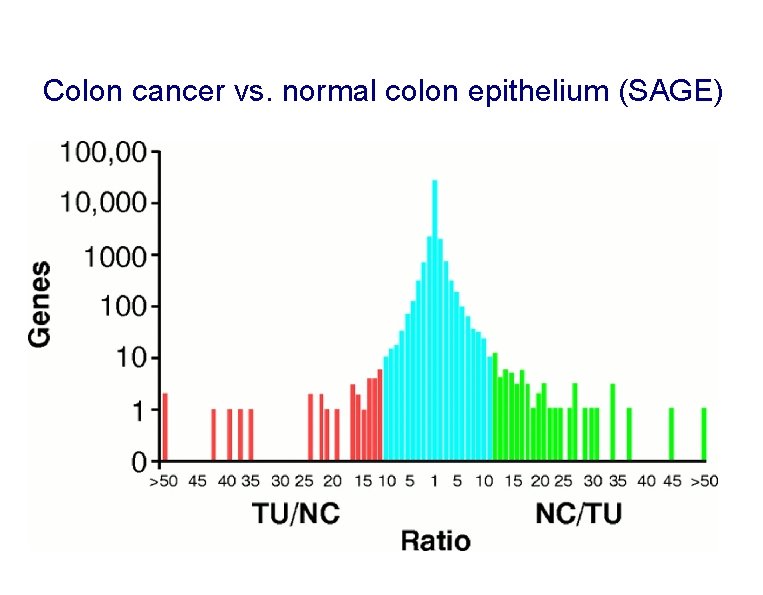
Colon cancer vs. normal colon epithelium (SAGE)

Microarray Hybridization • Determine gene expression by parallel hybridization of labeled c. DNA to DNA attached to a fixed support. • http: //cmgm. stanford. edu/pbrown/

Microarray Hybridization • Producing chips • Producing probes / reading arrays • Analyzing and interpreting data

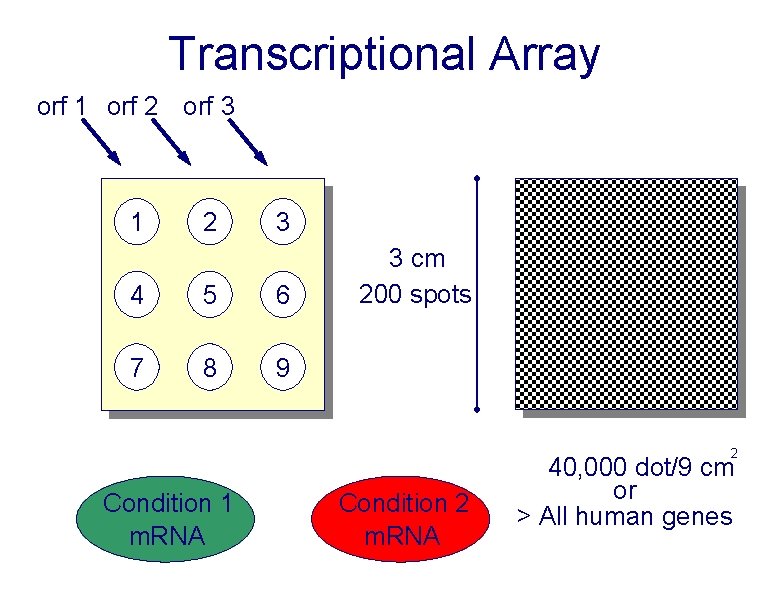
Transcriptional Array orf 1 orf 2 orf 3 1 2 3 4 5 6 7 8 9 3 cm 200 spots 2 Condition 1 m. RNA Condition 2 m. RNA 40, 000 dot/9 cm or > All human genes

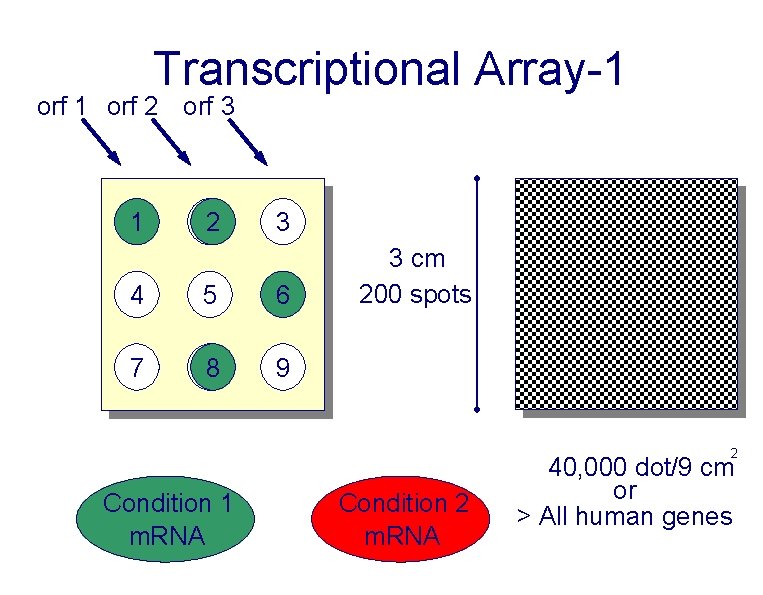
Transcriptional Array-1 orf 2 orf 3 1 22 3 4 5 6 7 88 9 3 cm 200 spots 2 Condition 1 m. RNA Condition 2 m. RNA 40, 000 dot/9 cm or > All human genes

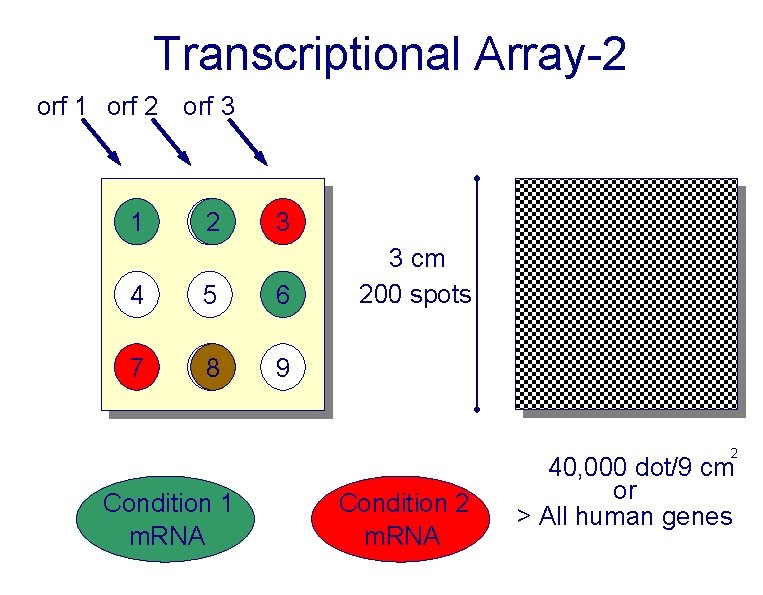
Transcriptional Array-2 orf 1 orf 2 orf 3 1 22 3 4 5 6 7 88 9 3 cm 200 spots 2 Condition 1 m. RNA Condition 2 m. RNA 40, 000 dot/9 cm or > All human genes

Microarray Technologies • Spotted arrays (Brown et al. ) – Spot arrays on glass slides – PCR fragments – Long (50 -70 bp) oligo arrays • Synthesis – Affymetrix (www. affymetrix. com) • High density array of 25 bp oligos • Made using light directed oligonucleotide synthesis and photolithography – Agilent, Combi. Matrix • Made using light directed oligonucleotide synthesis and mirrors.

Spotted Arrays

Print Quill

Spotted microarray image

Affymetrix photolithographic technology • Lithographic masks are used to either block or transmit light onto specific locations of the array. • The surface is then flooded with a solution containing either adenine, thymine, cytosine, or guanine, and coupling occurs only in those regions on the glass that have been deprotected through illumination. • The coupled nucleotide also bears a light-sensitive protecting group, so the cycle can be repeated. • Microarray is built as the probes are synthesized through repeated cycles of deprotection and coupling. • Typically ends at 25 bps. ) • Current arrays have 1. 3 million unique features per array.

Gene. Chip Expression Assay Design

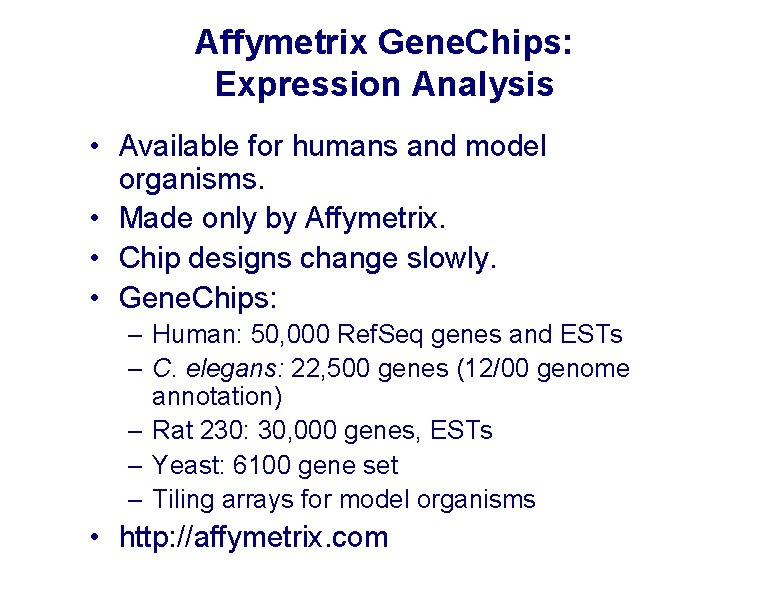
Affymetrix Gene. Chips: Expression Analysis • Available for humans and model organisms. • Made only by Affymetrix. • Chip designs change slowly. • Gene. Chips: – Human: 50, 000 Ref. Seq genes and ESTs – C. elegans: 22, 500 genes (12/00 genome annotation) – Rat 230: 30, 000 genes, ESTs – Yeast: 6100 gene set – Tiling arrays for model organisms • http: //affymetrix. com

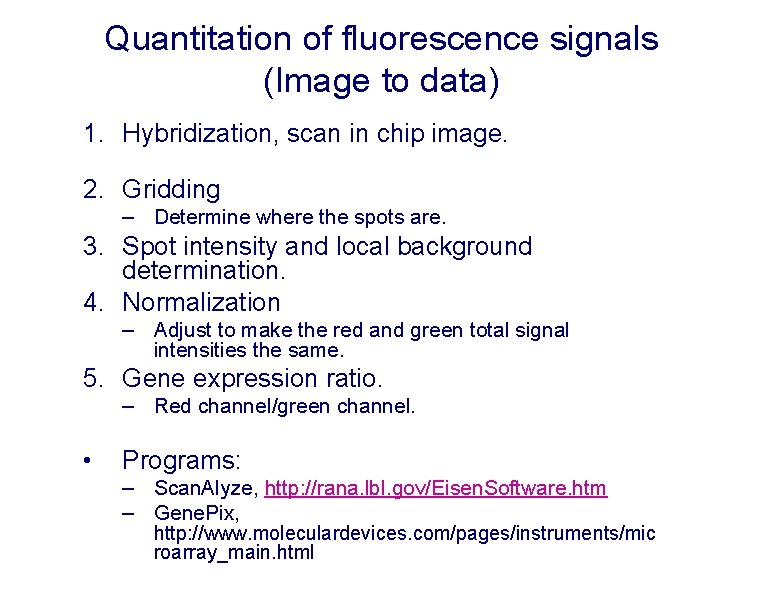
Quantitation of fluorescence signals (Image to data) 1. Hybridization, scan in chip image. 2. Gridding – Determine where the spots are. 3. Spot intensity and local background determination. 4. Normalization – Adjust to make the red and green total signal intensities the same. 5. Gene expression ratio. – Red channel/green channel. • Programs: – Scan. Alyze, http: //rana. lbl. gov/Eisen. Software. htm – Gene. Pix, http: //www. moleculardevices. com/pages/instruments/mic roarray_main. html

Microarray data Big tables of numbers!

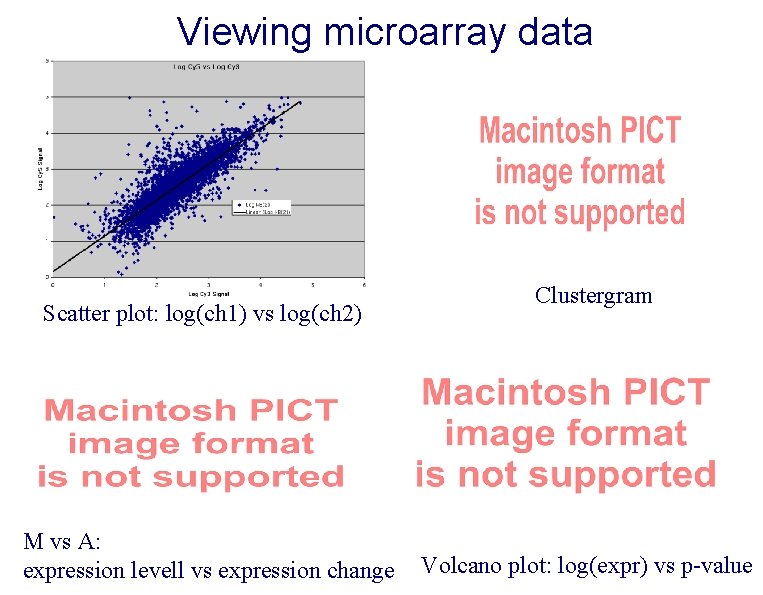
Viewing microarray data Scatter plot: log(ch 1) vs log(ch 2) M vs A: expression levell vs expression change Clustergram Volcano plot: log(expr) vs p-value