Regulation of Gene Expression Prokaryotic Gene expression producing



- Slides: 14

Regulation of Gene Expression: Prokaryotic • Gene expression, producing a protein from a gene, is complex • Lets start in bacteria • We will go into to detail here because the details of eukaryotic pathways are a bit over our heads at present • Viruses use similar regulation mechanisms

Gene Regulation in Bacteria and Viruses • Regulatory sequences – stretches of DNA that interact with regulatory proteins to control transcription • Regulatory genes produce regulatory proteins (or RNAs), whose function is to regulate the transcription of other genes • Both positive and negative control mechanisms regulate gene expression – Regulatory proteins can turn on, turn off, amplify or damp down gene expression


Inducers and Repressors • Trp is the repressor of the trp expression • Negative control - Repressors are small molecules that interact with regulatory proteins and/or sequences to repress transcription – they are not necessarily the end product of the pathway – Regulatory proteins inhibit gene expression by binding DNA and blocking transcription • Positive control - Inducers are any small molecule that interact with regulatory proteins and/or sequences to induce transcription – Regulatory proteins stimulate gene expression by binding DNA and stimulating transcription or binding to repressors to inactivate them. • Certain genes are continuously expressed; that is, they are always turned “on, ” e. g. the ribosomal genes.

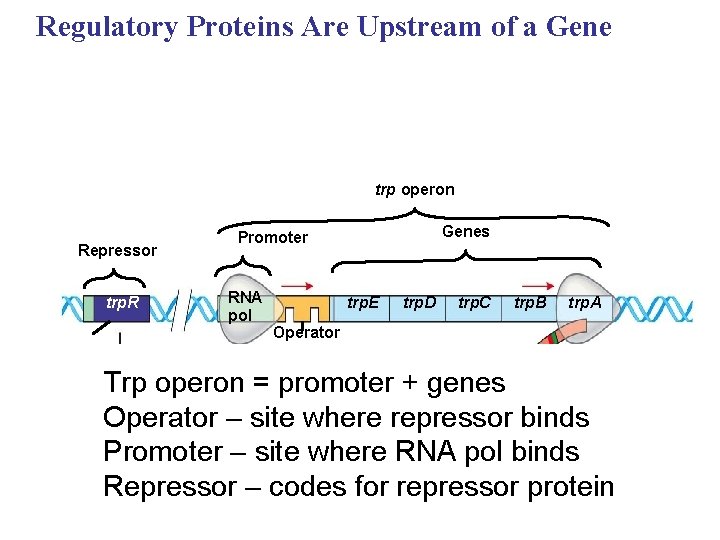
Regulatory Proteins Are Upstream of a Gene trp operon Repressor trp. R Genes Promoter RNA pol trp. E trp. D trp. C trp. B trp. A Operator Trp operon = promoter + genes Operator – site where repressor binds Promoter – site where RNA pol binds Repressor – codes for repressor protein

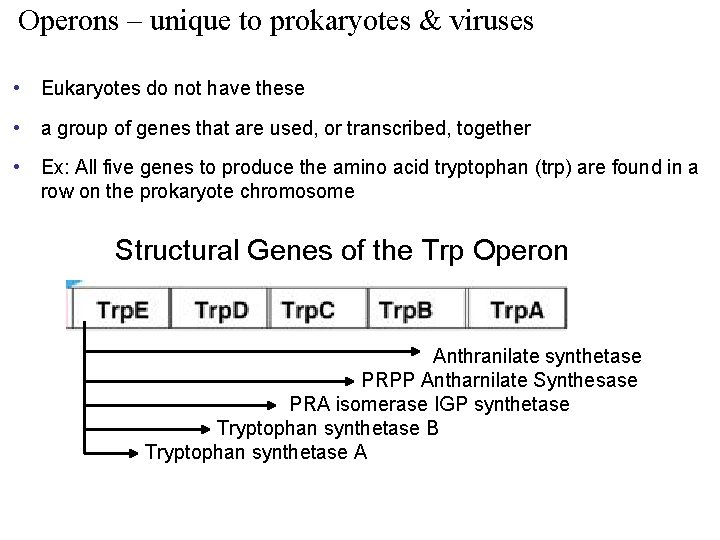
Operons – unique to prokaryotes & viruses • Eukaryotes do not have these • a group of genes that are used, or transcribed, together • Ex: All five genes to produce the amino acid tryptophan (trp) are found in a row on the prokaryote chromosome Structural Genes of the Trp Operon Anthranilate synthetase PRPP Antharnilate Synthesase PRA isomerase IGP synthetase Tryptophan synthetase B Tryptophan synthetase A

Ex: E. coli tryptophan synthesis • default - trp operon is on, genes to make tryptophan are transcribed • This makes sense because the cell is constantly making protein and needs amino acids • Sometimes, however, the cell makes more trp than it needs • Excess tryptophan binds and activates the trp repressor (trp. R) • This makes Tryptophan a corepressor https: //www. youtube. com/watch? v=10 YWgqm. AEs. Q Bozeman Science – operons in prokaryotes

Tryptophan absent, repressor inactive, operon on trp operon Repressor RNA pol trp. R Genes Promoter Operator trp. E trp. D trp. C trp. B trp. A B A m. RNA 3 m. RNA 5 5 E Protein Inactive repressor D C enzymes for tryptophan synthesis Operon is transcribed as one unit, but each gene is translated separately on a different ribosome. More later. Fig. 18 -3 a

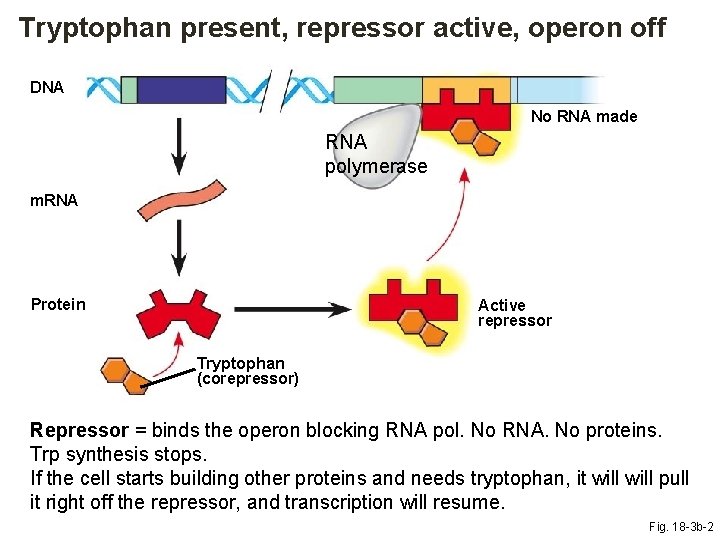
Tryptophan present, repressor active, operon off DNA No RNA made RNA polymerase m. RNA Protein Active repressor Tryptophan (corepressor) Repressor = binds the operon blocking RNA pol. No RNA. No proteins. Trp synthesis stops. If the cell starts building other proteins and needs tryptophan, it will pull it right off the repressor, and transcription will resume. Fig. 18 -3 b-2

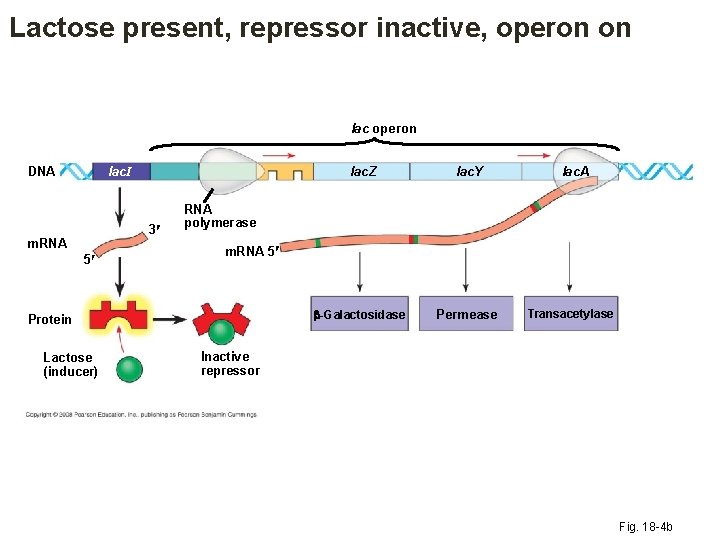
Types of Regulation • Repressible operon – default on; repressor is often an end product • anabolism, ex: trp operon • Inducible operon – default off; inducer inactivates repressor – catabolism, Ex: lac operon - codes enzymes for lactose hydrolysis – enables microbe to use lactose as an energy source – However, lactose is not the most common food source for most microbes.

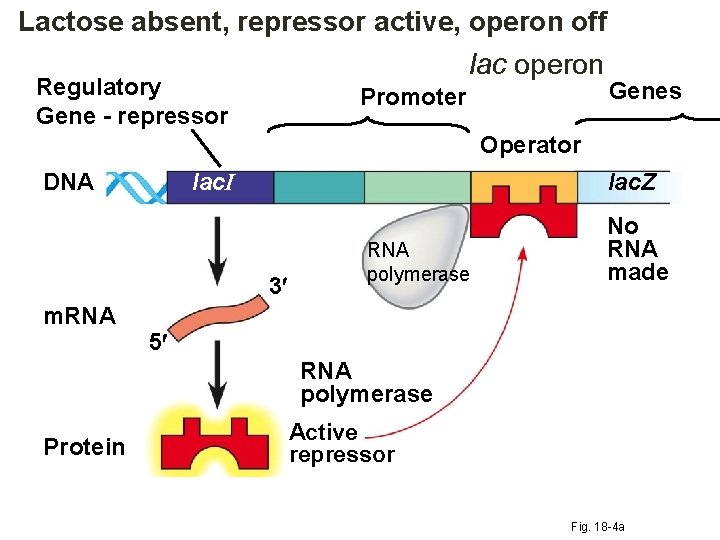
Lactose absent, repressor active, operon off lac operon Regulatory Gene - repressor Promoter Genes Operator lac. I DNA lac. Z 3 m. RNA polymerase No RNA made 5 RNA polymerase Protein Active repressor Fig. 18 -4 a

Lactose present, repressor inactive, operon on lac operon lac. I DNA 3 m. RNA 5 lac. Y -Galactosidase Permease lac. A RNA polymerase m. RNA 5 Protein Lactose (inducer) lac. Z Transacetylase Inactive repressor Fig. 18 -4 b

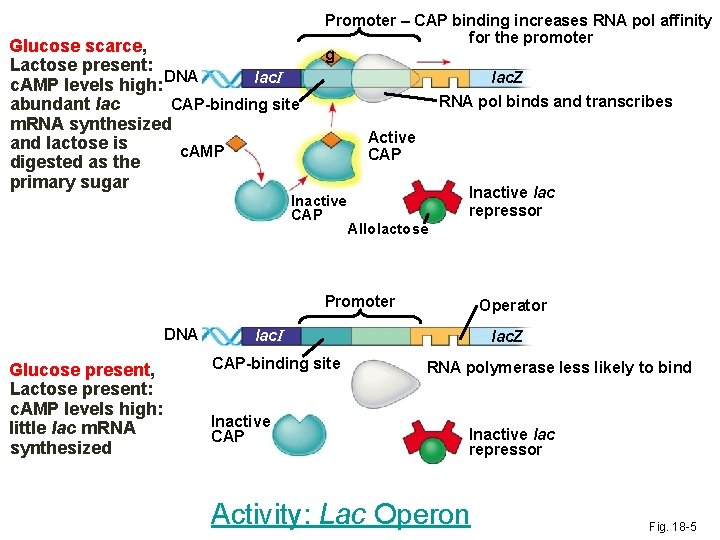
Positive Regulation • Glucose is the preferred food source for E. coli will use it if it is present. • When glucose is scarce, the catabolite activator protein (CAP), increases E. coli’s lactose metabolizism • cyclic AMP activates CAP which binds the lac promoter increasing its affinity for RNA pol, accelerating transcription • When glucose levels increase, CAP detaches from the lac operon and transcription returns to normal

Glucose scarce, Lactose present: lac. I c. AMP levels high: DNA CAP-binding site abundant lac m. RNA synthesized and lactose is c. AMP digested as the primary sugar Promoter – CAP binding increases RNA pol affinity for the promoter g lac. Z RNA pol binds and transcribes Active CAP Inactive lac repressor Allolactose Promoter DNA Glucose present, Lactose present: c. AMP levels high: little lac m. RNA synthesized Operator lac. I CAP-binding site Inactive CAP lac. Z RNA polymerase less likely to bind Inactive lac repressor Activity: Lac Operon Fig. 18 -5