Octet Training Part III Quantitation on the Octet







- Slides: 40

Octet Training Part III: Quantitation on the Octet Scott Zhou, North China FAS MB: 15810470035, Email: scott_zhou@ap. pall. com Mar. 20 th, 2013

Agenda l BLI Quantitation Workflow l BLI Quantitation Applications

BLI Quantitation Workflow

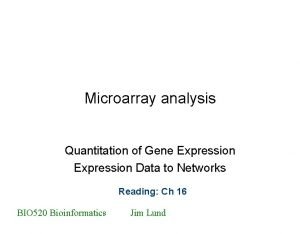
Sandwich ELISA Assay Procedure Typical read out could be fluorescence or luminescence Bottom of well (1) (2) (3) (4) (5) Plate is coated with a capture antibody Sample is added, and any antigen present binds to capture antibody Detecting antibody is added, and binds to antigen Enzyme-linked secondary antibody is added, and binds to detecting antibody Chromogen/substrate is added, and is converted by enzyme to detectable form Each step involves incubation time and wash steps in between. Manual ELISA can be an all day assay for just a few plates. Multiple Steps add to higher CV 5 -25%.

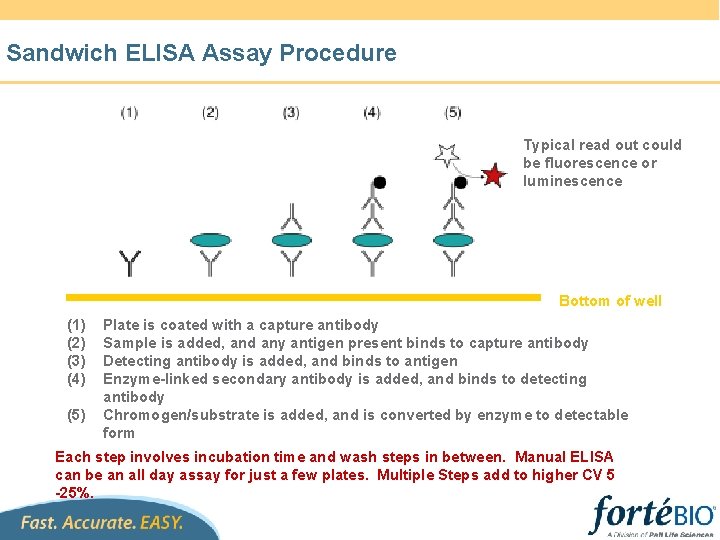
Biolayer Interferometry(BLI) 可实时检测到两个反射表面间距的改变 Relative Intensity 100% 0 Wavelength (nm) surfaces = ℓ Intensity λ = ƒ(λ, ℓ) nm shift Distance between the two reflecting Time

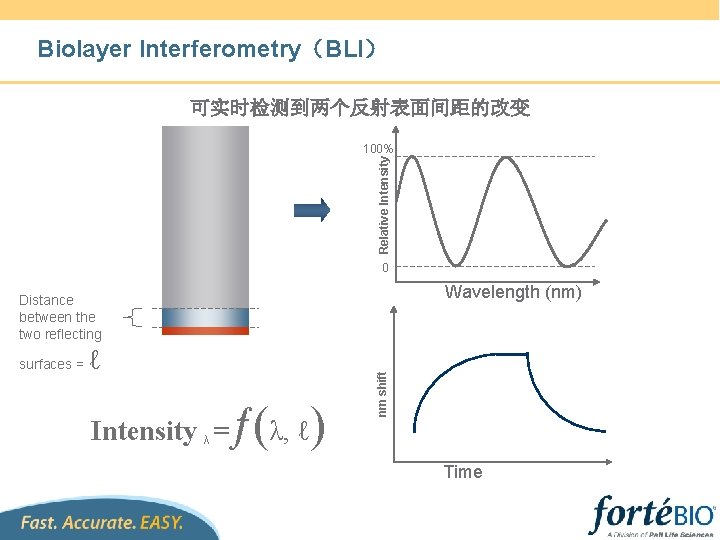
Biosensor selection According to application & sample type etc. 新开发了四种传感器 1. Anti-GST Biosensor: 用于含GST标签的蛋白 2. NTA Biosensor:用于含His标签的蛋白 3. Anti-human Fab-CH 1:用于人Fab, F(ab’)2及Ab 1~4 4. Anti-Flag biosensor:用于含Flag标签的蛋白

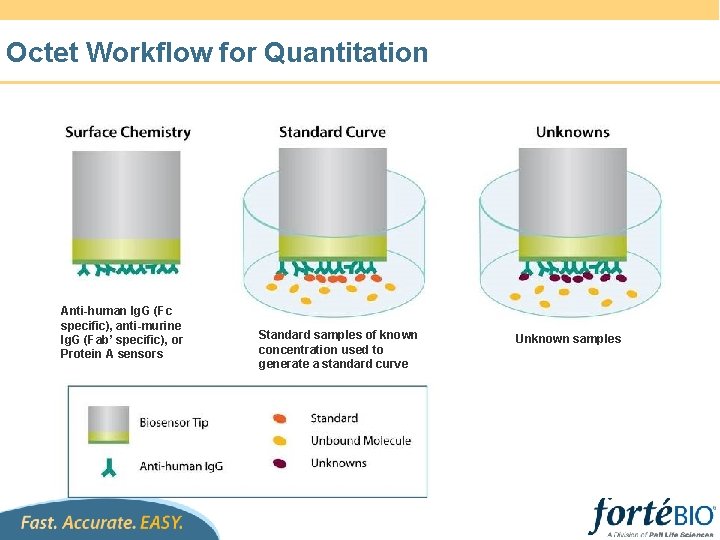
Octet Workflow for Quantitation Anti-human Ig. G (Fc specific), anti-murine Ig. G (Fab’ specific), or Protein A sensors Standard samples of known concentration used to generate a standard curve Unknown samples

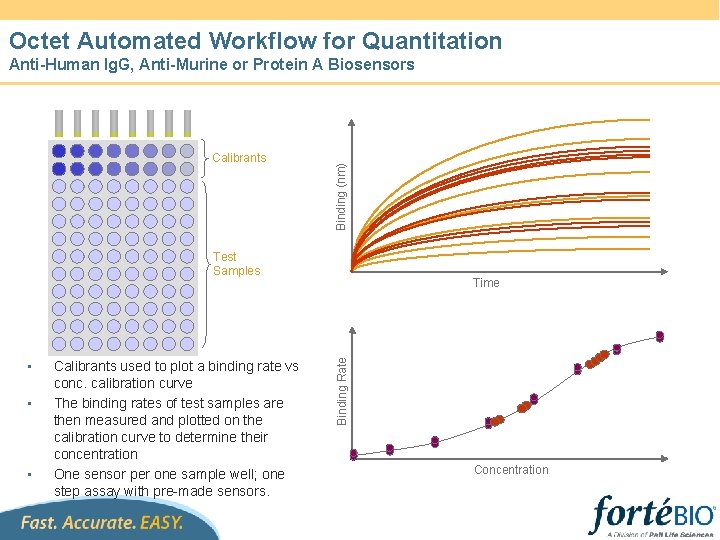
Octet Automated Workflow for Quantitation Calibrants Binding (nm) Anti-Human Ig. G, Anti-Murine or Protein A Biosensors Test Samples • • Calibrants used to plot a binding rate vs conc. calibration curve The binding rates of test samples are then measured and plotted on the calibration curve to determine their concentration One sensor per one sample well; one step assay with pre-made sensors. Binding Rate • Time Concentration

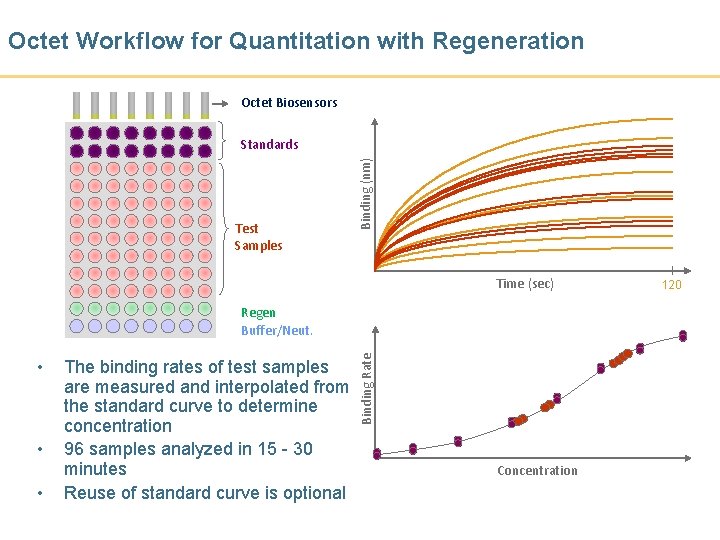
Octet Workflow for Quantitation with Regeneration Octet Biosensors Test Samples Binding (nm) Standards Time (sec) • • • The binding rates of test samples are measured and interpolated from the standard curve to determine concentration 96 samples analyzed in 15 - 30 minutes Reuse of standard curve is optional Binding Rate Regen Buffer/Neut. Concentration 120

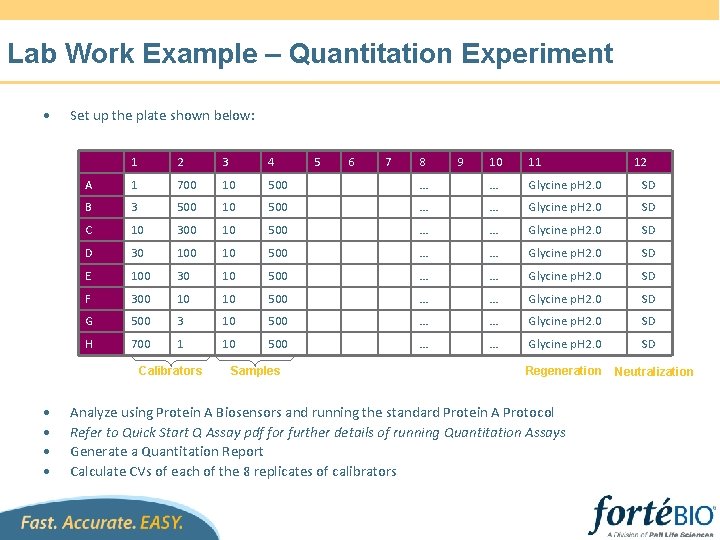
Lab Work Example – Quantitation Experiment • Set up the plate shown below: 1 2 3 4 A 1 700 10 500 B 3 500 10 C 10 300 D 30 E 10 11 … … Glycine p. H 2. 0 SD 500 … … Glycine p. H 2. 0 SD 100 10 500 … … Glycine p. H 2. 0 SD 100 30 10 500 … … Glycine p. H 2. 0 SD F 300 10 10 500 … … Glycine p. H 2. 0 SD G 500 3 10 500 … … Glycine p. H 2. 0 SD H 700 1 10 500 … … Glycine p. H 2. 0 SD Calibrators • • Samples 5 6 7 8 9 Regeneration Analyze using Protein A Biosensors and running the standard Protein A Protocol Refer to Quick Start Q Assay pdf for further details of running Quantitation Assays Generate a Quantitation Report Calculate CVs of each of the 8 replicates of calibrators 12 Neutralization

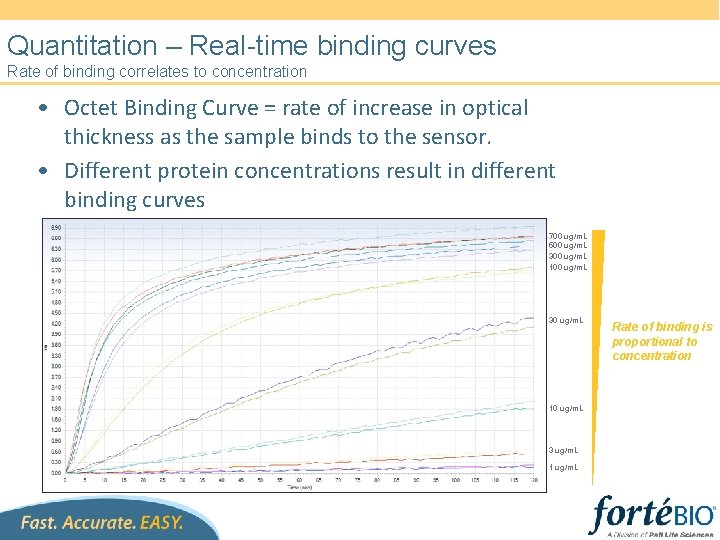
Quantitation – Real-time binding curves Rate of binding correlates to concentration • Octet Binding Curve = rate of increase in optical thickness as the sample binds to the sensor. • Different protein concentrations result in different binding curves 700 ug/m. L 500 ug/m. L 300 ug/m. L 100 ug/m. L 30 ug/m. L 10 ug/m. L 3 ug/m. L 1 ug/m. L Rate of binding is proportional to concentration

BLI Quantitation Applications

Fc-fused protein Quantitation Overview • • • Samples: 14 h. Fc-fused proteins in supernatant 1 Standards: Fc-fused proteins with original conc. of 92. 09 mg/ml Biosensors : Protein A biosensor Octet platform: RED 96 Other reagents & consumables : fresh medium, supernatant 2, PBS, 10 m. M p. H 1. 5 glycine, Greiner 96 -well micro-plate, pipettes , tips and etc. • Goal: CV% & Re%, throughput and etc. as to ELISA

Workflow for the Fc-fused protein quantitation Test for dilution factors Dilute standards and unknowns with diluted supernatant Enter sample information into software Bind Fc-fused protein to protein A biosensor Generate standard curve & Regenerate biosensors Bind known concentrations of Fc-fused proteins to regenerated sensor • Bind unknown samples & Regenerate biosensors • Interpolate samples from standard curve to determine active concentration • • •

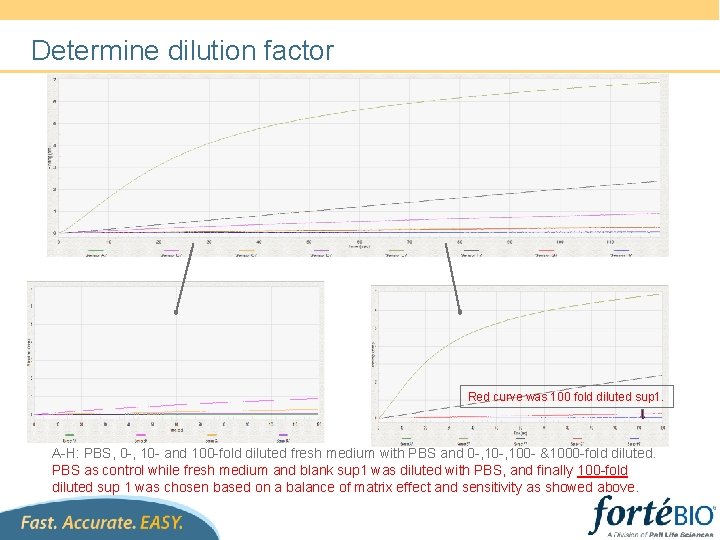
Determine dilution factor Red curve was 100 fold diluted sup 1. A-H: PBS, 0 -, 10 - and 100 -fold diluted fresh medium with PBS and 0 -, 100 - &1000 -fold diluted. PBS as control while fresh medium and blank sup 1 was diluted with PBS, and finally 100 -fold diluted sup 1 was chosen based on a balance of matrix effect and sensitivity as showed above.

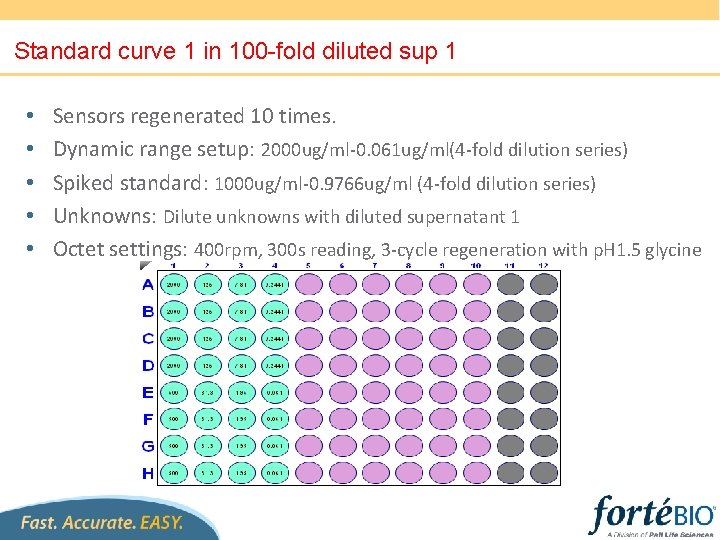
Standard curve 1 in 100 -fold diluted sup 1 • • • Sensors regenerated 10 times. Dynamic range setup: 2000 ug/ml-0. 061 ug/ml(4 -fold dilution series) Spiked standard: 1000 ug/ml-0. 9766 ug/ml (4 -fold dilution series) Unknowns: Dilute unknowns with diluted supernatant 1 Octet settings: 400 rpm, 300 s reading, 3 -cycle regeneration with p. H 1. 5 glycine

Standard curve 1 determined in 100 -fold diluted sup 1 • Analysis model: unweighted 4 PL • Effective Dynamic range setup: 2000 ug/ml-7. 8125 ug/ml(4 -fold dilution series) • Spiked standard: 1000 ug/ml-15. 625 ug/ml (4 -fold dilution series) • Octet settings: 400 rpm, 15 s reading

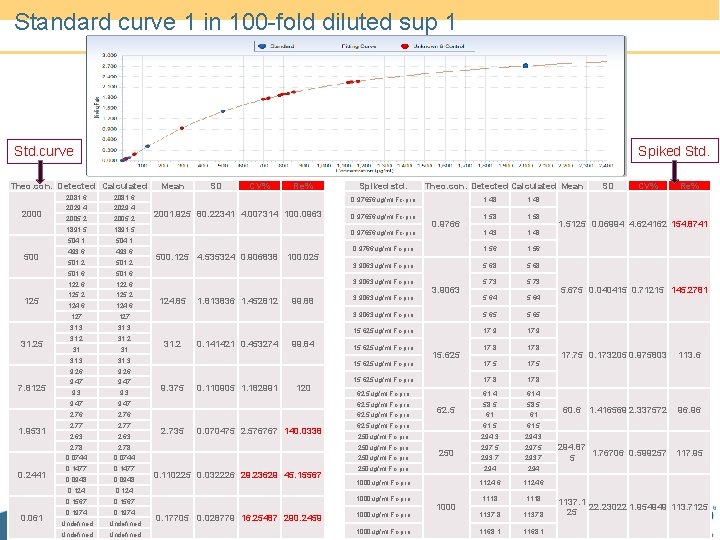
Standard curve 1 in 100 -fold diluted sup 1 Spiked Std. curve Theo. con. Detected Calculated 2000 500 125 31. 25 7. 8125 1. 9531 0. 2441 0. 061 2081. 6 2029. 4 2005. 2 1891. 5 504. 1 493. 6 501. 2 501. 6 122. 6 125. 2 124. 6 127 31. 3 31. 2 Mean SD CV% Re% Spiked std. Theo. con. Detected Calculated Mean 0. 97656 ug/ml Fc-pro 2001. 925 80. 22341 4. 007314 100. 0963 1. 48 1. 58 1. 43 0. 9766 ug/ml Fc-pro 1. 56 3. 9063 ug/ml Fc-pro 5. 68 5. 73 5. 64 3. 9063 ug/ml Fc-pro 5. 65 15. 625 ug/ml Fc-pro 17. 9 17. 8 17. 5 15. 625 ug/ml Fc-pro 17. 8 62. 5 ug/ml Fc-pro 61. 4 58. 5 61 61 0. 97656 ug/ml Fc-pro 500. 125 4. 535324 0. 906838 100. 025 3. 9063 ug/ml Fc-pro 124. 85 31. 2 1. 813836 1. 452812 0. 141421 0. 453274 99. 88 99. 84 3. 9063 ug/ml Fc-pro 15. 625 ug/ml Fc-pro 31 31 31. 3 9. 26 9. 47 9. 3 9. 47 62. 5 ug/ml Fc-pro 2. 76 62. 5 ug/ml Fc-pro 2. 77 2. 63 15. 625 ug/ml Fc-pro 9. 375 2. 735 0. 110905 1. 182991 120 0. 070475 2. 576767 140. 0338 15. 625 62. 5 61. 5 250 ug/ml Fc-pro 294. 3 297. 5 293. 7 250 ug/ml Fc-pro 294 1000 ug/ml Fc-pro 1124. 6 1118 1137. 8 1168. 1 2. 78 250 ug/ml Fc-pro 0. 0744 250 ug/ml Fc-pro 0. 1477 0. 0948 0. 124 0. 1567 1000 ug/ml Fc-pro 0. 1974 1000 ug/ml Fc-pro Undefined 0. 17705 0. 028779 16. 25487 290. 2459 3. 9063 62. 5 ug/ml Fc-pro 0. 0744 0. 110225 0. 032226 29. 23629 45. 15567 0. 9766 1000 ug/ml Fc-pro 250 1000 SD CV% Re% 1. 5125 0. 06994 4. 624162 154. 8741 5. 675 0. 040415 0. 71215 145. 2781 17. 75 0. 173205 0. 975803 113. 6 60. 6 1. 416569 2. 337572 96. 96 294. 87 1. 76706 0. 599257 117. 95 5 1137. 1 22. 23022 1. 954949 113. 7125 25

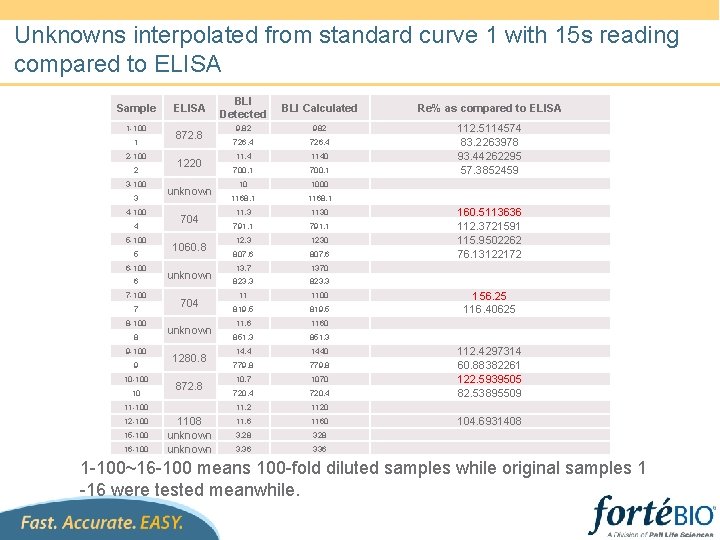
Unknowns interpolated from standard curve 1 with 15 s reading compared to ELISA Sample 1 -100 1 2 -100 2 3 -100 3 4 -100 4 5 -100 5 6 -100 6 7 -100 7 8 -100 8 9 -100 9 10 -100 10 ELISA 872. 8 1220 unknown 704 1060. 8 unknown 704 unknown 1280. 8 872. 8 11 -100 12 -100 15 -100 16 -100 1108 unknown BLI Detected BLI Calculated Re% as compared to ELISA 9. 82 982 726. 4 1140 700. 1 10 1000 1168. 1 11. 3 1130 791. 1 12. 3 1230 807. 6 13. 7 1370 823. 3 11 1100 819. 5 11. 6 1160 851. 3 14. 4 1440 779. 8 10. 7 1070 720. 4 11. 2 1120 11. 6 1160 3. 28 3. 36 336 112. 5114574 83. 2263978 93. 44262295 57. 3852459 160. 5113636 112. 3721591 115. 9502262 76. 13122172 156. 25 116. 40625 112. 4297314 60. 88382261 122. 5939505 82. 53895509 104. 6931408 1 -100~16 -100 means 100 -fold diluted samples while original samples 1 -16 were tested meanwhile.

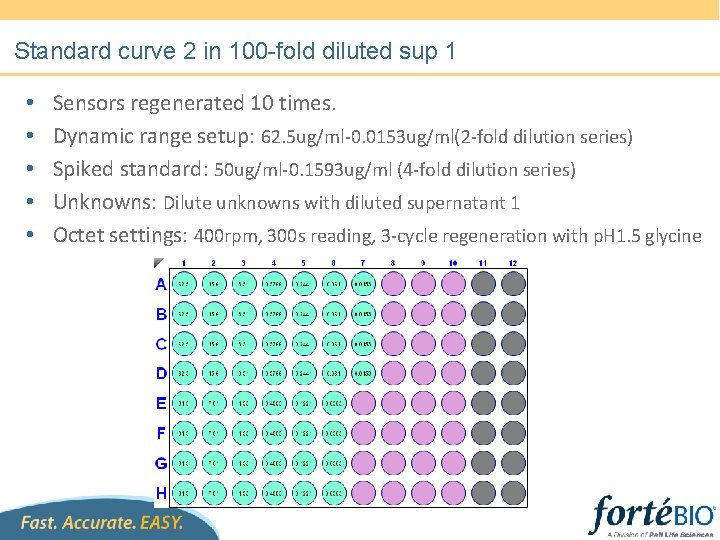
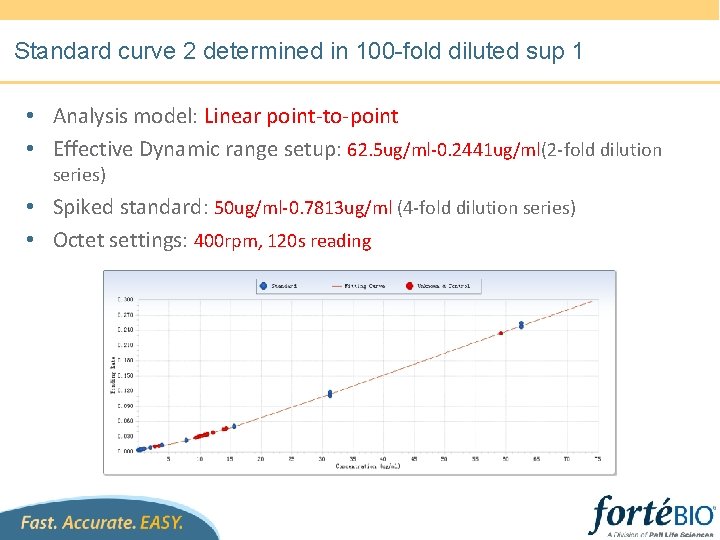
Standard curve 2 in 100 -fold diluted sup 1 • • • Sensors regenerated 10 times. Dynamic range setup: 62. 5 ug/ml-0. 0153 ug/ml(2 -fold dilution series) Spiked standard: 50 ug/ml-0. 1593 ug/ml (4 -fold dilution series) Unknowns: Dilute unknowns with diluted supernatant 1 Octet settings: 400 rpm, 300 s reading, 3 -cycle regeneration with p. H 1. 5 glycine

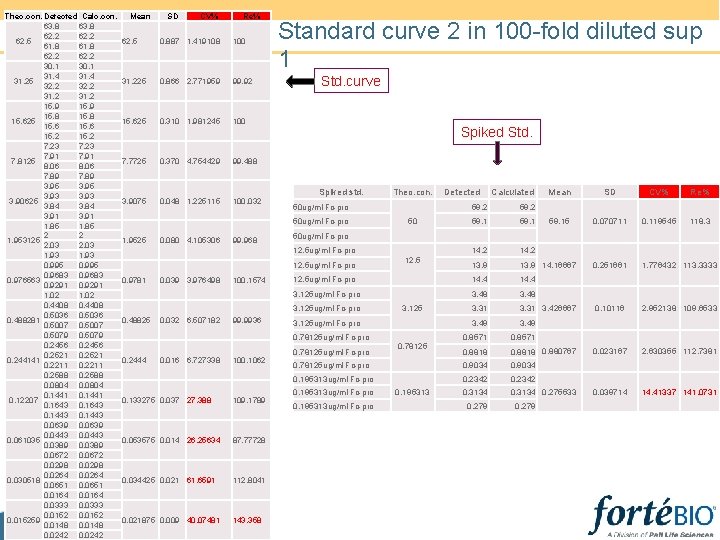
Standard curve 2 determined in 100 -fold diluted sup 1 • Analysis model: Linear point-to-point • Effective Dynamic range setup: 62. 5 ug/ml-0. 2441 ug/ml(2 -fold dilution series) • Spiked standard: 50 ug/ml-0. 7813 ug/ml (4 -fold dilution series) • Octet settings: 400 rpm, 120 s reading

Theo. con. Detected Calc. con. 63. 8 62. 2 62. 5 61. 8 62. 2 30. 1 31. 4 31. 25 32. 2 31. 2 15. 9 15. 8 15. 625 15. 6 15. 2 7. 23 7. 91 7. 8125 8. 06 7. 89 3. 95 3. 93 3. 90625 3. 84 3. 91 1. 85 2 2 1. 953125 2. 03 1. 93 0. 995 0. 9683 0. 976563 0. 9291 1. 02 0. 4408 0. 5036 0. 488281 0. 5007 0. 5079 0. 2456 0. 2521 0. 244141 0. 2211 0. 2588 0. 0804 0. 1441 0. 12207 0. 1643 0. 1443 0. 0639 0. 0443 0. 061035 0. 0389 0. 0672 0. 0298 0. 0264 0. 030518 0. 0651 0. 0164 0. 0333 0. 015259 0. 0148 0. 0242 Mean SD CV% Re% 62. 5 0. 887 1. 419108 100 31. 225 0. 866 2. 771959 99. 92 15. 625 0. 310 1. 981245 100 7. 7725 0. 370 4. 754429 99. 488 3. 9075 0. 048 1. 225115 100. 032 Standard curve 2 in 100 -fold diluted sup 1 Std. curve Spiked Std. Spiked std. 50 ug/ml Fc-pro 0. 080 4. 105306 99. 968 100. 1574 99. 9936 0. 016 6. 727338 100. 1062 0. 133275 0. 037 27. 388 109. 1789 0. 053575 0. 014 26. 25634 87. 77728 0. 034425 0. 021 61. 6591 112. 8041 0. 021875 0. 009 40. 07481 143. 358 Re% 59. 15 0. 070711 0. 119545 118. 3 0. 251661 1. 776432 113. 3333 0. 10116 2. 952138 109. 6533 0. 023167 2. 630355 112. 7381 0. 039714 14. 41337 141. 0731 14. 2 13. 9 14. 16667 14. 4 3. 48 3. 31 3. 426667 3. 49 0. 8571 0. 8818 0. 880767 0. 78125 ug/ml Fc-pro 0. 9034 0. 195313 ug/ml Fc-pro 0. 2342 0. 3134 0. 275533 12. 5 12. 5 ug/ml Fc-pro 3. 125 ug/ml Fc-pro 0. 78125 ug/ml Fc-pro 0. 2444 CV% 59. 1 3. 125 ug/ml Fc-pro 0. 032 6. 507182 SD 59. 1 50 3. 125 ug/ml Fc-pro 0. 48825 Mean 50 ug/ml Fc-pro 12. 5 ug/ml Fc-pro 0. 039 3. 976498 Calculated 59. 2 12. 5 ug/ml Fc-pro 0. 9781 Detected 59. 2 50 ug/ml Fc-pro 1. 9525 Theo. con. 0. 78125 ug/ml Fc-pro 0. 195313 ug/ml Fc-pro 0. 78125 0. 195313 0. 279

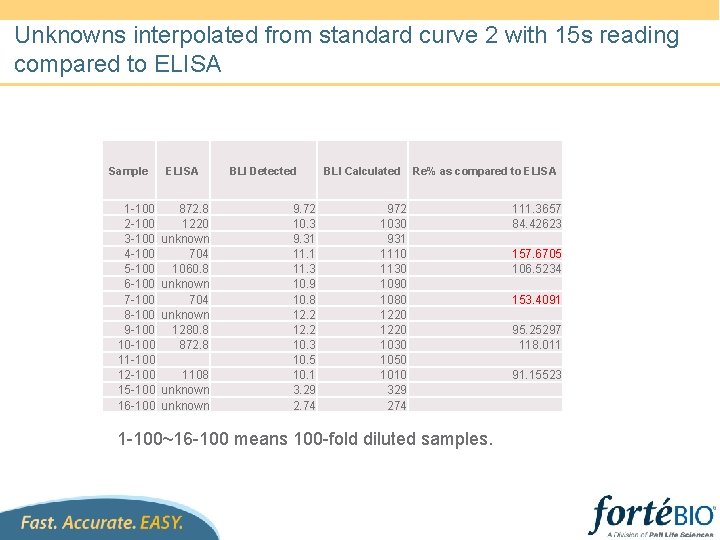
Unknowns interpolated from standard curve 2 with 15 s reading compared to ELISA Sample 1 -100 2 -100 3 -100 4 -100 5 -100 6 -100 7 -100 8 -100 9 -100 10 -100 11 -100 12 -100 15 -100 16 -100 ELISA 872. 8 1220 unknown 704 1060. 8 unknown 704 unknown 1280. 8 872. 8 1108 unknown BLI Detected 9. 72 10. 3 9. 31 11. 3 10. 9 10. 8 12. 2 10. 3 10. 5 10. 1 3. 29 2. 74 BLI Calculated Re% as compared to ELISA 972 1030 931 1110 1130 1090 1080 1220 1030 1050 1010 329 274 1 -100~16 -100 means 100 -fold diluted samples. 111. 3657 84. 42623 157. 6705 106. 5234 153. 4091 95. 25297 118. 011 91. 15523

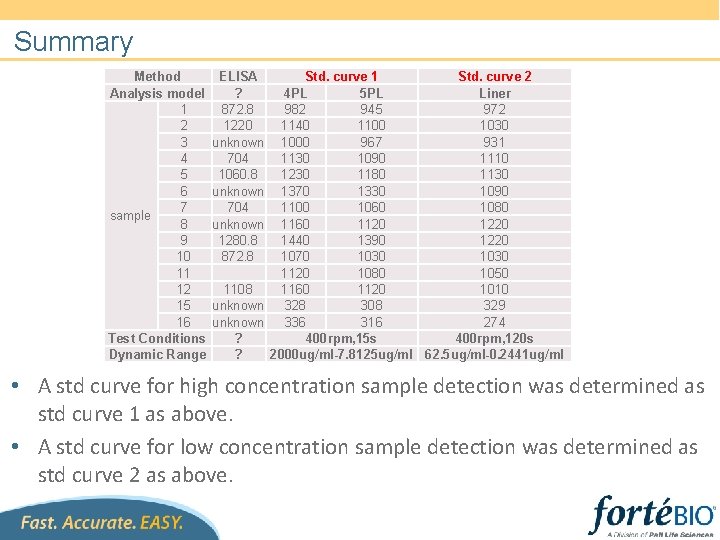
Summary Method Analysis model 1 2 3 4 5 6 7 sample 8 9 10 11 12 15 16 Test Conditions Dynamic Range ELISA Std. curve 1 Std. curve 2 ? 4 PL 5 PL Liner 872. 8 982 945 972 1220 1140 1100 1030 unknown 1000 967 931 704 1130 1090 1110 1060. 8 1230 1180 1130 unknown 1370 1330 1090 704 1100 1060 1080 unknown 1160 1120 1280. 8 1440 1390 1220 872. 8 1070 1030 1120 1080 1050 1108 1160 1120 1010 unknown 328 308 329 unknown 336 316 274 ? 400 rpm, 15 s 400 rpm, 120 s ? 2000 ug/ml-7. 8125 ug/ml 62. 5 ug/ml-0. 2441 ug/ml • A std curve for high concentration sample detection was determined as std curve 1 as above. • A std curve for low concentration sample detection was determined as std curve 2 as above.

Conclusion • A dilution factor of 100 for sup 1(samples in sup 1) was determined using PBS as control due to severe matrix effect of blank sup 1 as well as fresh medium. • A std curve with dynamic range 2000 ug/ml-7. 8125 ug/ml (4 -fold dilution series, unweighted 4 PL/5 PL analysis) was developed for high concentration samples under 400 rpm with 15 s reading, and samples were interpolated. • More sensitive method was developed under 400 rpm with 120 s 300 s reading and the dynamic range was 62. 5 ug/ml-0. 2441 ug/ml(2 fold dilution series, Linear point-to-point analysis). • Sensors could be regenerated well in 10 m. M p. H 1. 5 glycine buffer.

Crude sample detection: quantitation Case 1 l Object: quantitate pro in supernatant Solution : Pro A sensor with regeneration steps. (1000 rpm, Read. Time 120 s) u l Matrix: supernatant from CHO cells without centrifuge l Outcome: good data.

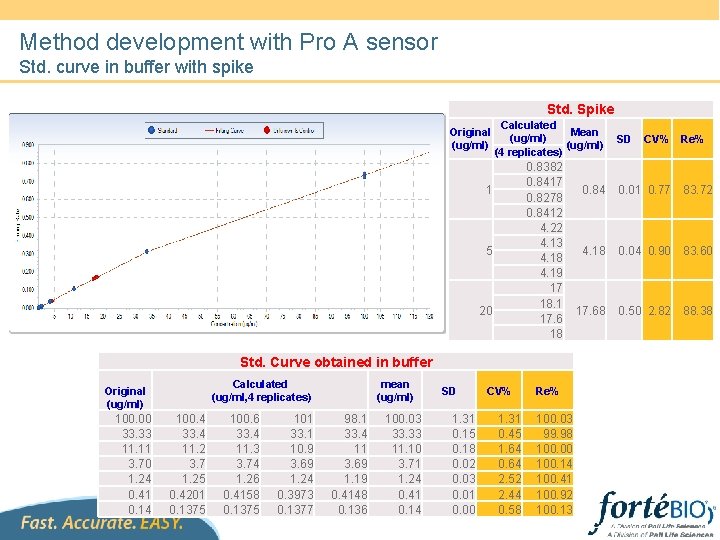
Method development with Pro A sensor Std. curve in buffer with spike Std. Spike Original (ug/ml) Calculated Mean (ug/ml) (4 replicates) 0. 8382 0. 8417 0. 8278 0. 8412 4. 22 4. 13 4. 18 4. 19 17 18. 1 17. 6 18 1 5 20 Std. Curve obtained in buffer Calculated (ug/ml, 4 replicates) Original (ug/ml) 100. 00 33. 33 11. 11 3. 70 1. 24 0. 41 0. 14 100. 4 33. 4 11. 2 3. 7 1. 25 0. 4201 0. 1375 100. 6 33. 4 11. 3 3. 74 1. 26 0. 4158 0. 1375 101 33. 1 10. 9 3. 69 1. 24 0. 3973 0. 1377 mean (ug/ml) 98. 1 33. 4 11 3. 69 1. 19 0. 4148 0. 136 100. 03 33. 33 11. 10 3. 71 1. 24 0. 41 0. 14 SD 1. 31 0. 15 0. 18 0. 02 0. 03 0. 01 0. 00 CV% 1. 31 0. 45 1. 64 0. 64 2. 52 2. 44 0. 58 Re% 100. 03 99. 98 100. 00 100. 14 100. 41 100. 92 100. 13 SD CV% Re% 0. 84 0. 01 0. 77 83. 72 4. 18 0. 04 0. 90 83. 60 17. 68 0. 50 2. 82 88. 38

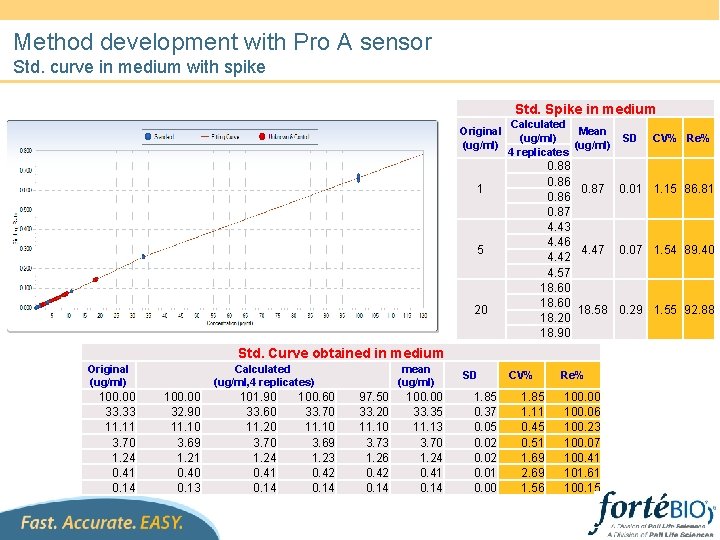
Method development with Pro A sensor Std. curve in medium with spike Std. Spike in medium Original (ug/ml) Calculated Mean (ug/ml) 4 replicates 5 20 Std. Curve obtained in medium Calculated (ug/ml, 4 replicates) 100. 00 33. 33 11. 11 3. 70 1. 24 0. 41 0. 14 100. 00 32. 90 11. 10 3. 69 1. 21 0. 40 0. 13 101. 90 33. 60 11. 20 3. 70 1. 24 0. 41 0. 14 100. 60 33. 70 11. 10 3. 69 1. 23 0. 42 0. 14 mean (ug/ml) 97. 50 33. 20 11. 10 3. 73 1. 26 0. 42 0. 14 100. 00 33. 35 11. 13 3. 70 1. 24 0. 41 0. 14 SD 1. 85 0. 37 0. 05 0. 02 0. 01 0. 00 CV% Re% 0. 88 0. 86 0. 87 0. 01 1. 15 86. 81 0. 86 0. 87 4. 43 4. 46 4. 47 0. 07 1. 54 89. 40 4. 42 4. 57 18. 60 18. 58 0. 29 1. 55 92. 88 18. 20 18. 90 1 Original (ug/ml) SD CV% 1. 85 1. 11 0. 45 0. 51 1. 69 2. 69 1. 56 Re% 100. 00 100. 06 100. 23 100. 07 100. 41 101. 61 100. 15

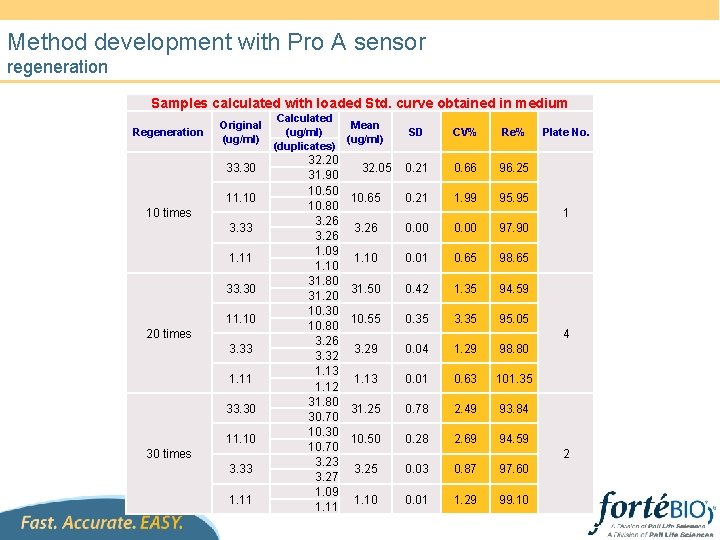
Method development with Pro A sensor regeneration Samples calculated with loaded Std. curve obtained in medium Regeneration Original (ug/ml) 33. 30 11. 10 10 times 3. 33 1. 11 33. 30 11. 10 20 times 3. 33 1. 11 33. 30 11. 10 30 times 3. 33 1. 11 Calculated (ug/ml) (duplicates) 32. 20 31. 90 10. 50 10. 80 3. 26 1. 09 1. 10 31. 80 31. 20 10. 30 10. 80 3. 26 3. 32 1. 13 1. 12 31. 80 30. 70 10. 30 10. 70 3. 23 3. 27 1. 09 1. 11 Mean (ug/ml) 32. 05 10. 65 SD CV% Re% 0. 21 0. 66 96. 25 0. 21 1. 99 95. 95 Plate No. 1 3. 26 0. 00 97. 90 1. 10 0. 01 0. 65 98. 65 31. 50 0. 42 1. 35 94. 59 10. 55 0. 35 3. 35 95. 05 4 3. 29 0. 04 1. 29 98. 80 1. 13 0. 01 0. 63 101. 35 31. 25 0. 78 2. 49 93. 84 10. 50 0. 28 2. 69 94. 59 2 3. 25 0. 03 0. 87 97. 60 1. 10 0. 01 1. 29 99. 10

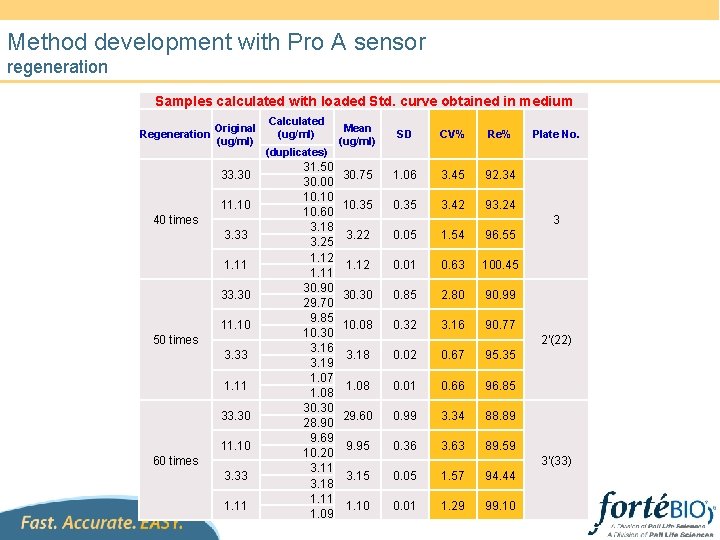
Method development with Pro A sensor regeneration Samples calculated with loaded Std. curve obtained in medium Regeneration Original (ug/ml) 33. 30 11. 10 40 times 3. 33 1. 11 33. 30 11. 10 50 times 3. 33 1. 11 33. 30 11. 10 60 times 3. 33 1. 11 Calculated (ug/ml) (duplicates) 31. 50 30. 00 10. 10 10. 60 3. 18 3. 25 1. 12 1. 11 30. 90 29. 70 9. 85 10. 30 3. 16 3. 19 1. 07 1. 08 30. 30 28. 90 9. 69 10. 20 3. 11 3. 18 1. 11 1. 09 Mean (ug/ml) SD CV% Re% 30. 75 1. 06 3. 45 92. 34 10. 35 3. 42 93. 24 Plate No. 3 3. 22 0. 05 1. 54 96. 55 1. 12 0. 01 0. 63 100. 45 30. 30 0. 85 2. 80 90. 99 10. 08 0. 32 3. 16 90. 77 3. 18 0. 02 0. 67 95. 35 1. 08 0. 01 0. 66 96. 85 29. 60 0. 99 3. 34 88. 89 9. 95 0. 36 3. 63 89. 59 3. 15 0. 05 1. 57 94. 44 1. 10 0. 01 1. 29 99. 10 2'(22) 3'(33)

Crude sample detection: quantitation Case 2 l Object: quantitate pro X in milk u Solution : AHC with regeneration steps. l Matrix: 100 fold dilution milk l Outcome: good data.

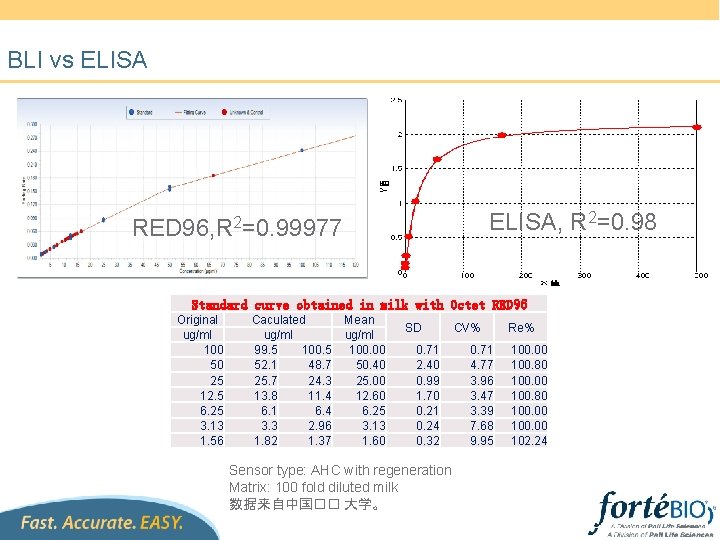
BLI vs ELISA, R 2=0. 98 RED 96, R 2=0. 99977 Standard curve obtained in milk with Octet RED 96 Original ug/ml 100 50 25 12. 5 6. 25 3. 13 1. 56 Caculated ug/ml 99. 5 100. 5 52. 1 48. 7 25. 7 24. 3 13. 8 11. 4 6. 1 6. 4 3. 3 2. 96 1. 82 1. 37 Mean ug/ml 100. 00 50. 40 25. 00 12. 60 6. 25 3. 13 1. 60 SD 0. 71 2. 40 0. 99 1. 70 0. 21 0. 24 0. 32 Sensor type: AHC with regeneration Matrix: 100 fold diluted milk 数据来自中国�� 大学。 CV% 0. 71 4. 77 3. 96 3. 47 3. 39 7. 68 9. 95 Re% 100. 00 100. 80 100. 00 102. 24

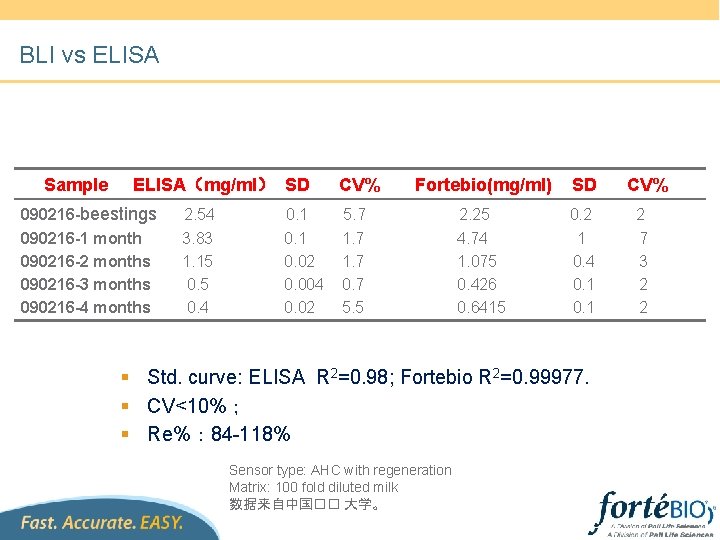
BLI vs ELISA Sample ELISA(mg/ml) SD 090216 -beestings 090216 -1 month 090216 -2 months 090216 -3 months 090216 -4 months 2. 54 3. 83 1. 15 0. 4 0. 1 0. 02 0. 004 0. 02 CV% Fortebio(mg/ml) SD 2. 25 4. 74 1. 075 0. 426 0. 6415 0. 2 1 0. 4 0. 1 5. 7 1. 7 0. 7 5. 5 § Std. curve: ELISA R 2=0. 98; Fortebio R 2=0. 99977. § CV<10%; § Re%: 84 -118% Sensor type: AHC with regeneration Matrix: 100 fold diluted milk 数据来自中国�� 大学。 CV% 2 7 3 2 2

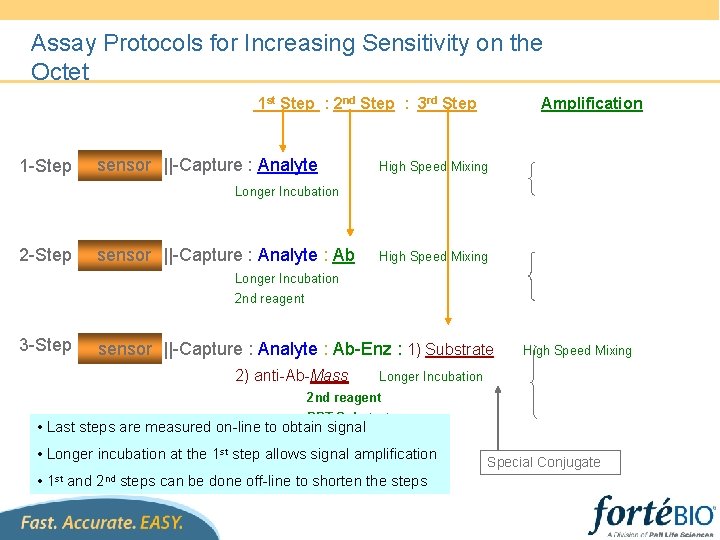
Assay Protocols for Increasing Sensitivity on the Octet 1 st Step : 2 nd Step : 3 rd Step 1 -Step sensor ||-Capture : Analyte Amplification High Speed Mixing Longer Incubation 2 -Step sensor ||-Capture : Analyte : Ab High Speed Mixing Longer Incubation 2 nd reagent 3 -Step sensor ||-Capture : Analyte : Ab-Enz : 1) Substrate 2) anti-Ab-Mass High Speed Mixing Longer Incubation 2 nd reagent PPT Substrate • Last steps are measured on-line to obtain signal • Longer incubation at the 1 st step allows signal amplification • 1 st and 2 nd steps can be done off-line to shorten the steps Special Conjugate

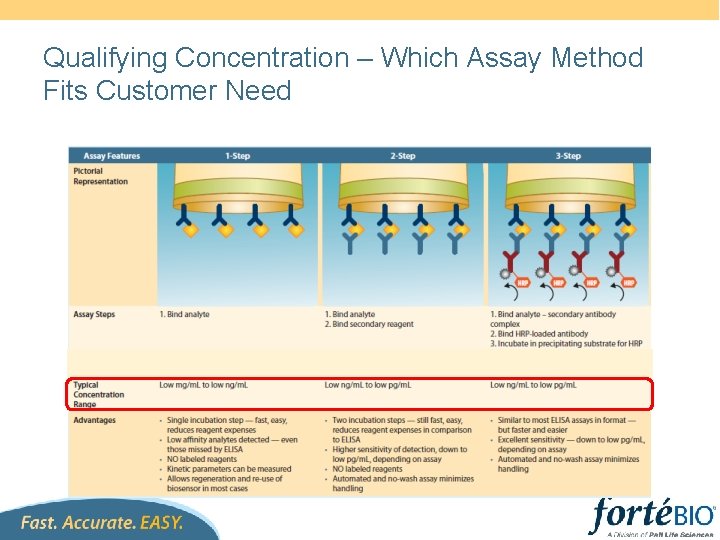
Qualifying Concentration – Which Assay Method Fits Customer Need

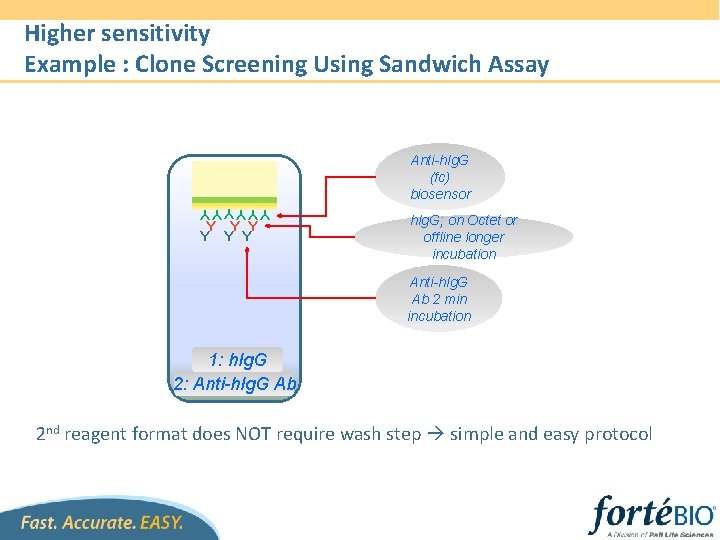
Higher sensitivity Example : Clone Screening Using Sandwich Assay Anti-h. Ig. G (fc) biosensor YYYYYY Y Y Y h. Ig. G; on Octet or offline longer incubation Anti-h. Ig. G Ab 2 min incubation 1: h. Ig. G 2: Anti-h. Ig. G Ab 2 nd reagent format does NOT require wash step simple and easy protocol

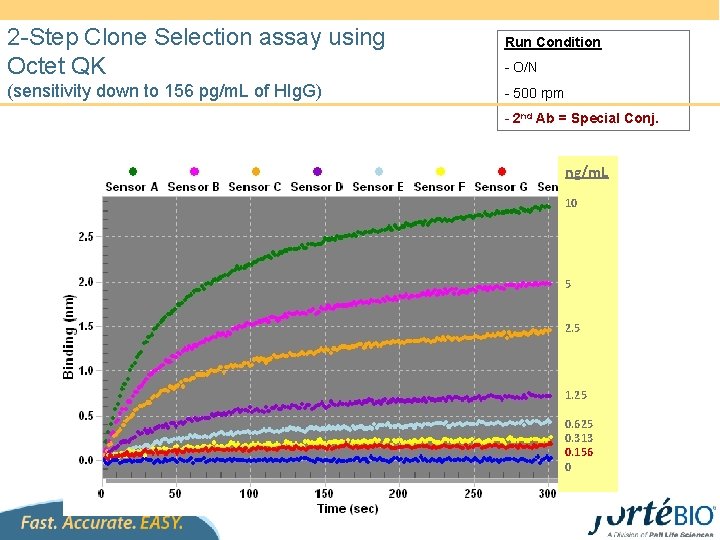
2 -Step Clone Selection assay using Octet QK (sensitivity down to 156 pg/m. L of HIg. G) Run Condition - O/N - 500 rpm - 2 nd Ab = Special Conj. ng/m. L 10 5 2. 5 1. 25 0. 625 0. 313 0. 156 0

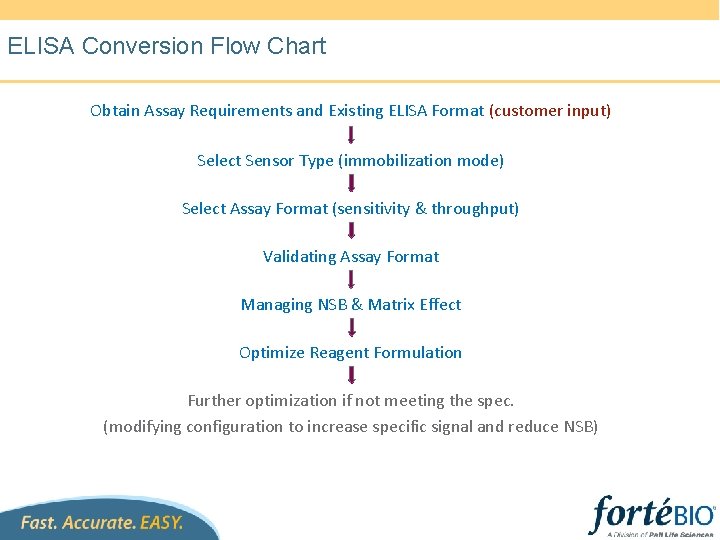
ELISA Conversion Flow Chart Obtain Assay Requirements and Existing ELISA Format (customer input) Select Sensor Type (immobilization mode) Select Assay Format (sensitivity & throughput) Validating Assay Format Managing NSB & Matrix Effect Optimize Reagent Formulation Further optimization if not meeting the spec. (modifying configuration to increase specific signal and reduce NSB)

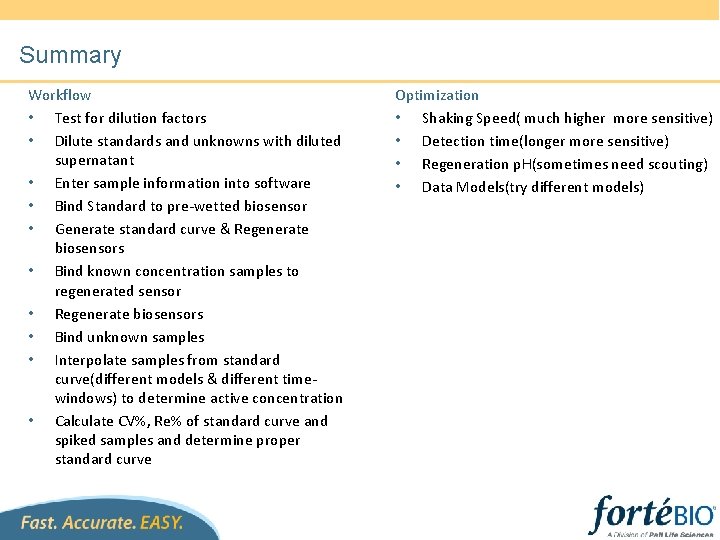
Summary Workflow • Test for dilution factors • Dilute standards and unknowns with diluted supernatant • Enter sample information into software • Bind Standard to pre-wetted biosensor • Generate standard curve & Regenerate biosensors • Bind known concentration samples to regenerated sensor • Regenerate biosensors • Bind unknown samples • Interpolate samples from standard curve(different models & different timewindows) to determine active concentration • Calculate CV%, Re% of standard curve and spiked samples and determine proper standard curve Optimization • Shaking Speed( much higher more sensitive) • Detection time(longer more sensitive) • Regeneration p. H(sometimes need scouting) • Data Models(try different models)

Pall Forte. Bio解决方案 u Label-free u Real time u Fluidics-free u Fast,Accurate,Easy. www. fortebio. com
 Octet quantitation protocol
Octet quantitation protocol Quantitation
Quantitation Hamlet act iii scene ii
Hamlet act iii scene ii Va handbook 5017 employee recognition and awards
Va handbook 5017 employee recognition and awards Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Voi kéo gỗ như thế nào
Voi kéo gỗ như thế nào Tư thế worm breton
Tư thế worm breton Bài hát chúa yêu trần thế alleluia
Bài hát chúa yêu trần thế alleluia Môn thể thao bắt đầu bằng từ chạy
Môn thể thao bắt đầu bằng từ chạy Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Công thức tính thế năng
Công thức tính thế năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân Phép trừ bù
Phép trừ bù Phản ứng thế ankan
Phản ứng thế ankan Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Thể thơ truyền thống
Thể thơ truyền thống Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Một số thể thơ truyền thống
Một số thể thơ truyền thống Bàn tay mà dây bẩn
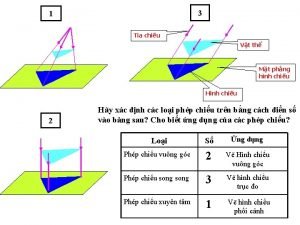
Bàn tay mà dây bẩn Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau Thế nào là sự mỏi cơ
Thế nào là sự mỏi cơ đặc điểm cơ thể của người tối cổ
đặc điểm cơ thể của người tối cổ Ví dụ giọng cùng tên
Ví dụ giọng cùng tên Vẽ hình chiếu đứng bằng cạnh của vật thể
Vẽ hình chiếu đứng bằng cạnh của vật thể Vẽ hình chiếu vuông góc của vật thể sau
Vẽ hình chiếu vuông góc của vật thể sau Thẻ vin
Thẻ vin đại từ thay thế
đại từ thay thế điện thế nghỉ
điện thế nghỉ Tư thế ngồi viết
Tư thế ngồi viết Diễn thế sinh thái là
Diễn thế sinh thái là Dạng đột biến một nhiễm là
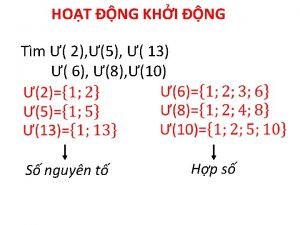
Dạng đột biến một nhiễm là Bảng số nguyên tố
Bảng số nguyên tố Tư thế ngồi viết
Tư thế ngồi viết Lời thề hippocrates
Lời thề hippocrates Thiếu nhi thế giới liên hoan
Thiếu nhi thế giới liên hoan ưu thế lai là gì
ưu thế lai là gì