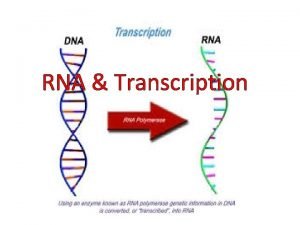
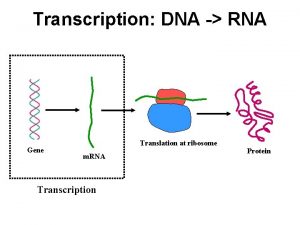
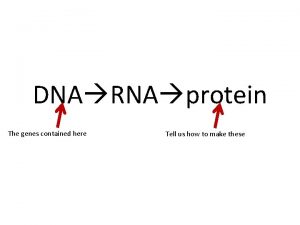
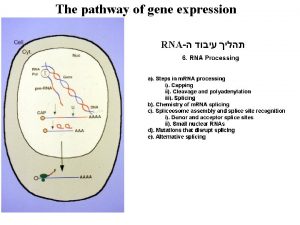
POLYADENYLATION Posttranscriptional Processes II Pre m RNA Polyadenylation













- Slides: 41

POLYADENYLATION

Post-transcriptional Processes II: Pre -m. RNA Polyadenylation • Most cytoplasmic m. RNAs have a poly. A tail (3’ end) of 50 -250 Adenylates – a notable exception is histone m. RNAs • Added post-transcriptionally by an enzyme, poly. A polymerase(s) • Turns over (recycles) in cytoplasm

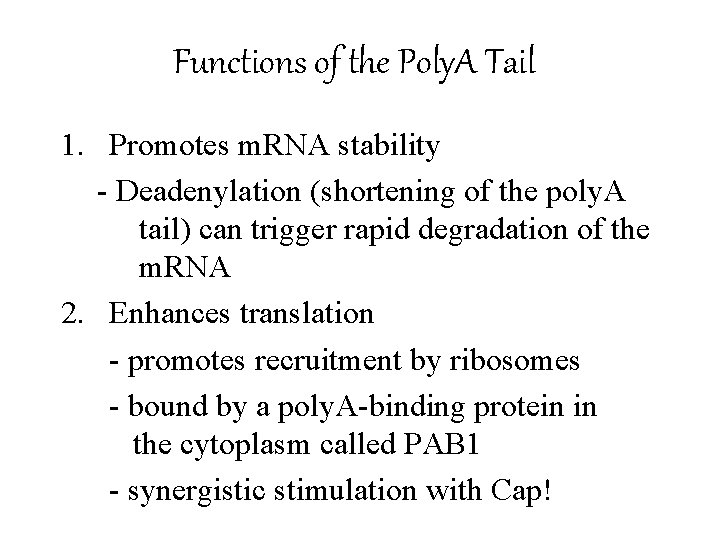
Functions of the Poly. A Tail 1. Promotes m. RNA stability - Deadenylation (shortening of the poly. A tail) can trigger rapid degradation of the m. RNA 2. Enhances translation - promotes recruitment by ribosomes - bound by a poly. A-binding protein in the cytoplasm called PAB 1 - synergistic stimulation with Cap!

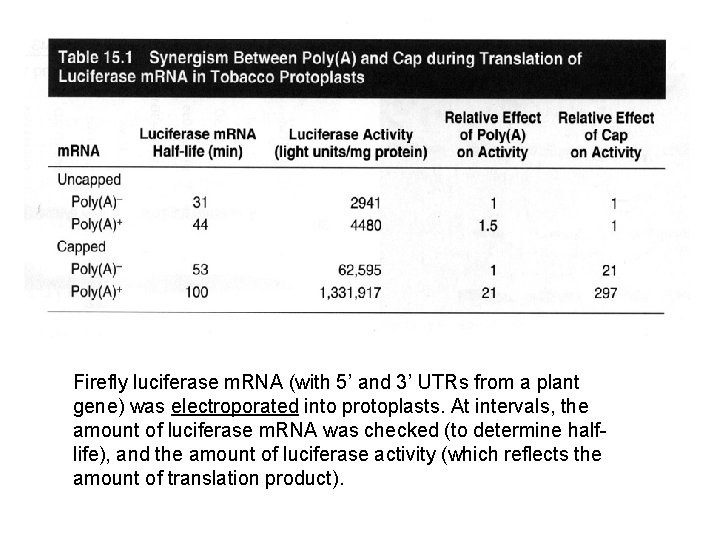
Firefly luciferase m. RNA (with 5’ and 3’ UTRs from a plant gene) was electroporated into protoplasts. At intervals, the amount of luciferase m. RNA was checked (to determine halflife), and the amount of luciferase activity (which reflects the amount of translation product).

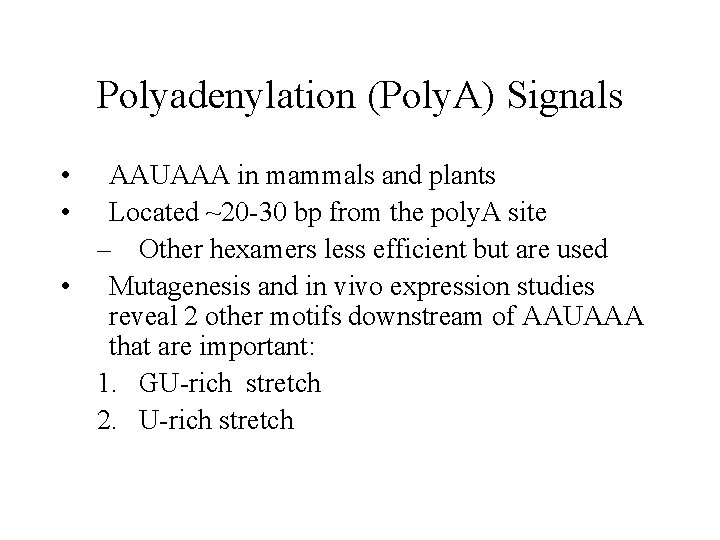
Polyadenylation (Poly. A) Signals • • AAUAAA in mammals and plants Located ~20 -30 bp from the poly. A site – Other hexamers less efficient but are used • Mutagenesis and in vivo expression studies reveal 2 other motifs downstream of AAUAAA that are important: 1. GU-rich stretch 2. U-rich stretch

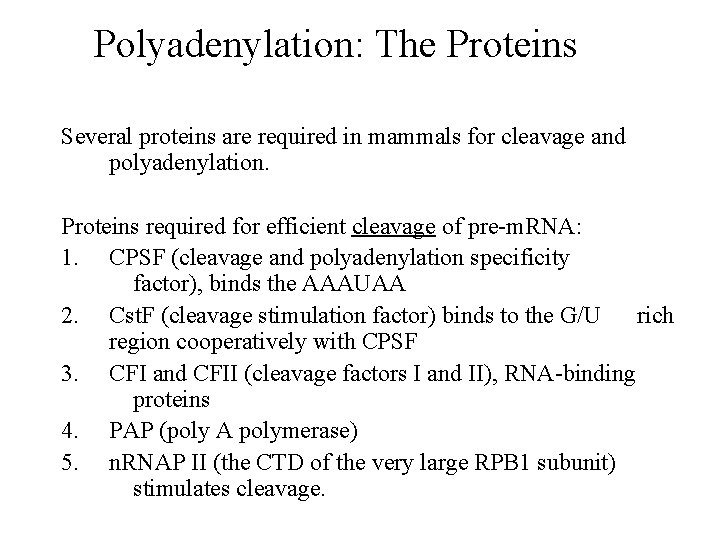
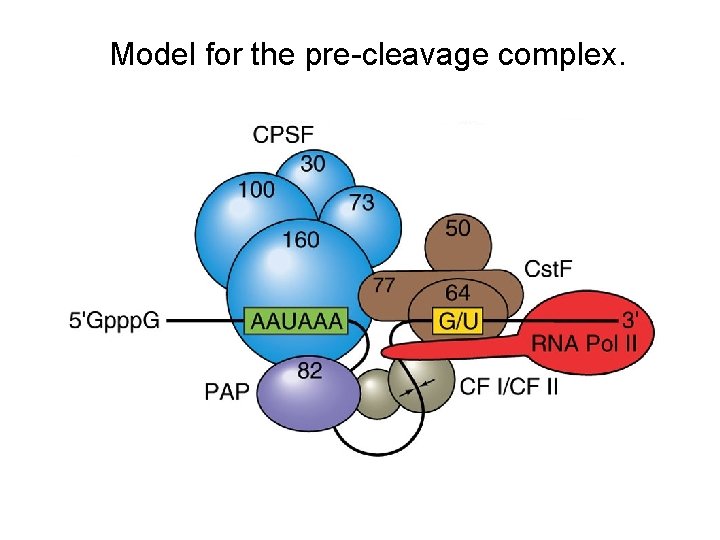
Polyadenylation: The Proteins Several proteins are required in mammals for cleavage and polyadenylation. Proteins required for efficient cleavage of pre-m. RNA: 1. CPSF (cleavage and polyadenylation specificity factor), binds the AAAUAA 2. Cst. F (cleavage stimulation factor) binds to the G/U rich region cooperatively with CPSF 3. CFI and CFII (cleavage factors I and II), RNA-binding proteins 4. PAP (poly A polymerase) 5. n. RNAP II (the CTD of the very large RPB 1 subunit) stimulates cleavage.

Model for the pre-cleavage complex.

Polyadenylation: Mechanism • Occurs in 2 phases – Phase 1: requires AAUAAA and ~8 nt downstream (3’) – Phase 2 : Once ~10 As are added, further adenylation does not require the AAUAAA

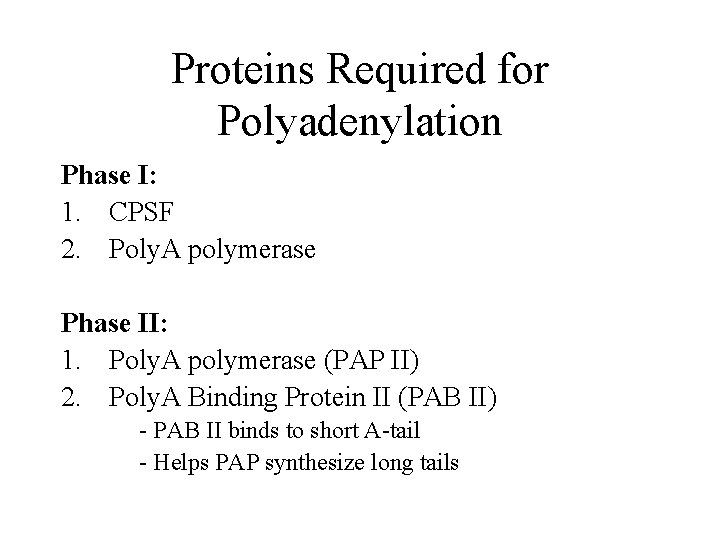
Proteins Required for Polyadenylation Phase I: 1. CPSF 2. Poly. A polymerase Phase II: 1. Poly. A polymerase (PAP II) 2. Poly. A Binding Protein II (PAB II) - PAB II binds to short A-tail - Helps PAP synthesize long tails

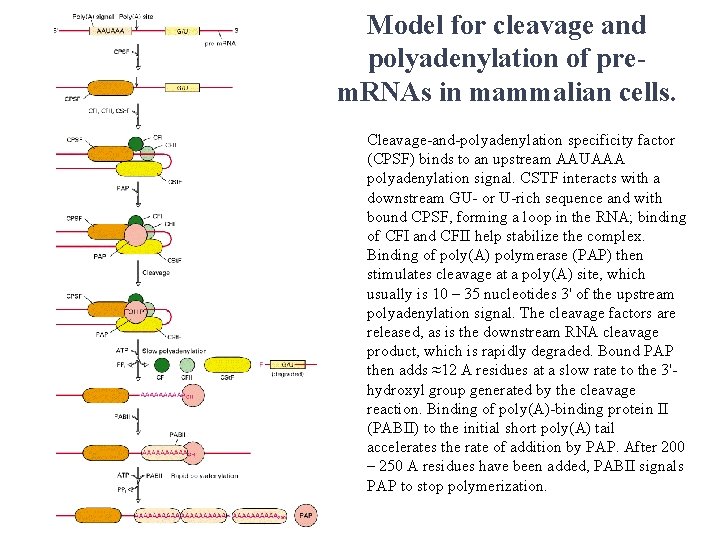
Model for cleavage and polyadenylation of prem. RNAs in mammalian cells. Cleavage-and-polyadenylation specificity factor (CPSF) binds to an upstream AAUAAA polyadenylation signal. CSTF interacts with a downstream GU- or U-rich sequence and with bound CPSF, forming a loop in the RNA; binding of CFI and CFII help stabilize the complex. Binding of poly(A) polymerase (PAP) then stimulates cleavage at a poly(A) site, which usually is 10 – 35 nucleotides 3' of the upstream polyadenylation signal. The cleavage factors are released, as is the downstream RNA cleavage product, which is rapidly degraded. Bound PAP then adds ≈12 A residues at a slow rate to the 3'hydroxyl group generated by the cleavage reaction. Binding of poly(A)-binding protein II (PABII) to the initial short poly(A) tail accelerates the rate of addition by PAP. After 200 – 250 A residues have been added, PABII signals PAP to stop polymerization.


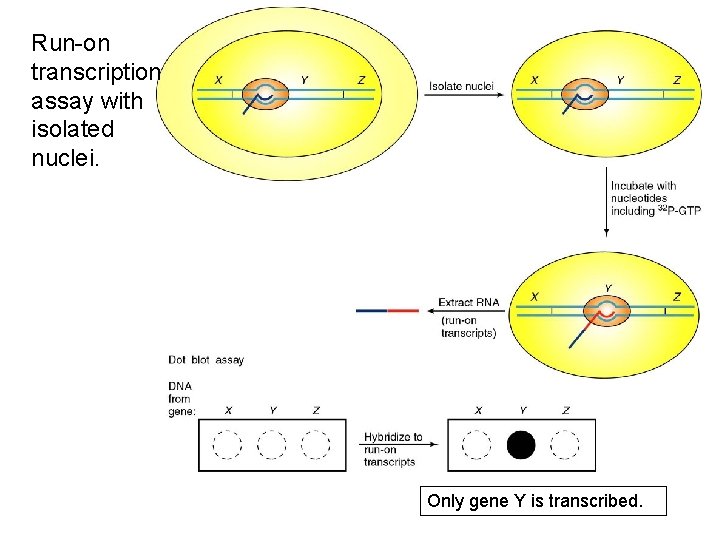
Run-on transcription assay with isolated nuclei. Only gene Y is transcribed.

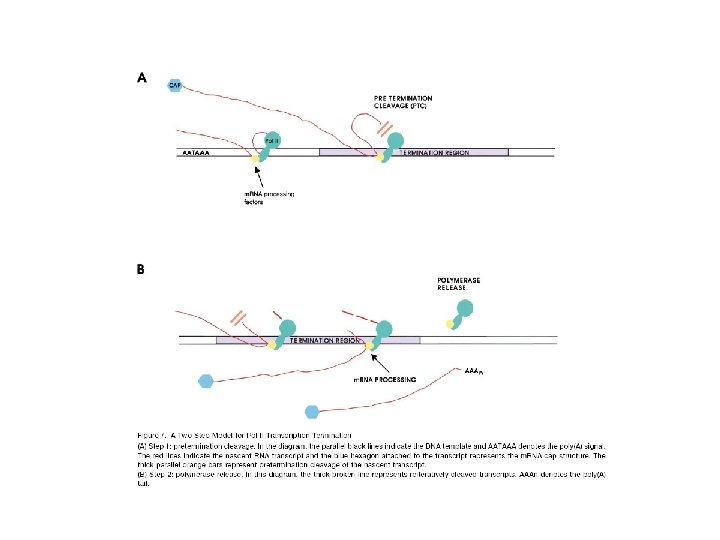
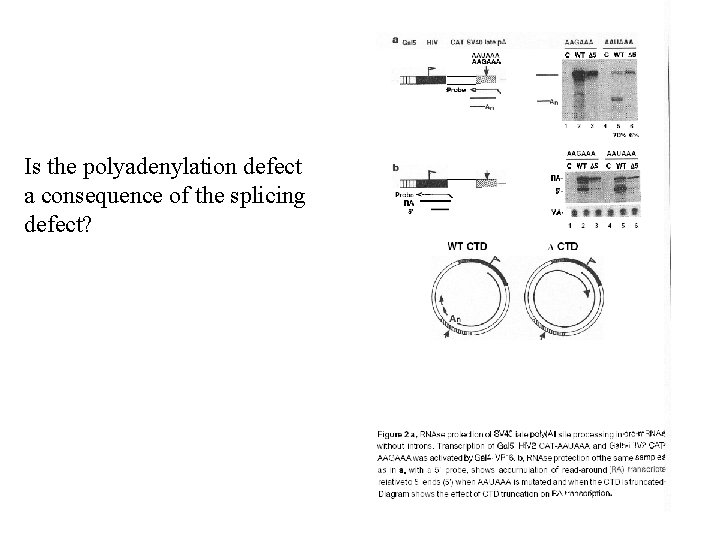
What happen with transcription after Cleavage site? Is the same in transient transfection?

Which region are the Important ones for Transcriptional Termination? ? Is this a consequence of Incorrect processing?

RT analysis 3’-5’ degradation? ? Cryptic promoter? Unstable regions? ? Autocatalytic? ?


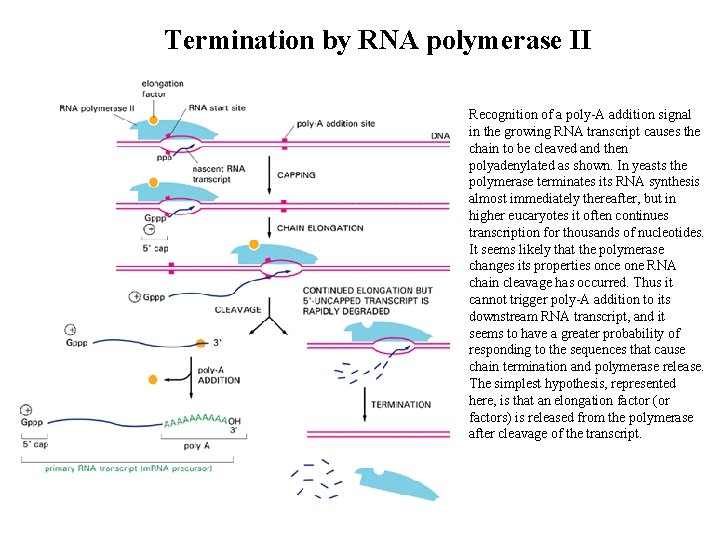
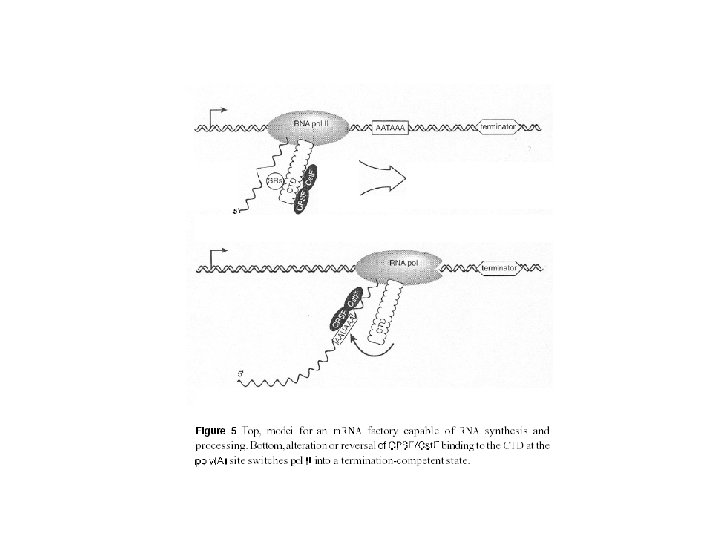
Termination by RNA polymerase II Recognition of a poly-A addition signal in the growing RNA transcript causes the chain to be cleaved and then polyadenylated as shown. In yeasts the polymerase terminates its RNA synthesis almost immediately thereafter, but in higher eucaryotes it often continues transcription for thousands of nucleotides. It seems likely that the polymerase changes its properties once one RNA chain cleavage has occurred. Thus it cannot trigger poly-A addition to its downstream RNA transcript, and it seems to have a greater probability of responding to the sequences that cause chain termination and polymerase release. The simplest hypothesis, represented here, is that an elongation factor (or factors) is released from the polymerase after cleavage of the transcript.

ALTERNATIVE SPLICING


Mutations that disrupt splicing • bo-thalassemia - no b-chain synthesis • b+-thalassemia - some b-chain synthesis Normal splice pattern: Exon 1 Exon 2 Exon 3 Intron 2 Intron 1 Donor site: /GU Acceptor site: AG/ Intron 2 acceptor site bo mutation: no use of mutant site; use of cryptic splice site in intron 2 Exon 1 Exon 2 Intron 1 Intron 2 cryptic acceptor site: UUUCAG/G mutant site: GG/ Translation of the retained portion of intron 2 results in premature termination of translation due to a stop codon within the intron, 15 codons from the cryptic splice site

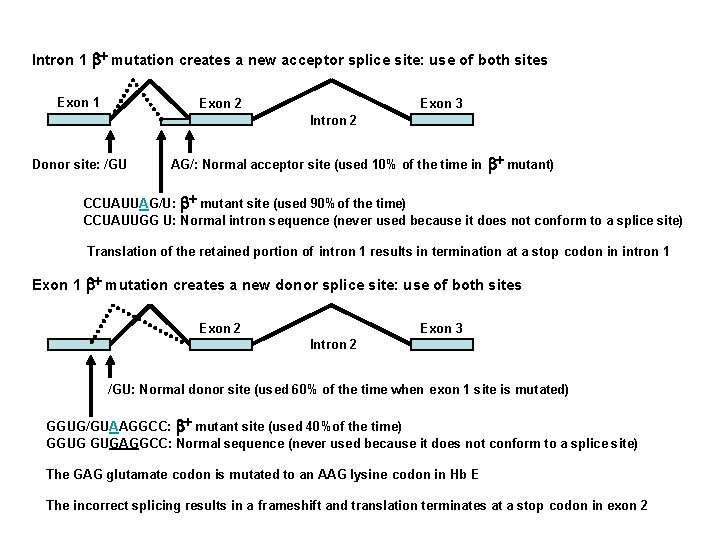
Intron 1 b+ mutation creates a new acceptor splice site: use of both sites Exon 1 Exon 2 Exon 3 Intron 2 Donor site: /GU AG/: Normal acceptor site (used 10% of the time in b+ mutant) CCUAUUAG/U: b+ mutant site (used 90%of the time) CCUAUUGG U: Normal intron sequence (never used because it does not conform to a splice site) Translation of the retained portion of intron 1 results in termination at a stop codon in intron 1 Exon 1 b+ mutation creates a new donor splice site: use of both sites Exon 2 Exon 3 Intron 2 /GU: Normal donor site (used 60% of the time when exon 1 site is mutated) GGUG/GUAAGGCC: b+ mutant site (used 40%of the time) GGUG GUGAGGCC: Normal sequence (never used because it does not conform to a splice site) The GAG glutamate codon is mutated to an AAG lysine codon in Hb E The incorrect splicing results in a frameshift and translation terminates at a stop codon in exon 2

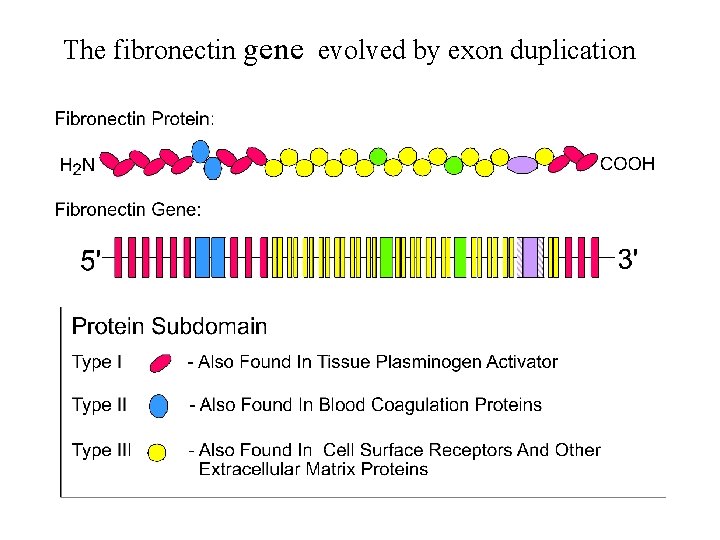
The fibronectin gene evolved by exon duplication


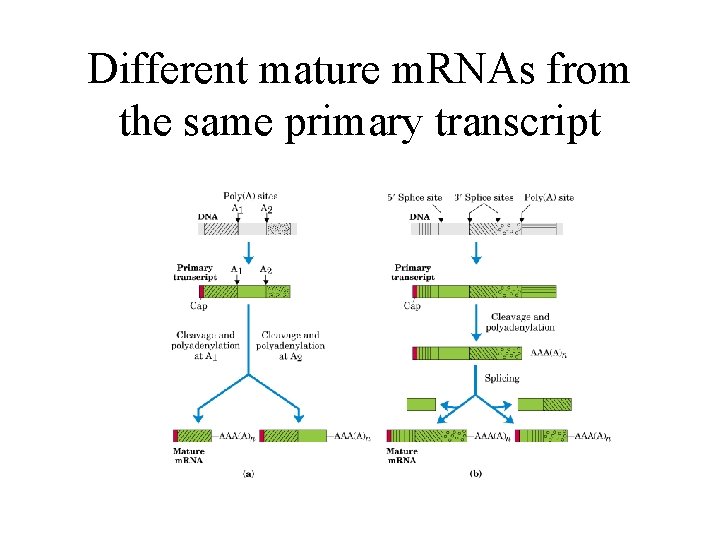
Different mature m. RNAs from the same primary transcript

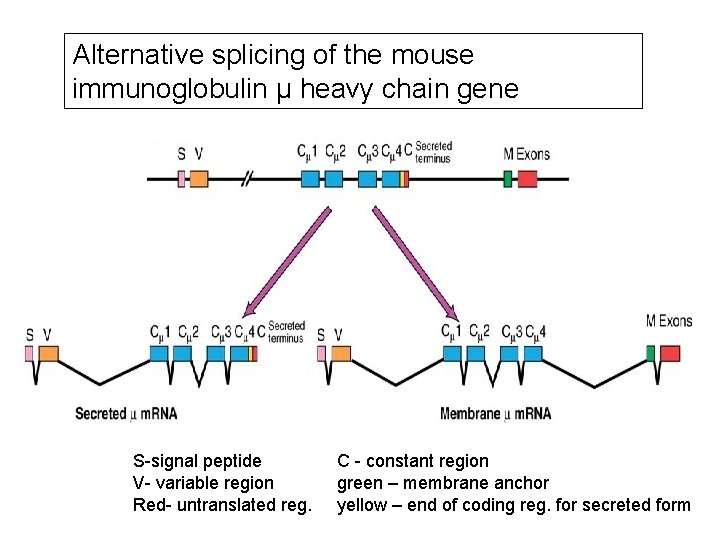
Alternative splicing of the mouse immunoglobulin μ heavy chain gene S-signal peptide V- variable region Red- untranslated reg. C - constant region green – membrane anchor yellow – end of coding reg. for secreted form

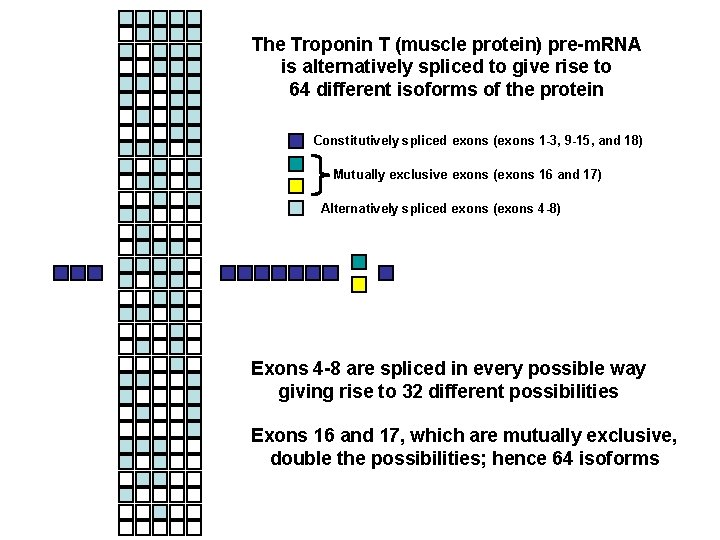
The Troponin T (muscle protein) pre-m. RNA is alternatively spliced to give rise to 64 different isoforms of the protein Constitutively spliced exons (exons 1 -3, 9 -15, and 18) Mutually exclusive exons (exons 16 and 17) Alternatively spliced exons (exons 4 -8) Exons 4 -8 are spliced in every possible way giving rise to 32 different possibilities Exons 16 and 17, which are mutually exclusive, double the possibilities; hence 64 isoforms

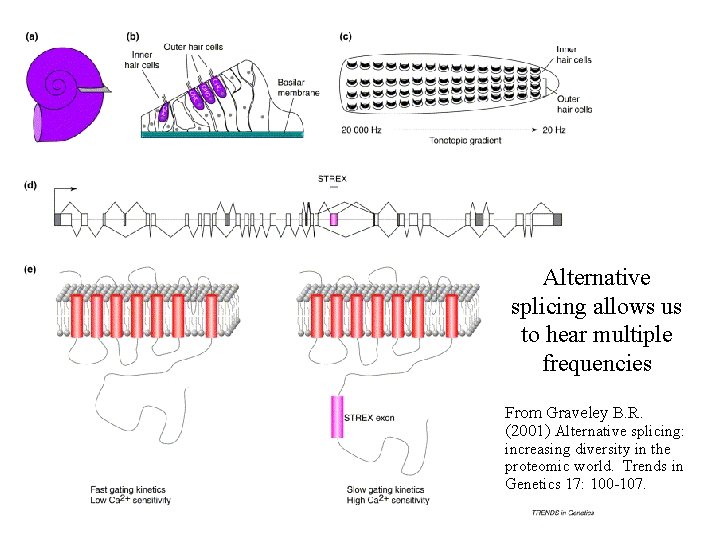
Alternative splicing allows us to hear multiple frequencies From Graveley B. R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics 17: 100 -107.

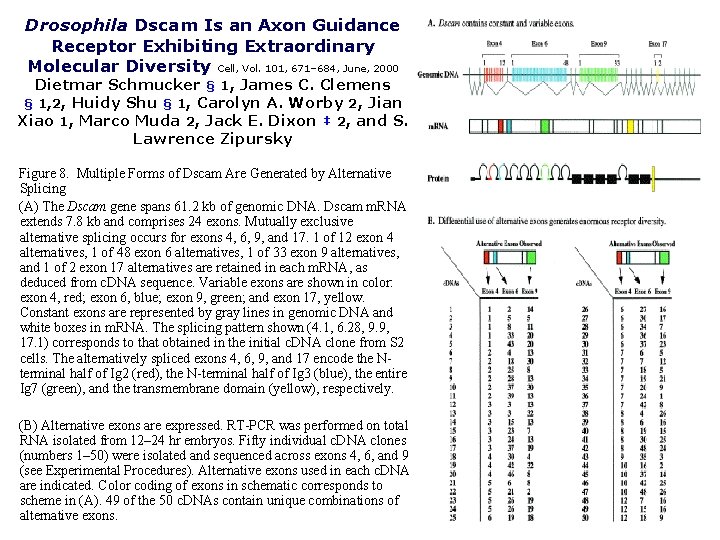
Drosophila Dscam Is an Axon Guidance Receptor Exhibiting Extraordinary Molecular Diversity Cell, Vol. 101, 671– 684, June, 2000 Dietmar Schmucker § 1, James C. Clemens § 1, 2, Huidy Shu § 1, Carolyn A. Worby 2, Jian Xiao 1, Marco Muda 2, Jack E. Dixon ‡ 2, and S. Lawrence Zipursky Figure 8. Multiple Forms of Dscam Are Generated by Alternative Splicing (A) The Dscam gene spans 61. 2 kb of genomic DNA. Dscam m. RNA extends 7. 8 kb and comprises 24 exons. Mutually exclusive alternative splicing occurs for exons 4, 6, 9, and 17. 1 of 12 exon 4 alternatives, 1 of 48 exon 6 alternatives, 1 of 33 exon 9 alternatives, and 1 of 2 exon 17 alternatives are retained in each m. RNA, as deduced from c. DNA sequence. Variable exons are shown in color: exon 4, red; exon 6, blue; exon 9, green; and exon 17, yellow. Constant exons are represented by gray lines in genomic DNA and white boxes in m. RNA. The splicing pattern shown (4. 1, 6. 28, 9. 9, 17. 1) corresponds to that obtained in the initial c. DNA clone from S 2 cells. The alternatively spliced exons 4, 6, 9, and 17 encode the Nterminal half of Ig 2 (red), the N-terminal half of Ig 3 (blue), the entire Ig 7 (green), and the transmembrane domain (yellow), respectively. (B) Alternative exons are expressed. RT-PCR was performed on total RNA isolated from 12– 24 hr embryos. Fifty individual c. DNA clones (numbers 1– 50) were isolated and sequenced across exons 4, 6, and 9 (see Experimental Procedures). Alternative exons used in each c. DNA are indicated. Color coding of exons in schematic corresponds to scheme in (A). 49 of the 50 c. DNAs contain unique combinations of alternative exons.

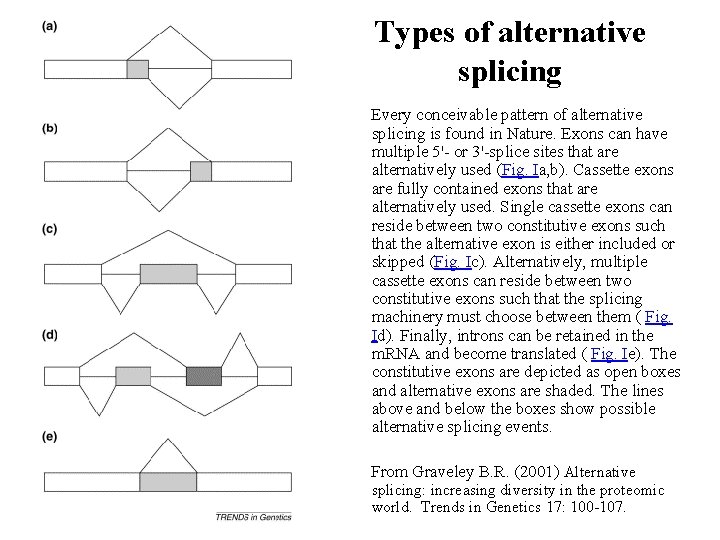
Types of alternative splicing Every conceivable pattern of alternative splicing is found in Nature. Exons can have multiple 5'- or 3'-splice sites that are alternatively used (Fig. Ia, b). Cassette exons are fully contained exons that are alternatively used. Single cassette exons can reside between two constitutive exons such that the alternative exon is either included or skipped (Fig. Ic). Alternatively, multiple cassette exons can reside between two constitutive exons such that the splicing machinery must choose between them ( Fig. Id). Finally, introns can be retained in the m. RNA and become translated ( Fig. Ie). The constitutive exons are depicted as open boxes and alternative exons are shaded. The lines above and below the boxes show possible alternative splicing events. From Graveley B. R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends in Genetics 17: 100 -107.

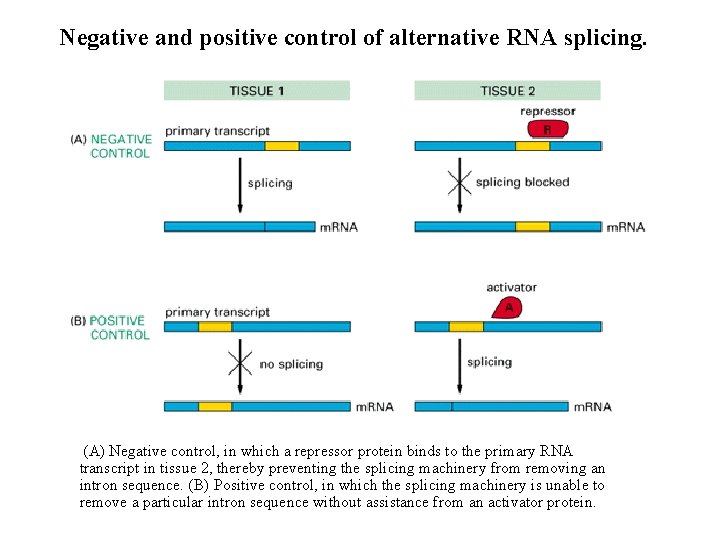
Negative and positive control of alternative RNA splicing. (A) Negative control, in which a repressor protein binds to the primary RNA transcript in tissue 2, thereby preventing the splicing machinery from removing an intron sequence. (B) Positive control, in which the splicing machinery is unable to remove a particular intron sequence without assistance from an activator protein.

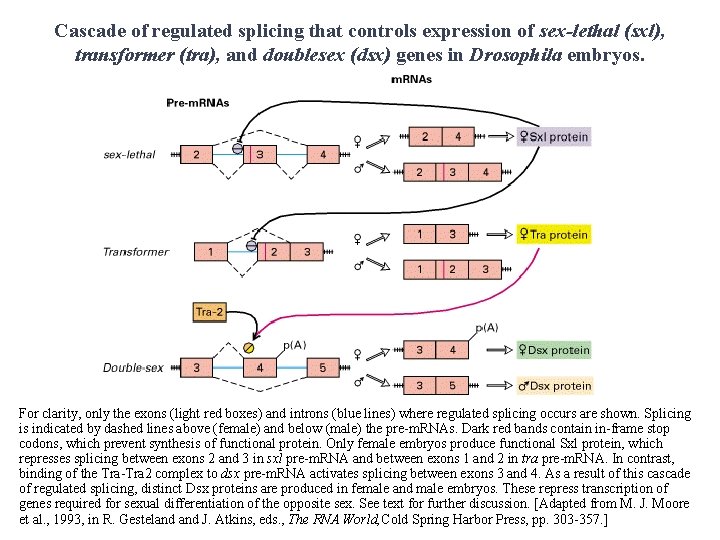
Cascade of regulated splicing that controls expression of sex-lethal (sxl), transformer (tra), and doublesex (dsx) genes in Drosophila embryos. For clarity, only the exons (light red boxes) and introns (blue lines) where regulated splicing occurs are shown. Splicing is indicated by dashed lines above (female) and below (male) the pre-m. RNAs. Dark red bands contain in-frame stop codons, which prevent synthesis of functional protein. Only female embryos produce functional Sxl protein, which represses splicing between exons 2 and 3 in sxl pre-m. RNA and between exons 1 and 2 in tra pre-m. RNA. In contrast, binding of the Tra-Tra 2 complex to dsx pre-m. RNA activates splicing between exons 3 and 4. As a result of this cascade of regulated splicing, distinct Dsx proteins are produced in female and male embryos. These repress transcription of genes required for sexual differentiation of the opposite sex. See text for further discussion. [Adapted from M. J. Moore et al. , 1993, in R. Gesteland J. Atkins, eds. , The RNA World, Cold Spring Harbor Press, pp. 303 -357. ]

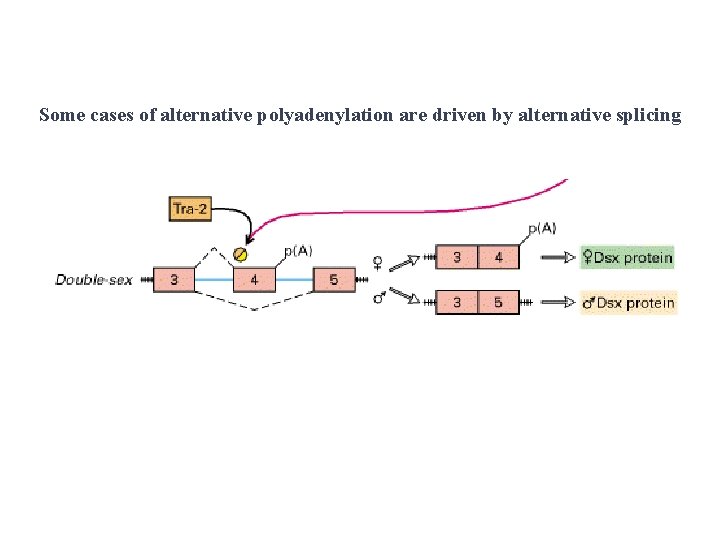
Some cases of alternative polyadenylation are driven by alternative splicing

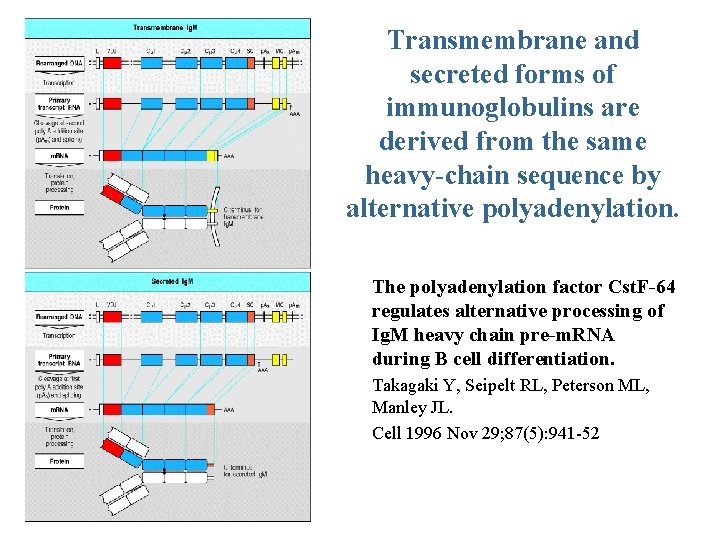
Transmembrane and secreted forms of immunoglobulins are derived from the same heavy-chain sequence by alternative polyadenylation. The polyadenylation factor Cst. F-64 regulates alternative processing of Ig. M heavy chain pre-m. RNA during B cell differentiation. Takagaki Y, Seipelt RL, Peterson ML, Manley JL. Cell 1996 Nov 29; 87(5): 941 -52

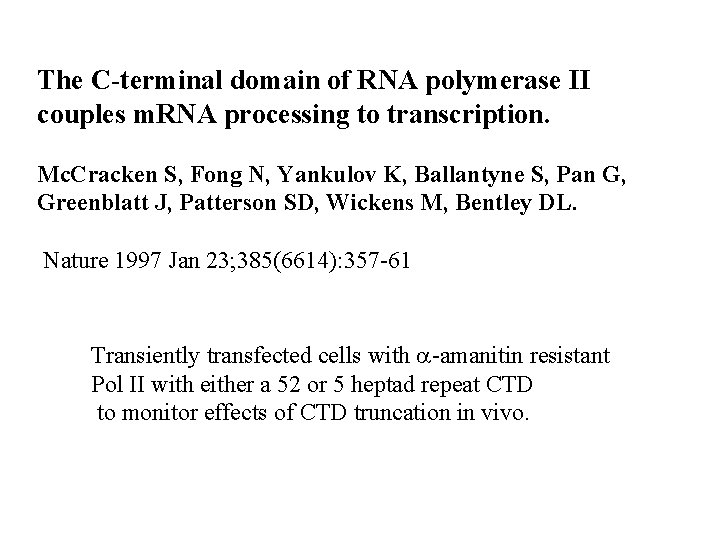
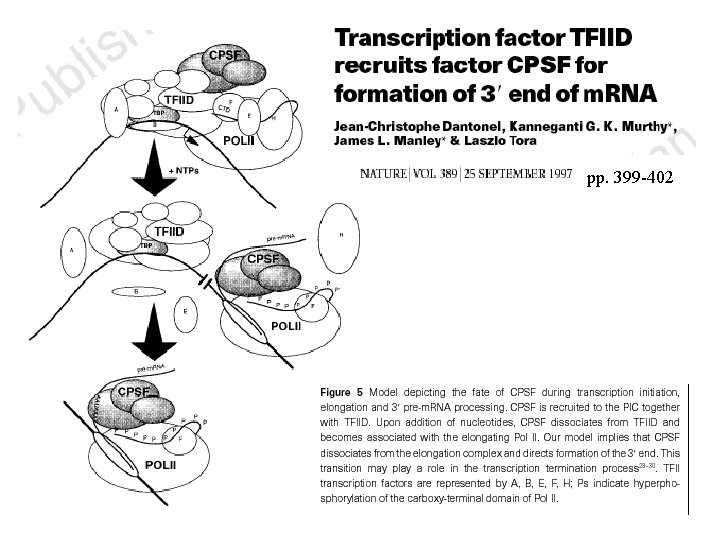
The C-terminal domain of RNA polymerase II couples m. RNA processing to transcription. Mc. Cracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. Nature 1997 Jan 23; 385(6614): 357 -61 Transiently transfected cells with a-amanitin resistant Pol II with either a 52 or 5 heptad repeat CTD to monitor effects of CTD truncation in vivo.


Is the polyadenylation defect a consequence of the splicing defect?

Affinity columns of GST, GST-mut. CTD, GST-Hiper. PCTD


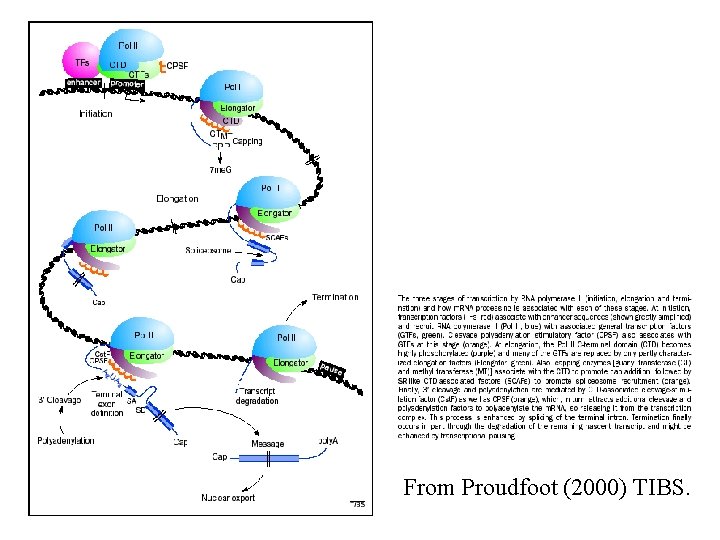
From Proudfoot (2000) TIBS.

pp. 399 -402

From Proudfoot (2000) TIBS.
 Concurrent in os
Concurrent in os Osnowa dokumentu
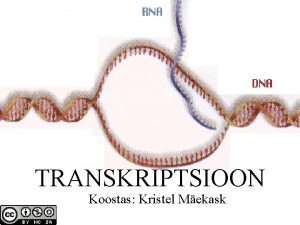
Osnowa dokumentu B-dna structure
B-dna structure Dna and rna coloring worksheet
Dna and rna coloring worksheet Biosinteza e acideve nukleike
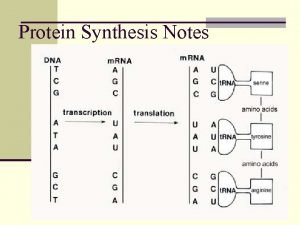
Biosinteza e acideve nukleike Dna and rna
Dna and rna Dna to rna rules
Dna to rna rules Per base sequence quality
Per base sequence quality Tradução
Tradução Who is this
Who is this Rna mensageiro
Rna mensageiro Ribosomal rna
Ribosomal rna Messenger rna codons
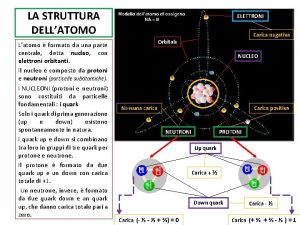
Messenger rna codons La struttura dell'atomo mappa concettuale
La struttura dell'atomo mappa concettuale Che cos'è rna
Che cos'è rna Unlike dna, rna contains
Unlike dna, rna contains Rna polimerasi
Rna polimerasi Rna ehitus
Rna ehitus What type of rna
What type of rna Transcription and translation practice worksheet answer key
Transcription and translation practice worksheet answer key A260/280 ratio for rna
A260/280 ratio for rna Dna rna and proteins study guide answers
Dna rna and proteins study guide answers Rna world
Rna world Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Stop codons
Stop codons Explain rna
Explain rna Rna
Rna Ss rna
Ss rna Messenger rna sequence
Messenger rna sequence Rna polymerase
Rna polymerase Rna brainpop
Rna brainpop Dna protein synthesis study guide answers
Dna protein synthesis study guide answers Chapter 12 dna and rna
Chapter 12 dna and rna Polymeric gene interaction
Polymeric gene interaction Ribosomal rna
Ribosomal rna Mikro rna nedir
Mikro rna nedir Rna virus
Rna virus Dna rna protein
Dna rna protein Rna
Rna Compare and contrast dna and rna.
Compare and contrast dna and rna. Rna çeşitleri
Rna çeşitleri Rna codon chart
Rna codon chart