Tutorial 7 Gene expression analysis 1 Gene expression











- Slides: 57

Tutorial 7 Gene expression analysis 1

Gene expression analysis • Expression data – GEO – UCSC – Array. Express • General clustering methods – Unsupervised Clustering • Hierarchical clustering • K-means clustering • Tools for clustering – EPCLUST – Mev • Functional analysis – Go annotation 2

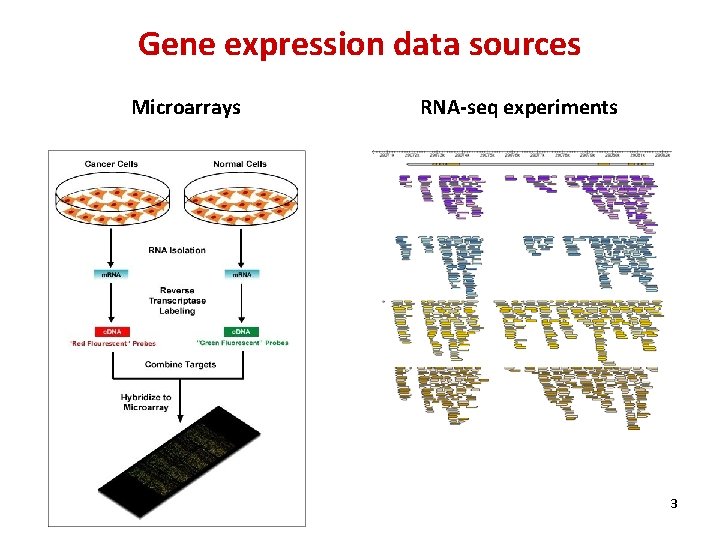
Gene expression data sources Microarrays RNA-seq experiments 3

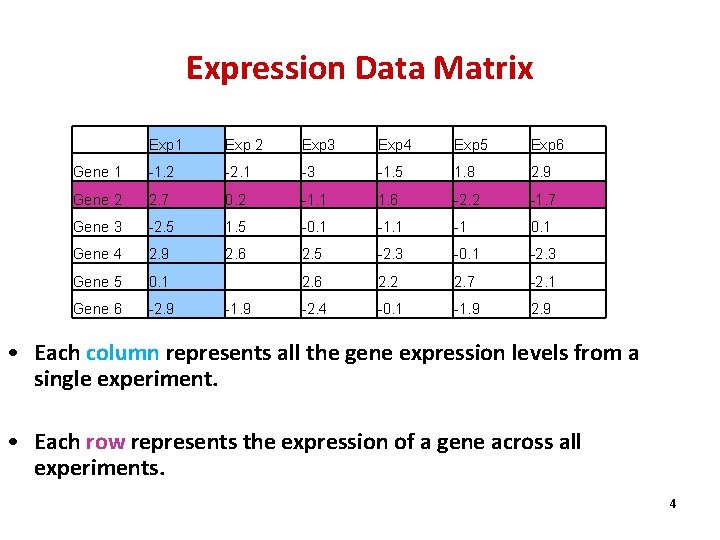
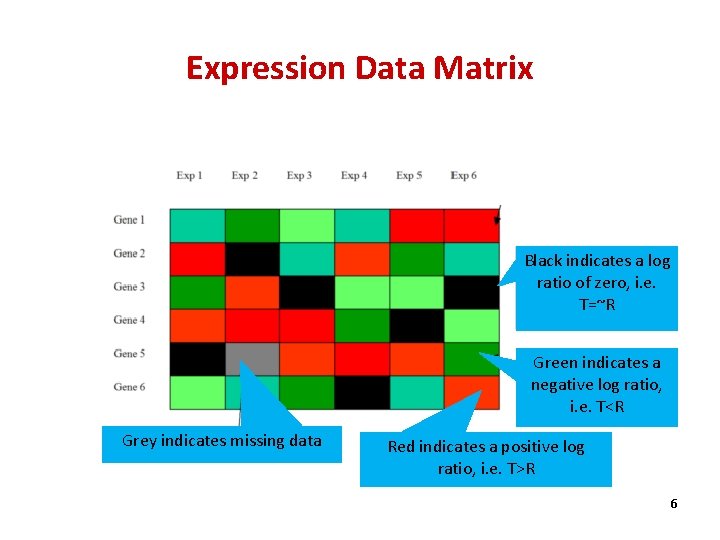
Expression Data Matrix Exp 1 Exp 2 Exp 3 Exp 4 Exp 5 Exp 6 Gene 1 -1. 2 -2. 1 -3 -1. 5 1. 8 2. 9 Gene 2 2. 7 0. 2 -1. 1 1. 6 -2. 2 -1. 7 Gene 3 -2. 5 1. 5 -0. 1 -1 0. 1 Gene 4 2. 9 2. 6 2. 5 -2. 3 -0. 1 -2. 3 Gene 5 0. 1 2. 6 2. 2 2. 7 -2. 1 Gene 6 -2. 9 -2. 4 -0. 1 -1. 9 2. 9 -1. 9 • Each column represents all the gene expression levels from a single experiment. • Each row represents the expression of a gene across all experiments. 4

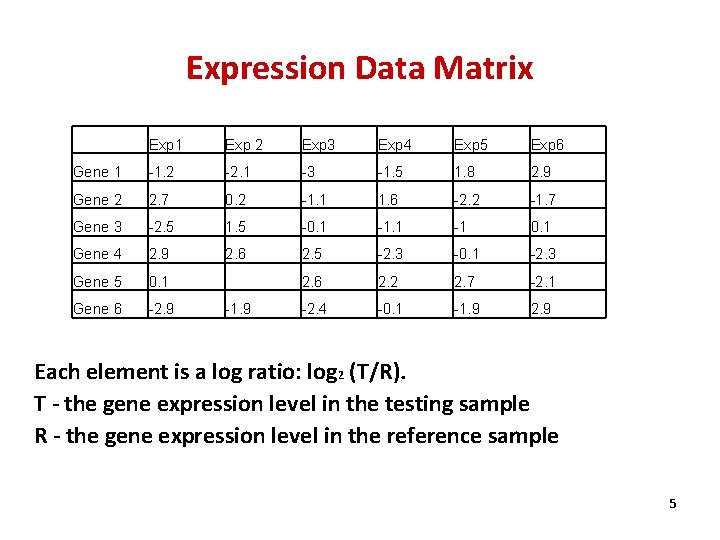
Expression Data Matrix Exp 1 Exp 2 Exp 3 Exp 4 Exp 5 Exp 6 Gene 1 -1. 2 -2. 1 -3 -1. 5 1. 8 2. 9 Gene 2 2. 7 0. 2 -1. 1 1. 6 -2. 2 -1. 7 Gene 3 -2. 5 1. 5 -0. 1 -1 0. 1 Gene 4 2. 9 2. 6 2. 5 -2. 3 -0. 1 -2. 3 Gene 5 0. 1 2. 6 2. 2 2. 7 -2. 1 Gene 6 -2. 9 -2. 4 -0. 1 -1. 9 2. 9 -1. 9 Each element is a log ratio: log 2 (T/R). T - the gene expression level in the testing sample R - the gene expression level in the reference sample 5

Expression Data Matrix Black indicates a log ratio of zero, i. e. T=~R Green indicates a negative log ratio, i. e. T<R Grey indicates missing data Red indicates a positive log ratio, i. e. T>R 6

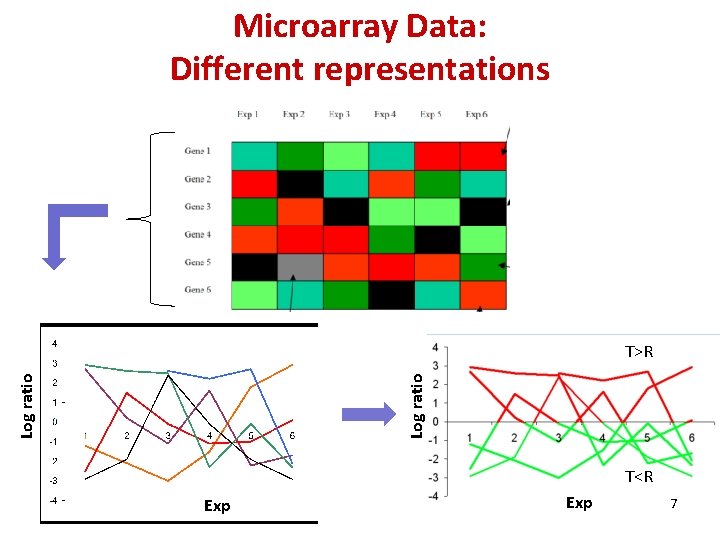
Microarray Data: Different representations Log ratio T>R T<R Exp 7

How to search for expression profiles • GEO (Gene Expression Omnibus) http: //www. ncbi. nlm. nih. gov/geo/ • Human genome browser http: //genome. ucsc. edu/ • Array. Express http: //www. ebi. ac. uk/arrayexpress/ 8

9

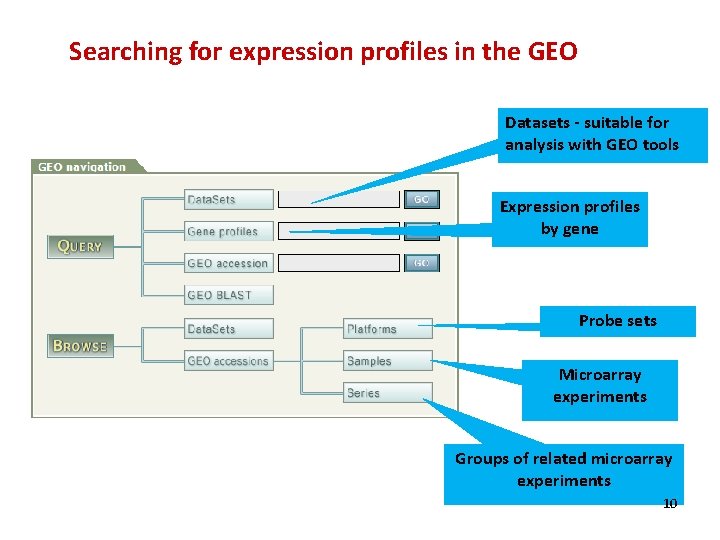
Searching for expression profiles in the GEO Datasets - suitable for analysis with GEO tools Expression profiles by gene Probe sets Microarray experiments Groups of related microarray experiments 10

Clustering Statistic analysis Download dataset 11

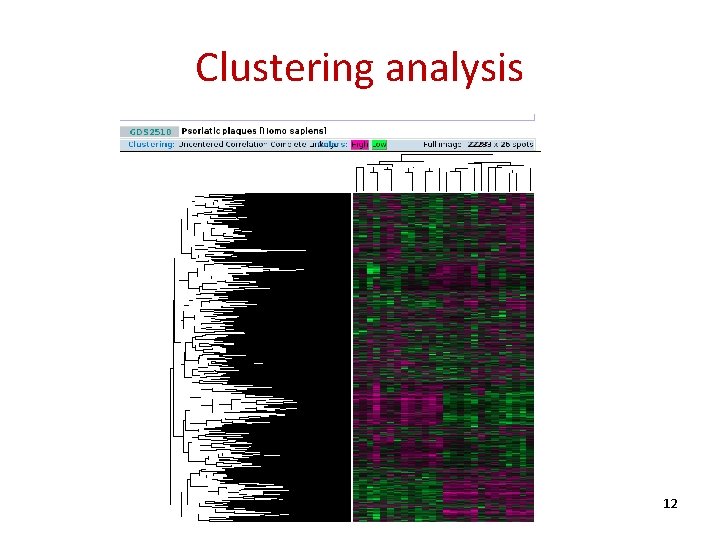
Clustering analysis 12

Clustering Statistic analysis Download dataset 13

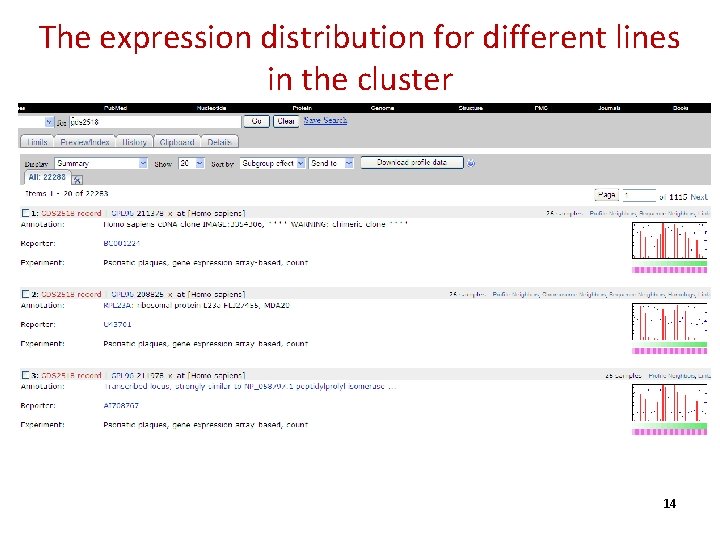
The expression distribution for different lines in the cluster 14

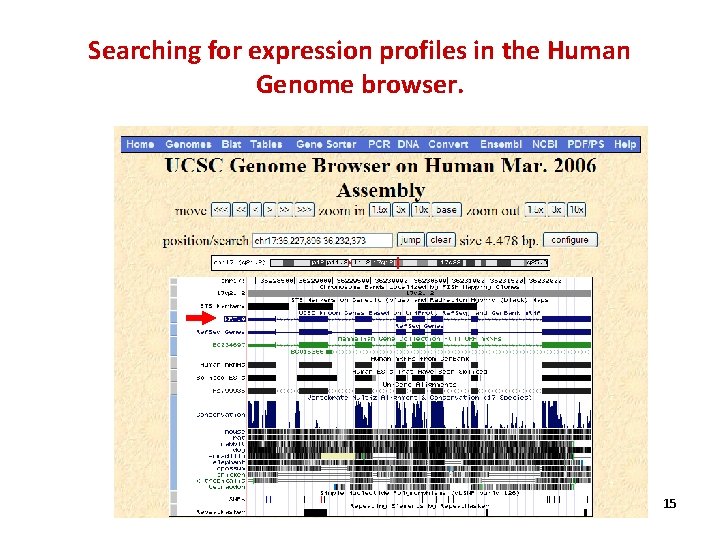
Searching for expression profiles in the Human Genome browser. 15

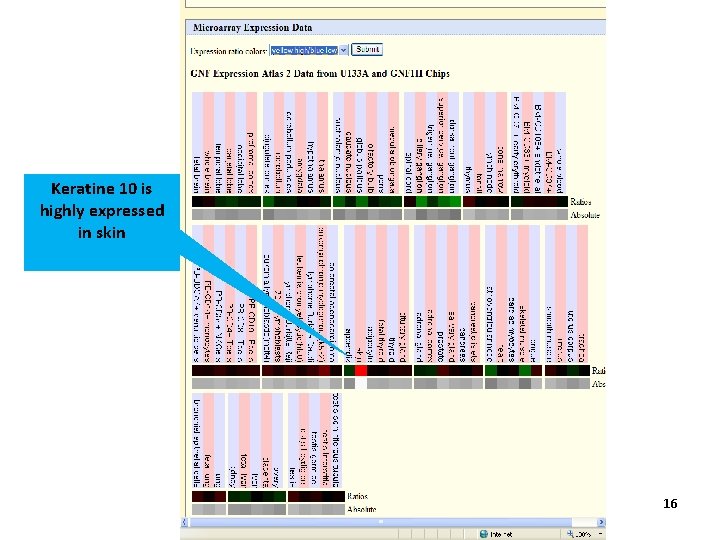
Keratine 10 is highly expressed in skin 16

Array. Express http: //www. ebi. ac. uk/arrayexpress/ 17

18

19

20

21

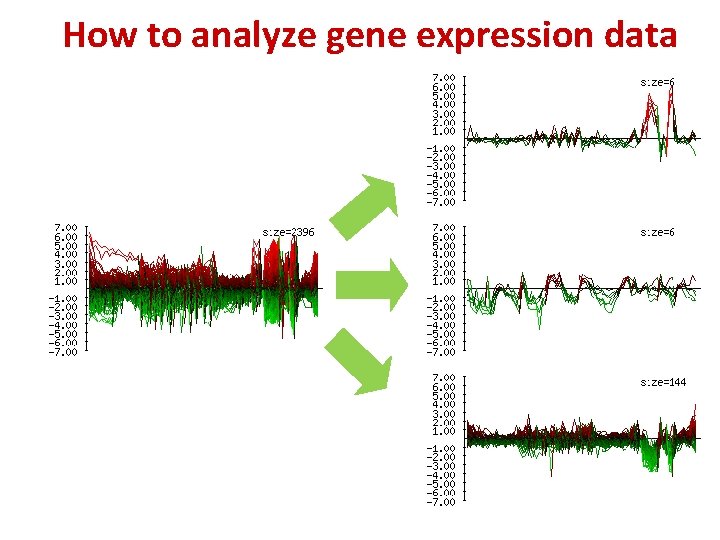
How to analyze gene expression data 22

Unsupervised Clustering - Hierarchical Clustering 23

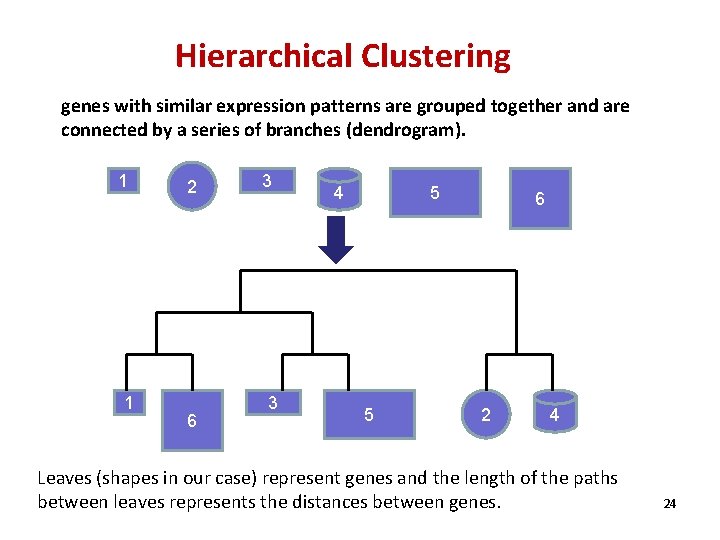
Hierarchical Clustering genes with similar expression patterns are grouped together and are connected by a series of branches (dendrogram). 1 1 2 6 3 3 5 4 5 6 2 4 Leaves (shapes in our case) represent genes and the length of the paths between leaves represents the distances between genes. 24

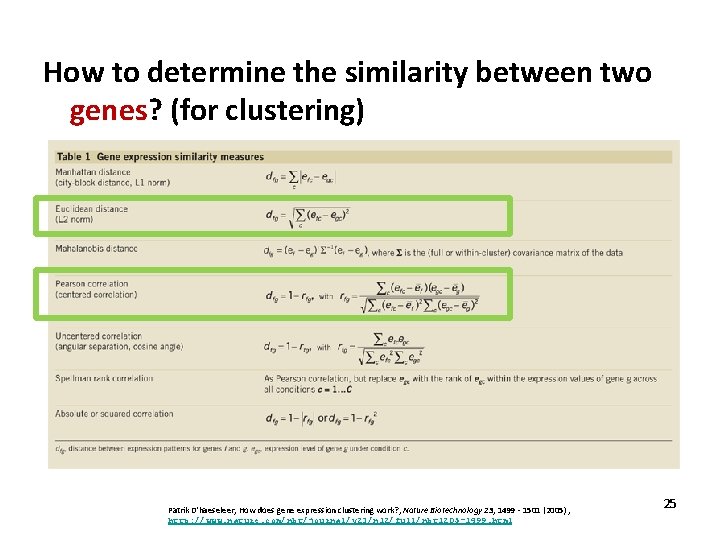
How to determine the similarity between two genes? (for clustering) Patrik D'haeseleer, How does gene expression clustering work? , Nature Biotechnology 23, 1499 - 1501 (2005) , http: //www. nature. com/nbt/journal/v 23/n 12/full/nbt 1205 -1499. html 25

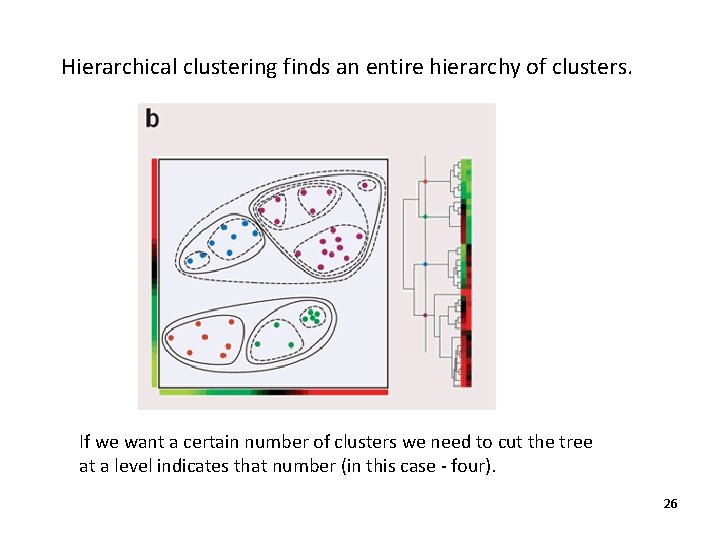
Hierarchical clustering finds an entire hierarchy of clusters. If we want a certain number of clusters we need to cut the tree at a level indicates that number (in this case - four). 26

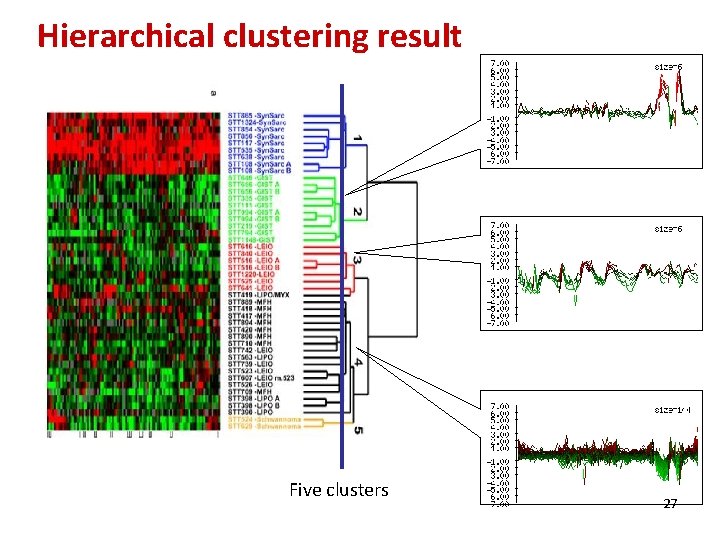
Hierarchical clustering result Five clusters 27

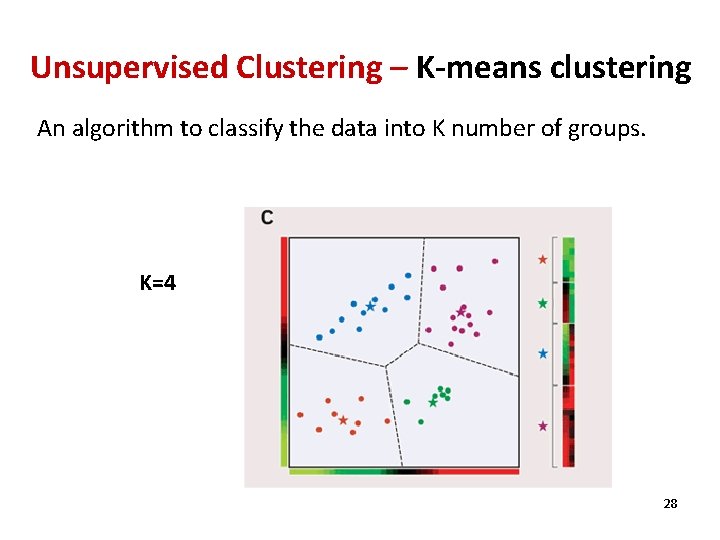
Unsupervised Clustering – K-means clustering An algorithm to classify the data into K number of groups. K=4 28

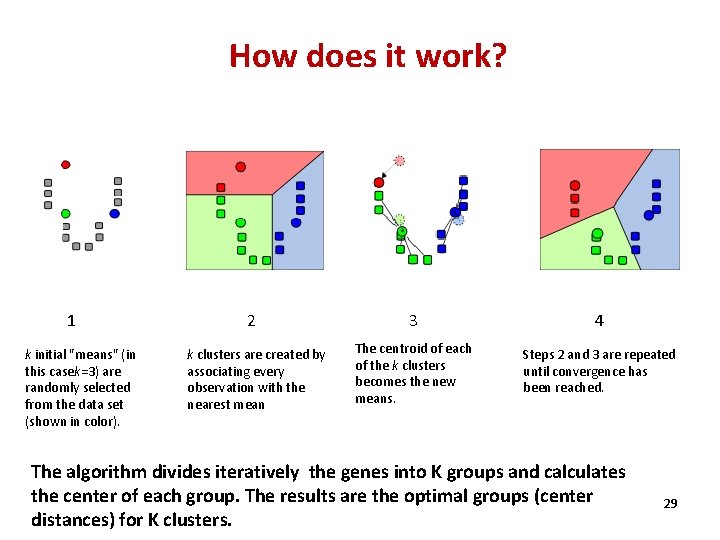
How does it work? 1 k initial "means" (in this casek=3) are randomly selected from the data set (shown in color). 2 k clusters are created by associating every observation with the nearest mean 3 4 The centroid of each of the k clusters becomes the new means. Steps 2 and 3 are repeated until convergence has been reached. The algorithm divides iteratively the genes into K groups and calculates the center of each group. The results are the optimal groups (center distances) for K clusters. 29

How should we determine K? • Trial and error • Take K as square root of gene number 30

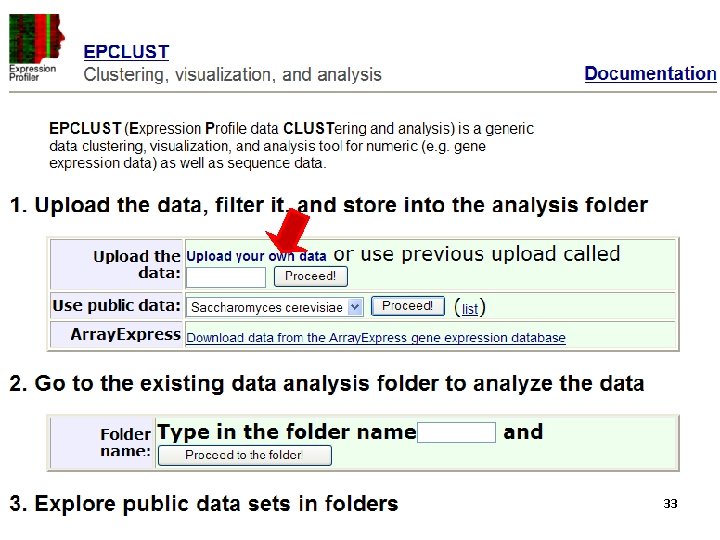
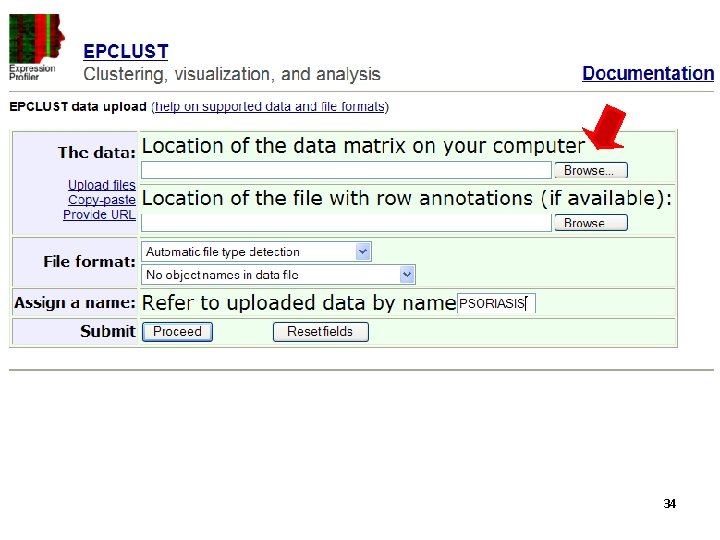
Tools for clustering - EPclust http: //www. bioinf. ebc. ee/EP/EP/EPCLUST/ 31

32

33

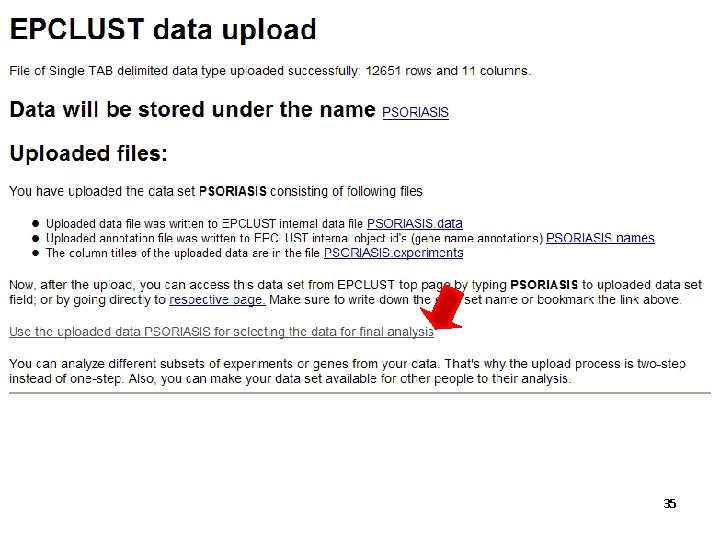
34

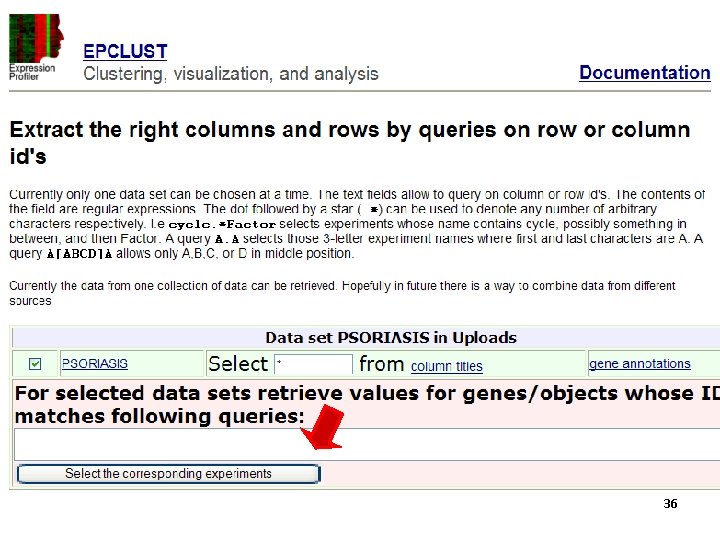
35

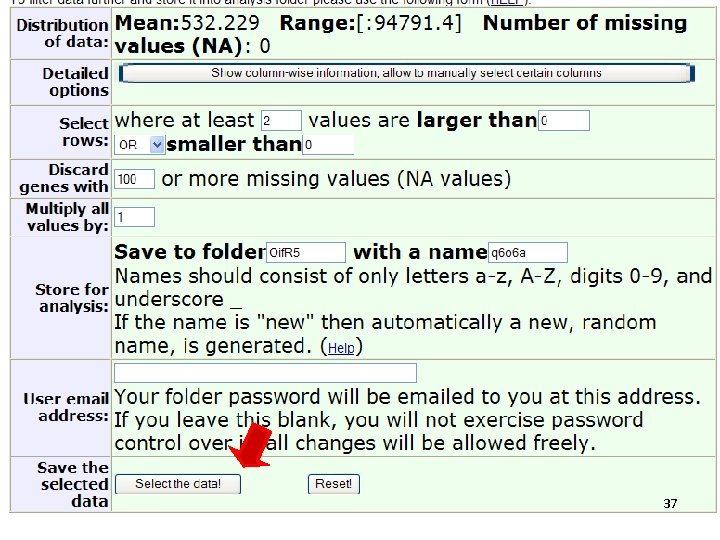
36

37

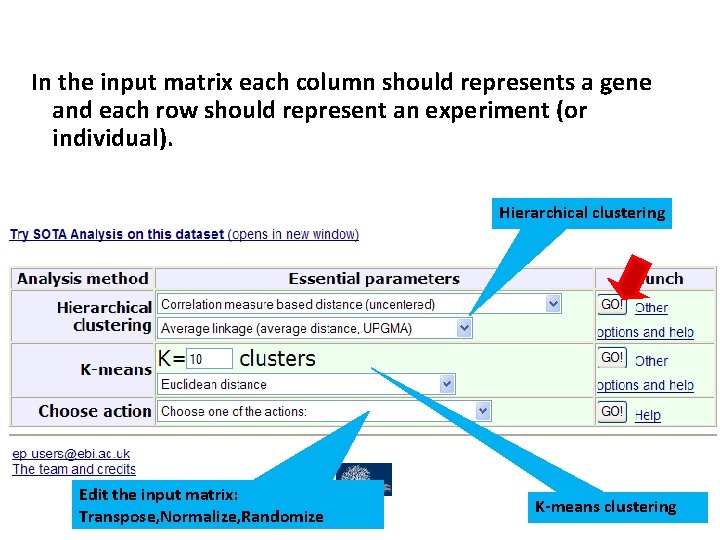
In the input matrix each column should represents a gene and each row should represent an experiment (or individual). Hierarchical clustering Edit the input matrix: Transpose, Normalize, Randomize 38 K-means clustering

In the input matrix each column should represents a gene and each row should represent an experiment (or individual). Hierarchical clustering 39

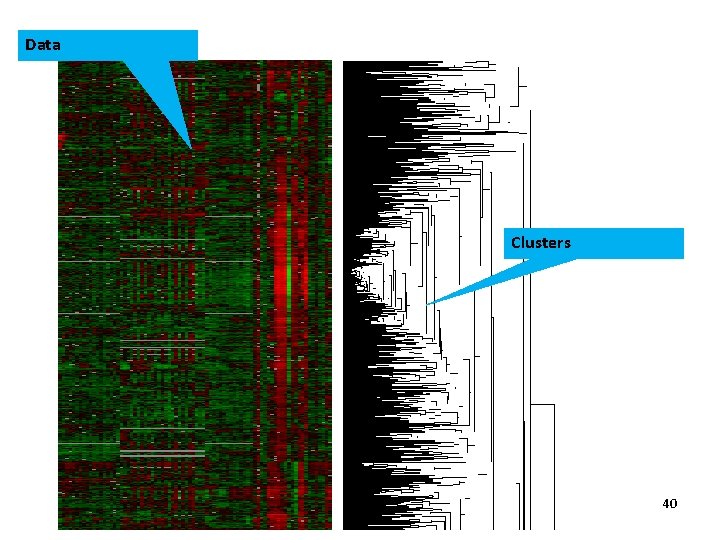
Data Clusters 40

In the input matrix each column should represents a gene and each row should represent an experiment (or individual). K-means clustering 41

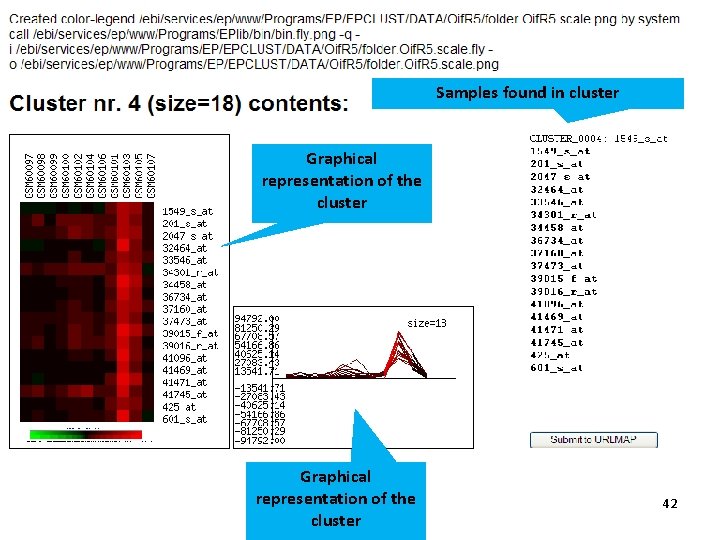
Samples found in cluster Graphical representation of the cluster 42

10 clusters, as requested 43

Tools for clustering - Me. V http: //www. tm 4. org/mev/ 44

Gene expression function analysis 1007_s_at 1053_at 117_at 121_at 1255_g_at 1294_at 1316_at 1320_at 1405_i_at 1431_at 1438_at 1487_at 1494_f_at 1598_g_at What can we learn from clusters? 45

Gene Ontology (GO) http: //www. geneontology. org/ The Gene Ontology project provides an ontology of defined terms representing gene product properties. The ontology covers three domains:

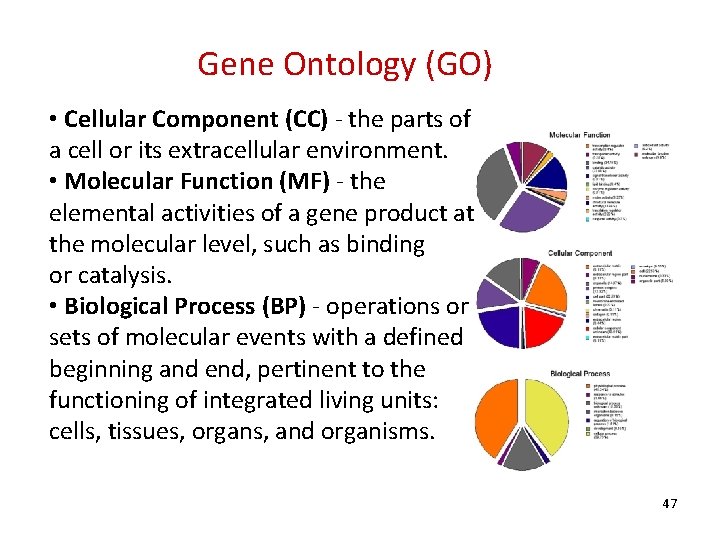
Gene Ontology (GO) • Cellular Component (CC) - the parts of a cell or its extracellular environment. • Molecular Function (MF) - the elemental activities of a gene product at the molecular level, such as binding or catalysis. • Biological Process (BP) - operations or sets of molecular events with a defined beginning and end, pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. 47

The GO tree

GO sources ISS Inferred from Sequence/Structural Similarity IDA Inferred from Direct Assay IPI Inferred from Physical Interaction TAS Traceable Author Statement NAS Non-traceable Author Statement IMP Inferred from Mutant Phenotype IGI Inferred from Genetic Interaction IEP Inferred from Expression Pattern IC Inferred by Curator ND No Data available IEA Inferred from electronic annotation

Search by Ami. GO

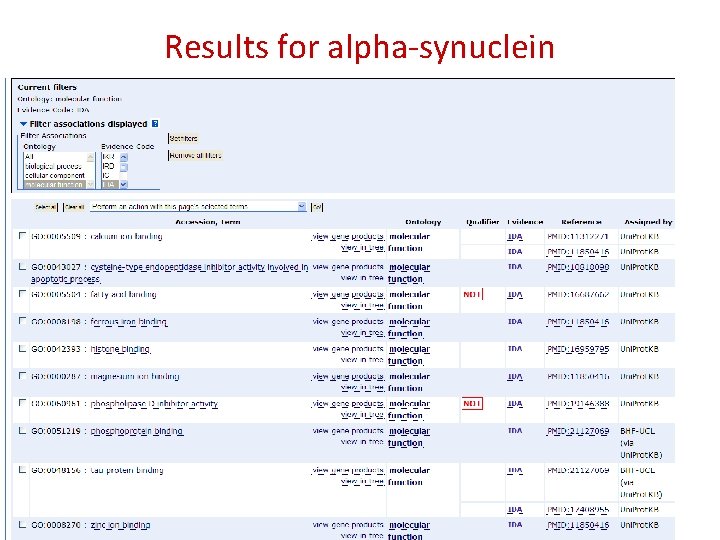
Results for alpha-synuclein

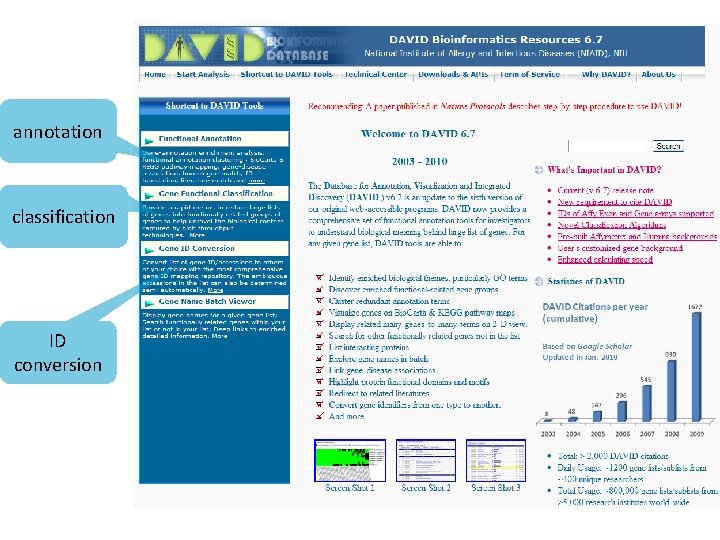
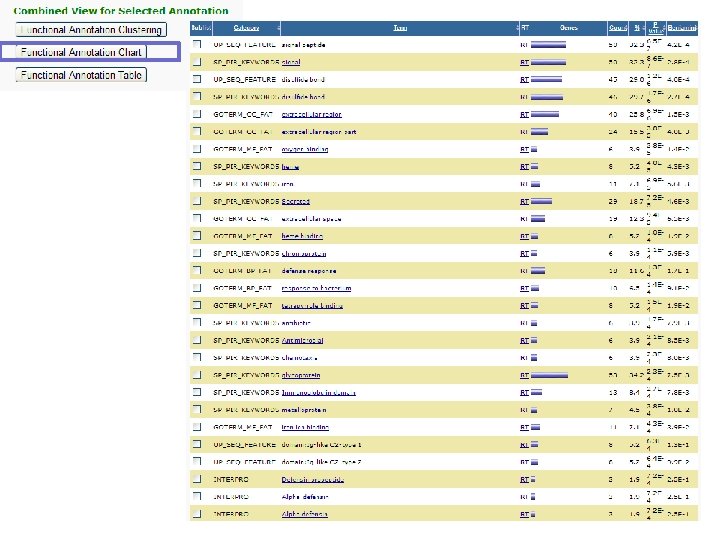
DAVID http: //david. abcc. ncifcrf. gov/ Functional Annotation Bioinformatics Microarray Analysis • Identify enriched biological themes, particularly GO terms • Discover enriched functional-related gene/protein groups • Cluster redundant annotation terms • Explore gene names in batch

annotation classification ID conversion

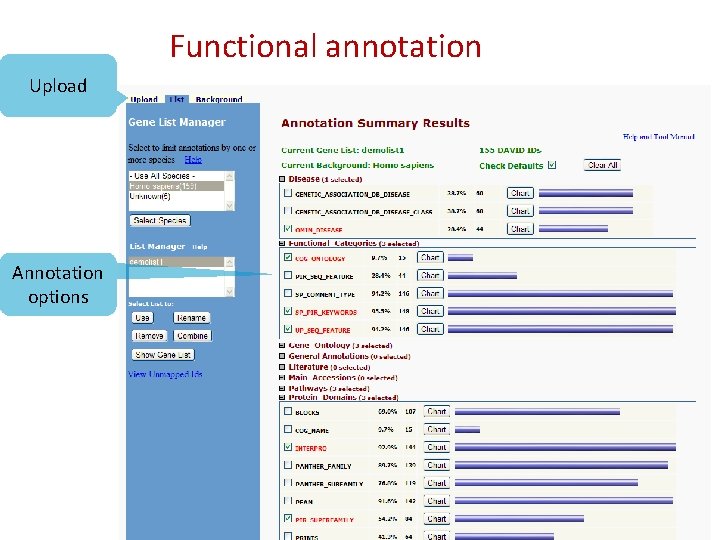
Functional annotation Upload Annotation options


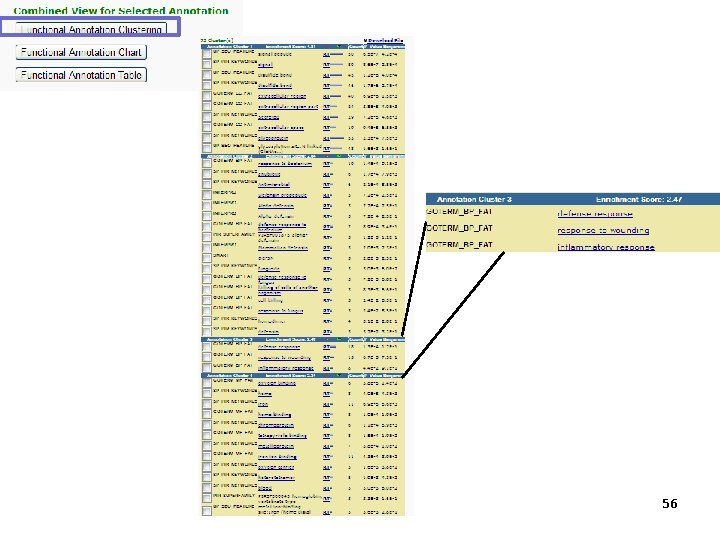
56

Gene expression analysis • Expression data – GEO – UCSC – Array. Express • General clustering methods – Unsupervised Clustering • Hierarchical clustering • K-means clustering • Tools for clustering – EPCLUST – Mev • Functional analysis – Go annotation 57