Chapter 18 Regulation of Gene Expression Overview Conducting


- Slides: 56

Chapter 18 Regulation of Gene Expression

Overview: Conducting the Genetic Orchestra • Prokaryotes and eukaryotes alter gene expression in response to their changing environment • In multicellular eukaryotes, gene expression regulates development and is responsible for differences in cell types • RNA molecules play many roles in regulating gene expression in eukaryotes © 2011 Pearson Education, Inc.

Concept 18. 1: Bacteria respond to environmental change by regulating transcription • Natural selection favors bacteria that produce only the products needed by that cell • Regulation – feedback inhibition – gene regulation • Gene expression in bacteria is controlled by the operon model © 2011 Pearson Education, Inc.

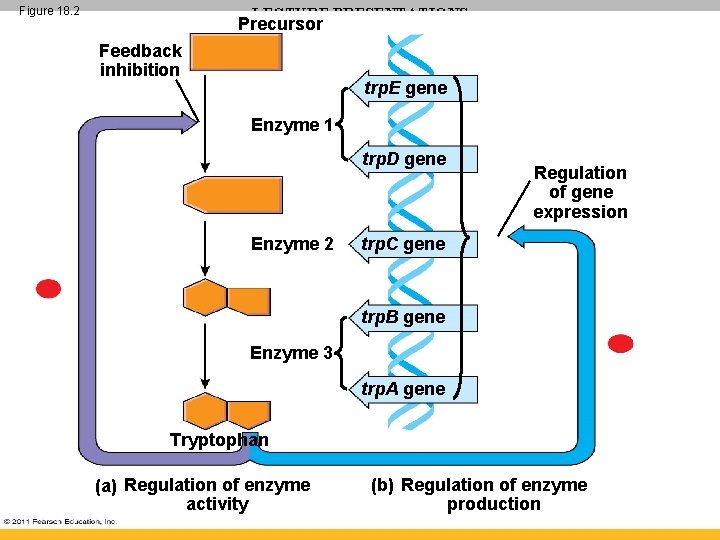
Figure 18. 2 LECTURE PRESENTATIONS Precursor For CAMPBELL BIOLOGY, NINTH EDITION Jane B. Reece, Lisa A. Urry, Michael L. Cain, Steven A. Wasserman, Peter V. Minorsky, Robert B. Jackson Feedback inhibition trp. E gene Enzyme 1 trp. D gene Enzyme 2 Regulation of gene expression trp. C gene trp. B gene Enzyme 3 trp. A gene Tryptophan (a) Regulation of enzyme activity Lectures by Erin Barley (b) Regulation of enzyme Kathleen Fitzpatrick production

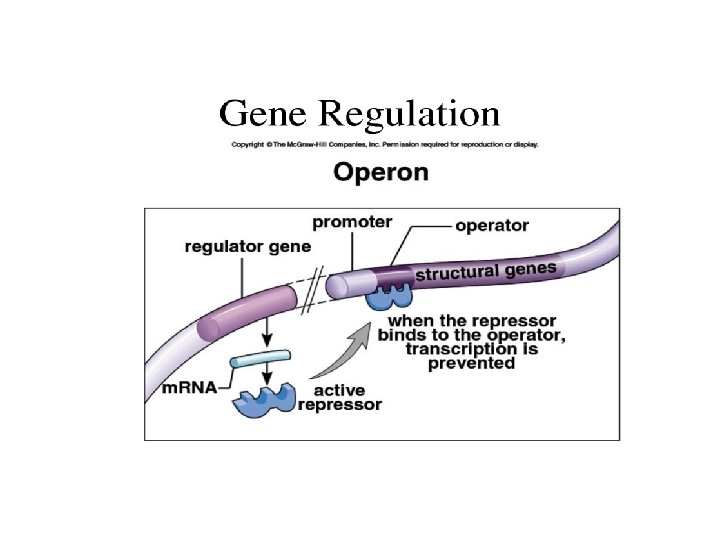
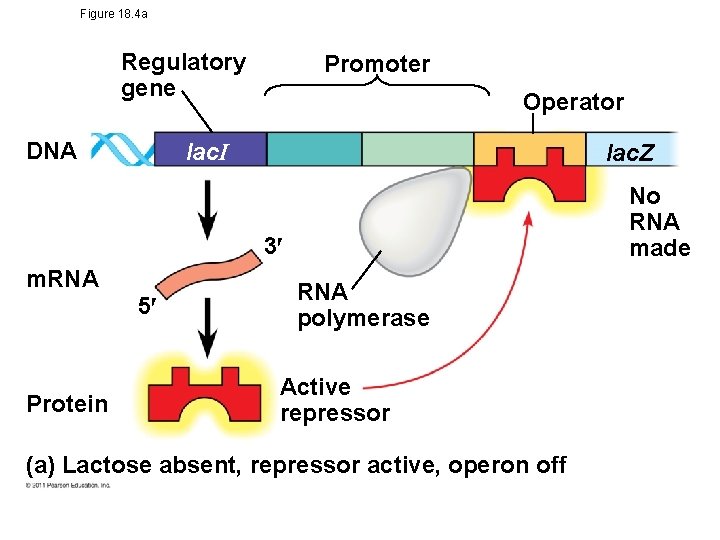
Operons: The Basic Concept • An operon -entire stretch of DNA that includes the operator, the promoter, and the genes that they control • A cluster of functionally related genes can be under coordinated control by a single “on-off switch” • “switch” is a segment of DNA called an operator © 2011 Pearson Education, Inc.


• The operon can be switched off by a protein repressor – prevents gene transcription by binding to the operator and blocking RNA polymerase • Made by separate regulatory gene © 2011 Pearson Education, Inc.

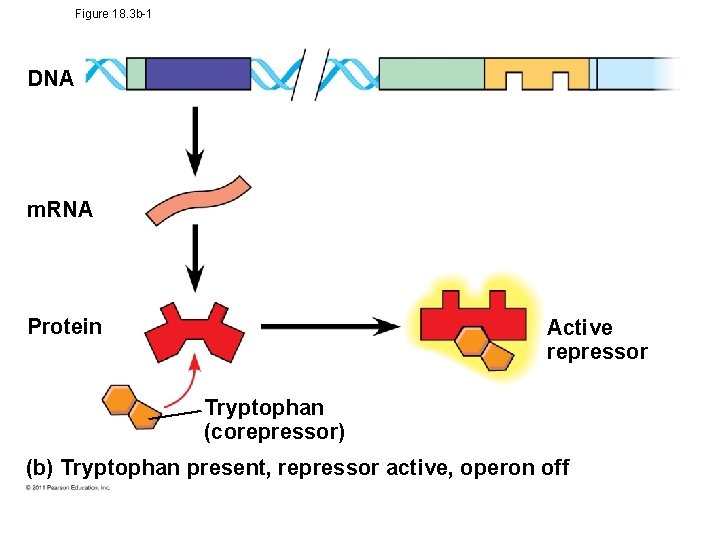
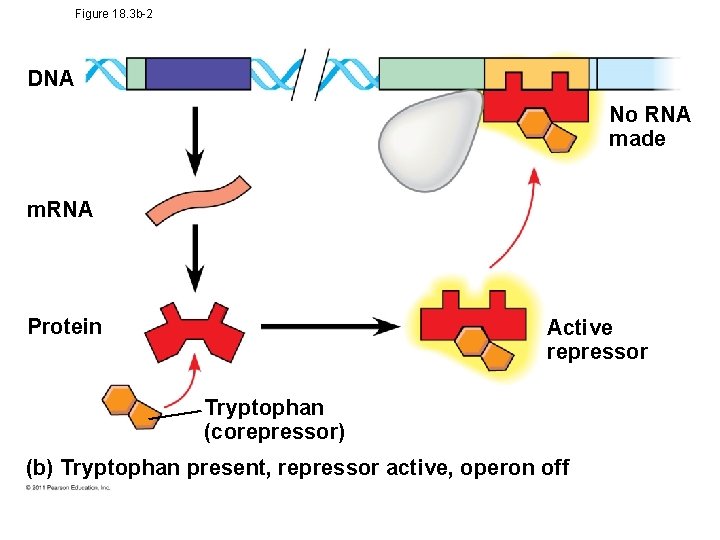
Operon Model • The repressor can be in an active or inactive form, depending on the presence of other molecules • A corepressor is a molecule that cooperates with a repressor protein to switch an operon off • For example, E. coli can synthesize the amino acid tryptophan © 2011 Pearson Education, Inc.

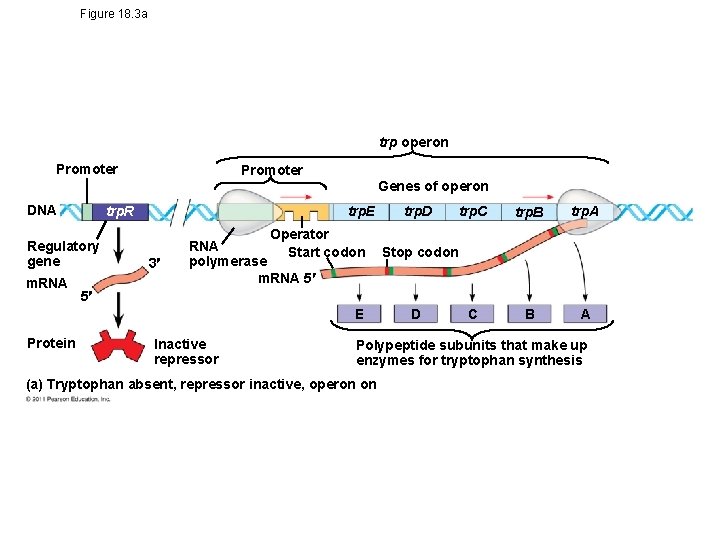
Tryptophan Operon • trp operon is on unless switched off by repressor • Anabolic- tryptophan needed • When tryptophan is present, it binds to the trp repressor protein, which turns the operon off • thus the trp operon is turned off (repressed) if tryptophan levels are high (saves cell energy) © 2011 Pearson Education, Inc.

Figure 18. 3 a trp operon Promoter Genes of operon DNA trp. R Regulatory gene m. RNA trp. E 3 Operator RNA Start codon polymerase m. RNA 5 trp. C trp. B trp. A C B A Stop codon 5 E Protein trp. D Inactive repressor D Polypeptide subunits that make up enzymes for tryptophan synthesis (a) Tryptophan absent, repressor inactive, operon on

Figure 18. 3 b-1 DNA m. RNA Protein Active repressor Tryptophan (corepressor) (b) Tryptophan present, repressor active, operon off

Figure 18. 3 b-2 DNA No RNA made m. RNA Protein Active repressor Tryptophan (corepressor) (b) Tryptophan present, repressor active, operon off

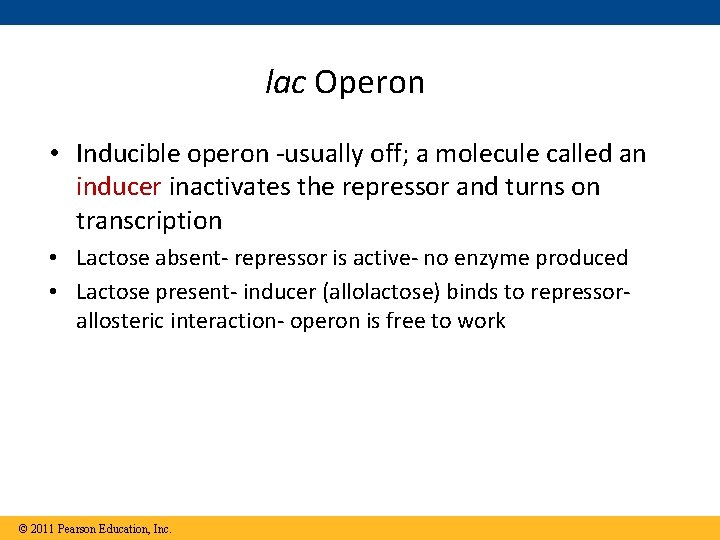
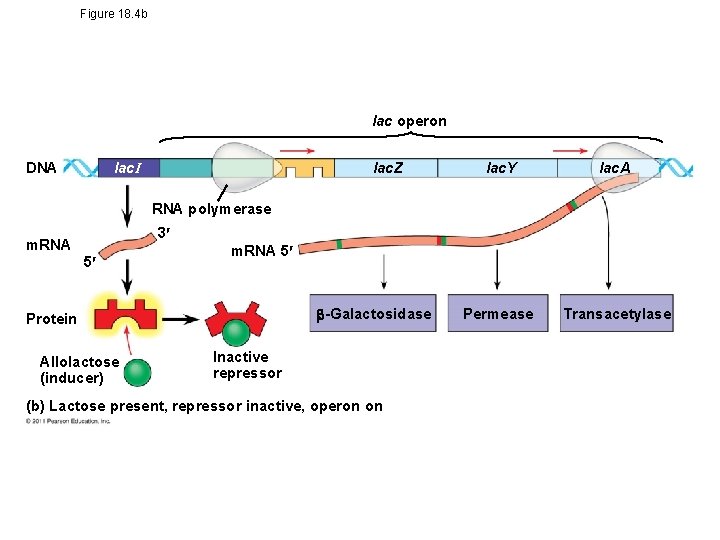
lac Operon • Inducible operon -usually off; a molecule called an inducer inactivates the repressor and turns on transcription • Lactose absent- repressor is active- no enzyme produced • Lactose present- inducer (allolactose) binds to repressorallosteric interaction- operon is free to work © 2011 Pearson Education, Inc.

Figure 18. 4 a Regulatory gene DNA Promoter Operator lac. I lac. Z No RNA made 3 m. RNA 5 Protein RNA polymerase Active repressor (a) Lactose absent, repressor active, operon off

Figure 18. 4 b lac operon lac. I DNA lac. Z lac. Y lac. A Permease Transacetylase RNA polymerase 3 m. RNA 5 -Galactosidase Protein Allolactose (inducer) Inactive repressor (b) Lactose present, repressor inactive, operon on

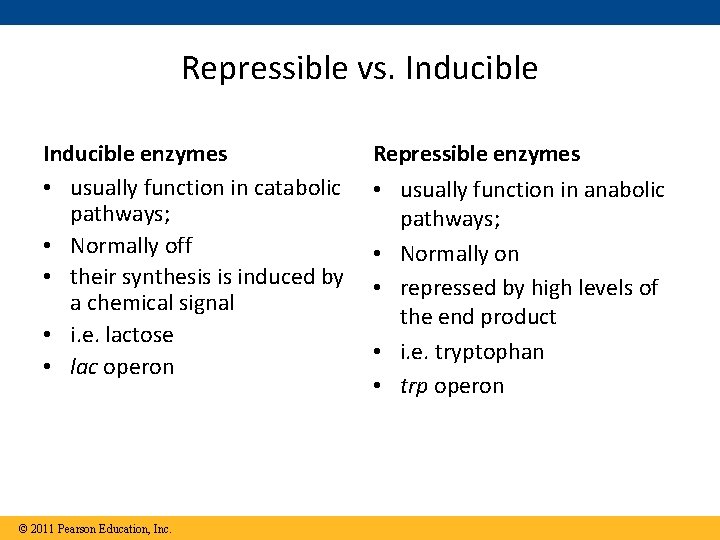
Repressible vs. Inducible enzymes • usually function in catabolic pathways; • Normally off • their synthesis is induced by a chemical signal • i. e. lactose • lac operon © 2011 Pearson Education, Inc. Repressible enzymes • usually function in anabolic pathways; • Normally on • repressed by high levels of the end product • i. e. tryptophan • trp operon

Negative Gene Regulation • Operon is off with active for of a repressor

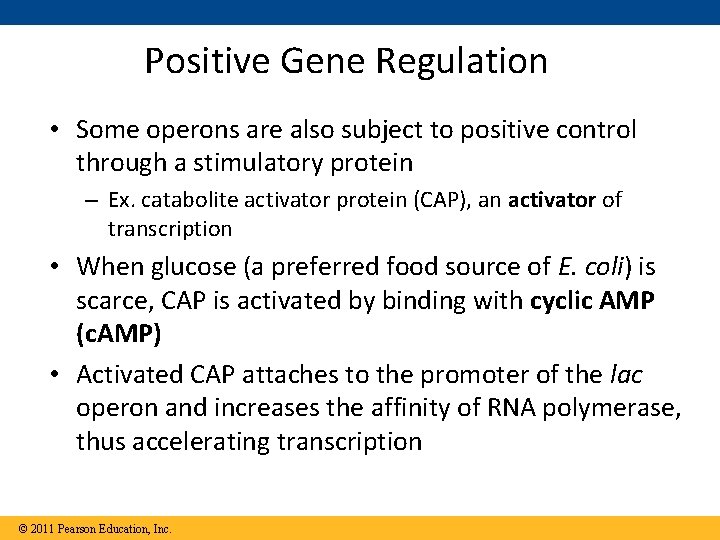
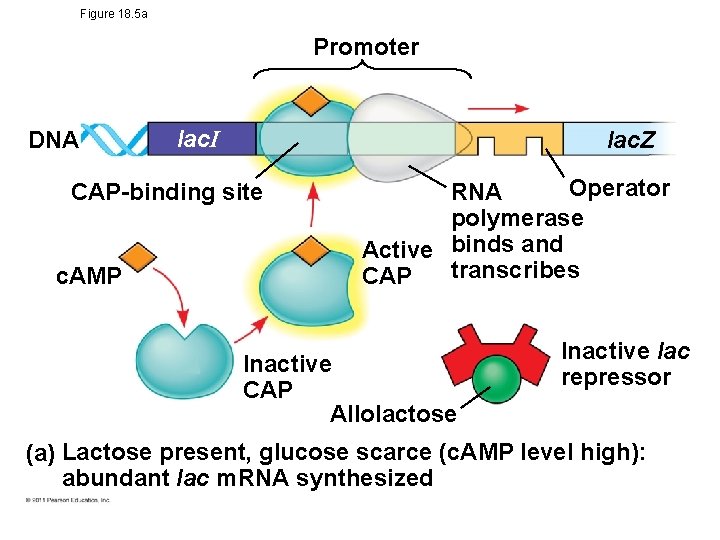
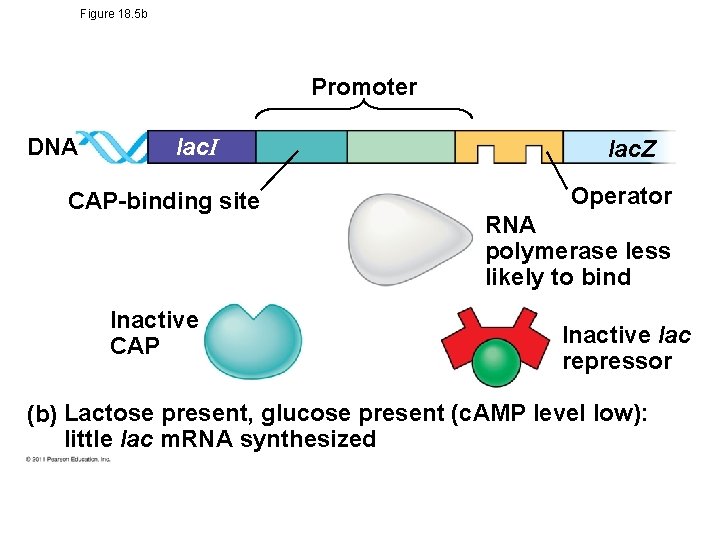
Positive Gene Regulation • Some operons are also subject to positive control through a stimulatory protein – Ex. catabolite activator protein (CAP), an activator of transcription • When glucose (a preferred food source of E. coli) is scarce, CAP is activated by binding with cyclic AMP (c. AMP) • Activated CAP attaches to the promoter of the lac operon and increases the affinity of RNA polymerase, thus accelerating transcription © 2011 Pearson Education, Inc.

Figure 18. 5 a Promoter DNA lac. I lac. Z CAP-binding site c. AMP Operator RNA polymerase Active binds and transcribes CAP Inactive CAP Allolactose Inactive lac repressor (a) Lactose present, glucose scarce (c. AMP level high): abundant lac m. RNA synthesized

Figure 18. 5 b Promoter DNA lac. I CAP-binding site Inactive CAP lac. Z Operator RNA polymerase less likely to bind Inactive lac repressor (b) Lactose present, glucose present (c. AMP level low): little lac m. RNA synthesized

Concept 18. 2: Eukaryotic gene expression is regulated at many stages • In multicellular organisms regulation of gene expression is essential for cell specialization • differential gene expression, the expression of different genes by cells with the same genome – Results in different cell types © 2011 Pearson Education, Inc.

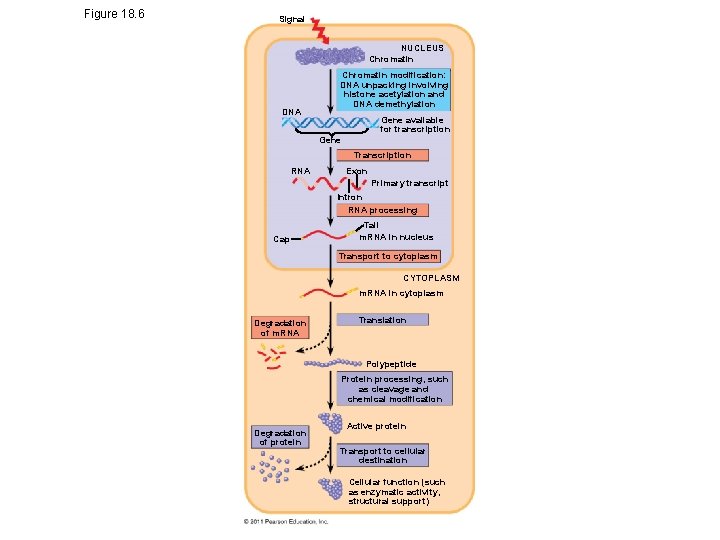
Figure 18. 6 Signal NUCLEUS Chromatin DNA Chromatin modification: DNA unpacking involving histone acetylation and DNA demethylation Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Cap Tail m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing, such as cleavage and chemical modification Degradation of protein Active protein Transport to cellular destination Cellular function (such as enzymatic activity, structural support)

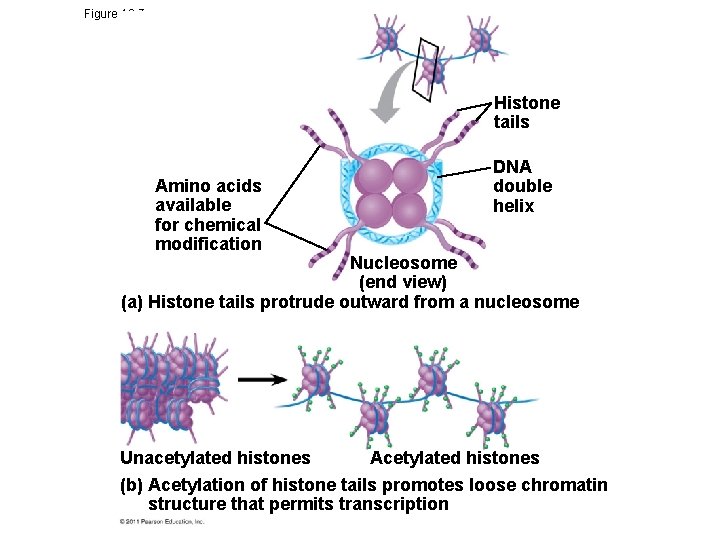
Regulation of Chromatin Structure • Genes within highly packed heterochromatin are usually not expressed • Chemical modifications to histones – histone acetylation, acetyl groups are attached to positively charged lysines in histone tails • loosens chromatin structure, promotes the initiation of transcription • addition of methyl groups (methylation) can condense chromatin; and DNA of chromatin influence both chromatin structure and gene expression © 2011 Pearson Education, Inc.

Figure 18. 7 Histone tails Amino acids available for chemical modification DNA double helix Nucleosome (end view) (a) Histone tails protrude outward from a nucleosome Acetylated histones Unacetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription

DNA Methylation • DNA methylation, the addition of methyl groups to certain bases in DNA, is associated with reduced transcription in some species • DNA methylation cause long-term inactivation of genes in cellular differentiation • In genomic imprinting, methylation regulates expression of either the maternal or paternal alleles of certain genes at the start of development © 2011 Pearson Education, Inc.

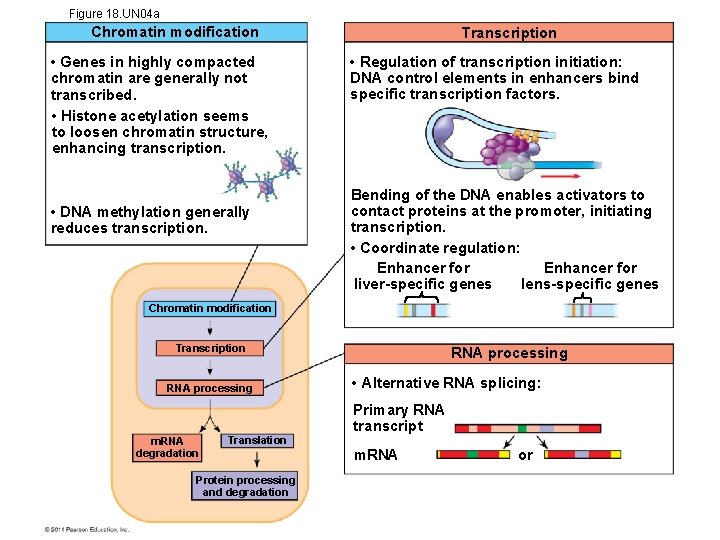
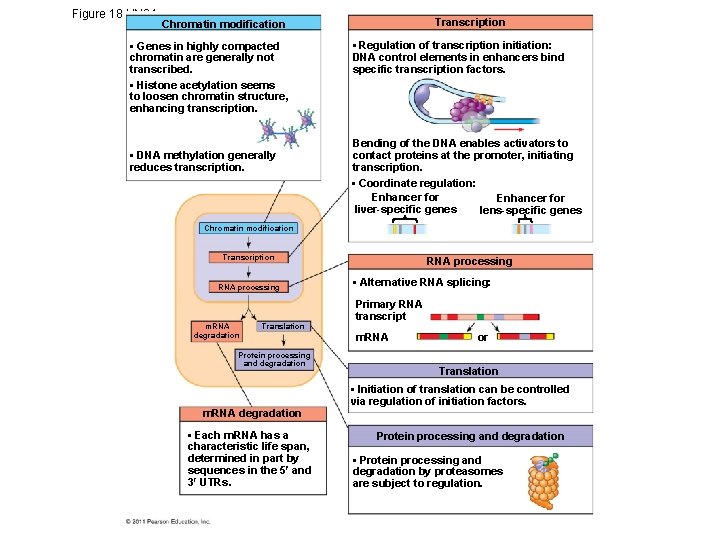
Figure 18. UN 04 a Chromatin modification • Genes in highly compacted chromatin are generally not transcribed. • Histone acetylation seems to loosen chromatin structure, enhancing transcription. • DNA methylation generally reduces transcription. Transcription • Regulation of transcription initiation: DNA control elements in enhancers bind specific transcription factors. Bending of the DNA enables activators to contact proteins at the promoter, initiating transcription. • Coordinate regulation: Enhancer for lens-specific genes liver-specific genes Chromatin modification Transcription RNA processing m. RNA degradation Translation Protein processing and degradation RNA processing • Alternative RNA splicing: Primary RNA transcript m. RNA or

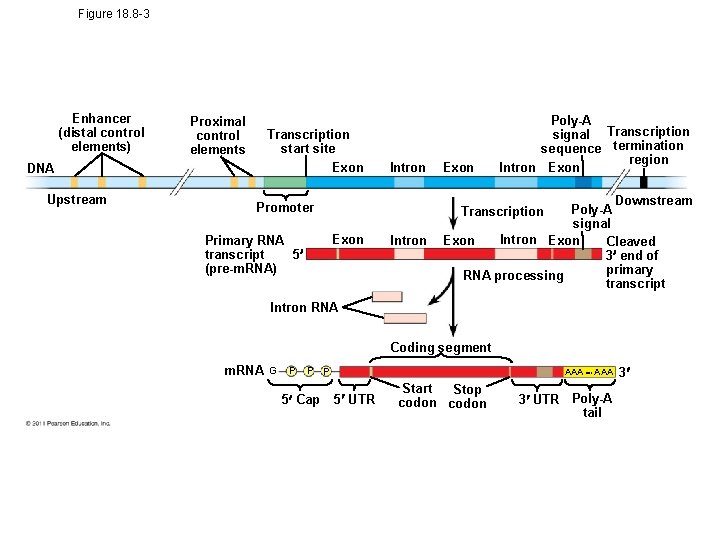
Regulation of Transcription Initiation • Chromatin-modifying enzymes provide initial control of gene expression by making a region of DNA either more or less able to bind the transcription machinery • Associated with most eukaryotic genes are multiple control elements, segments of noncoding DNA that serve as binding sites for transcription factors that help regulate transcription • Control elements and the transcription factors they bind are critical to the precise regulation of gene expression in different cell types © 2011 Pearson Education, Inc.

Figure 18. 8 -3 Enhancer (distal control elements) Proximal control elements Transcription start site Exon DNA Upstream Intron Exon Intron Downstream Poly-A signal Intron Exon Cleaved 3 end of primary RNA processing transcript Promoter Transcription Exon Primary RNA 5 transcript (pre-m. RNA) Poly-A signal Transcription sequence termination region Intron Exon Intron RNA Coding segment m. RNA G P P P 5 Cap AAA 5 UTR Start Stop codon 3 UTR Poly-A tail 3

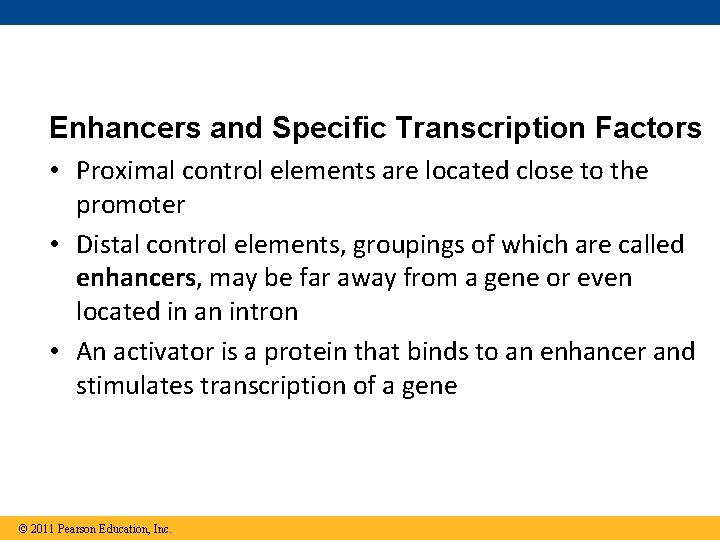
Enhancers and Specific Transcription Factors • Proximal control elements are located close to the promoter • Distal control elements, groupings of which are called enhancers, may be far away from a gene or even located in an intron • An activator is a protein that binds to an enhancer and stimulates transcription of a gene © 2011 Pearson Education, Inc.

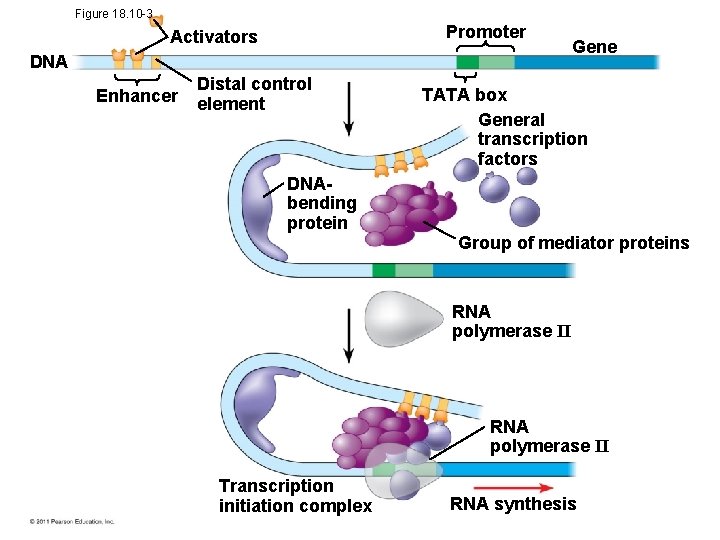
Figure 18. 10 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNAbending protein Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis

Coordinately Controlled Genes in Eukaryotes • Unlike the genes of a prokaryotic operon, each of the co-expressed eukaryotic genes has a promoter and control elements • These genes can be scattered over different chromosomes, but each has the same combination of control elements • Copies of the activators recognize specific control elements and promote simultaneous transcription of the genes © 2011 Pearson Education, Inc.

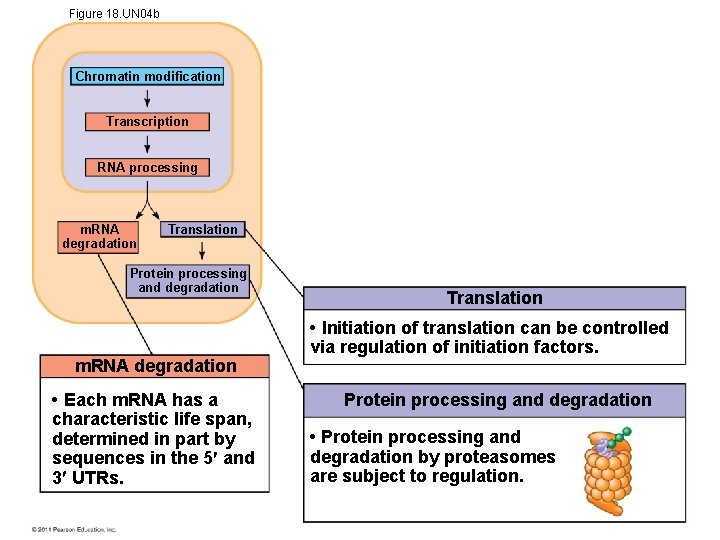
Figure 18. UN 04 b Chromatin modification Transcription RNA processing m. RNA degradation Translation Protein processing and degradation m. RNA degradation • Each m. RNA has a characteristic life span, determined in part by sequences in the 5 and 3 UTRs. Translation • Initiation of translation can be controlled via regulation of initiation factors. Protein processing and degradation • Protein processing and degradation by proteasomes are subject to regulation.

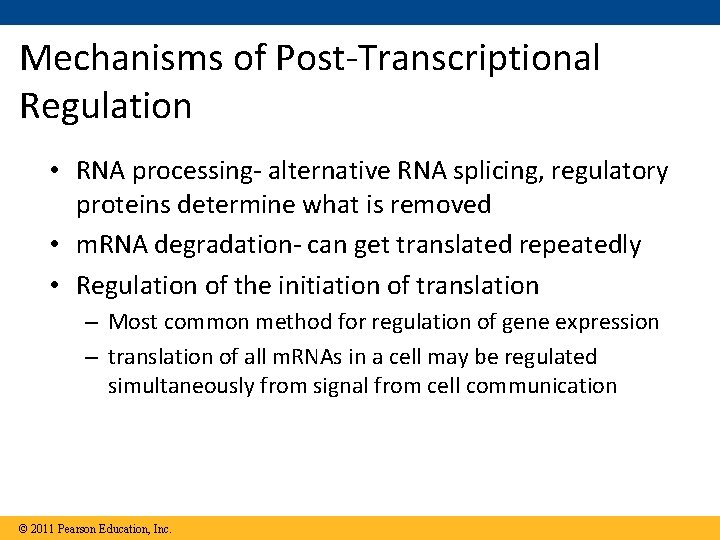
Mechanisms of Post-Transcriptional Regulation • RNA processing- alternative RNA splicing, regulatory proteins determine what is removed • m. RNA degradation- can get translated repeatedly • Regulation of the initiation of translation – Most common method for regulation of gene expression – translation of all m. RNAs in a cell may be regulated simultaneously from signal from cell communication © 2011 Pearson Education, Inc.

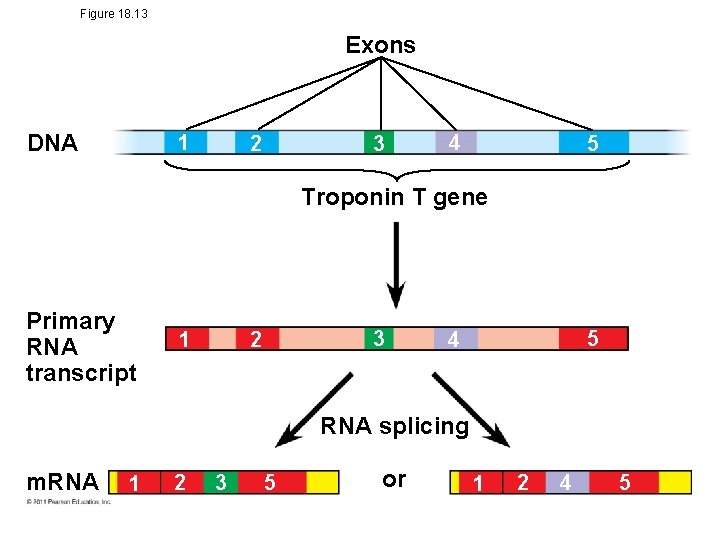
Figure 18. 13 Exons DNA 1 3 2 4 5 Troponin T gene Primary RNA transcript 3 2 1 5 4 RNA splicing m. RNA 1 2 3 5 or 1 2 4 5

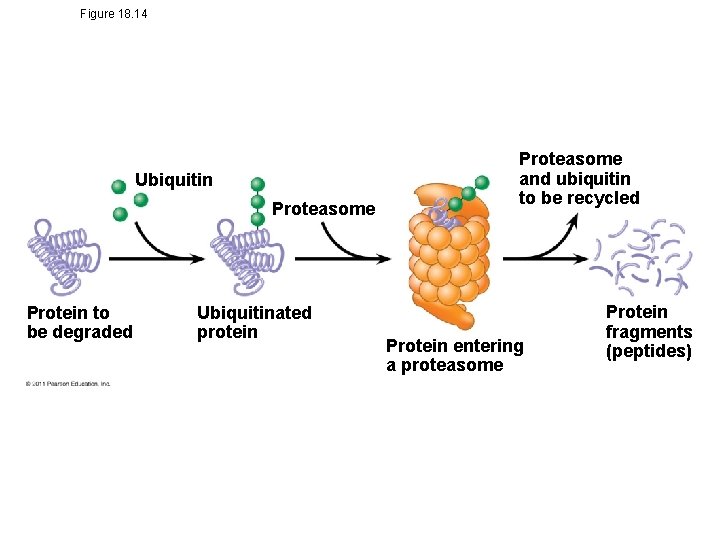
Protein Processing and Degradation • After translation, various types of protein processing, including cleavage and the addition of chemical groups, are subject to control • Proteasomes are giant protein complexes that bind protein molecules and degrade them © 2011 Pearson Education, Inc.

Figure 18. 14 Ubiquitin Proteasome Protein to be degraded Ubiquitinated protein Proteasome and ubiquitin to be recycled Protein entering a proteasome Protein fragments (peptides)

Concept 18. 3: Noncoding RNAs play multiple roles in controlling gene expression • A significant amount of the genome may be transcribed into noncoding RNAs (nc. RNAs) • Noncoding RNAs regulate gene expression at two points: m. RNA translation and chromatin configuration © 2011 Pearson Education, Inc.

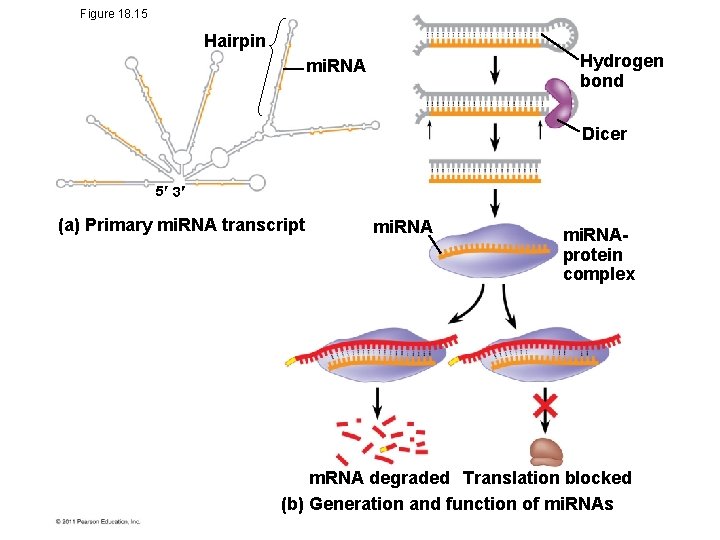
Effects on m. RNAs by Micro. RNAs and Small Interfering RNAs • Micro. RNAs (mi. RNAs) are small single-stranded RNA molecules that can bind to m. RNA • These can degrade m. RNA or block its translation © 2011 Pearson Education, Inc.

Figure 18. 15 Hairpin Hydrogen bond mi. RNA Dicer 5 3 (a) Primary mi. RNA transcript mi. RNAprotein complex m. RNA degraded Translation blocked (b) Generation and function of mi. RNAs

• The phenomenon of inhibition of gene expression by RNA molecules is called RNA interference (RNAi) • RNAi is caused by small interfering RNAs (si. RNAs) • si. RNAs and mi. RNAs are similar but form from different RNA precursors © 2011 Pearson Education, Inc.

Concept 18. 4: A program of differential gene expression leads to the different cell types in a multicellular organism • During embryonic development, a fertilized egg gives rise to many different cell types • Cell types are organized successively into tissues, organ systems, and the whole organism • Gene expression orchestrates the developmental programs of animals © 2011 Pearson Education, Inc.

A Genetic Program for Embryonic Development • The transformation from zygote to adult results from cell division, cell differentiation, and morphogenesis – Cell differentiation is the process by which cells become specialized in structure and function – The physical processes that give an organism its shape constitute morphogenesis © 2011 Pearson Education, Inc.

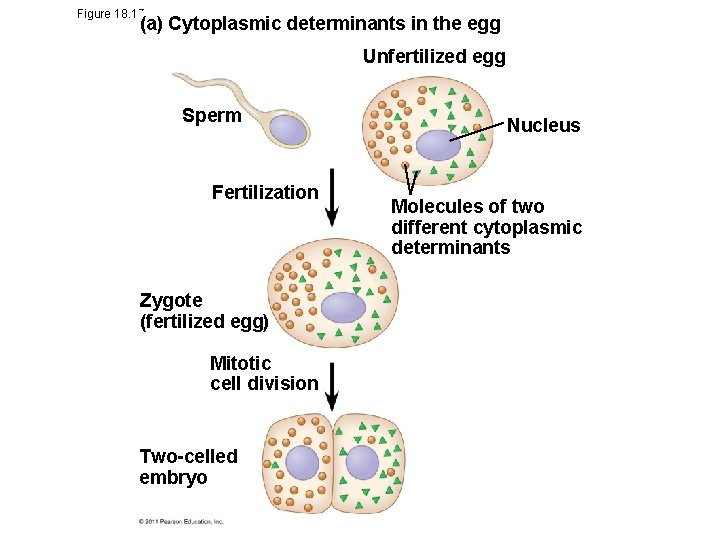
Cytoplasmic Determinants and Inductive Signals • An egg’s cytoplasm contains RNA, proteins, and other substances that are distributed unevenly in the unfertilized egg • Cytoplasmic determinants are maternal substances in the egg that influence early development © 2011 Pearson Education, Inc.

Figure 18. 17 a (a) Cytoplasmic determinants in the egg Unfertilized egg Sperm Fertilization Zygote (fertilized egg) Mitotic cell division Two-celled embryo Nucleus Molecules of two different cytoplasmic determinants

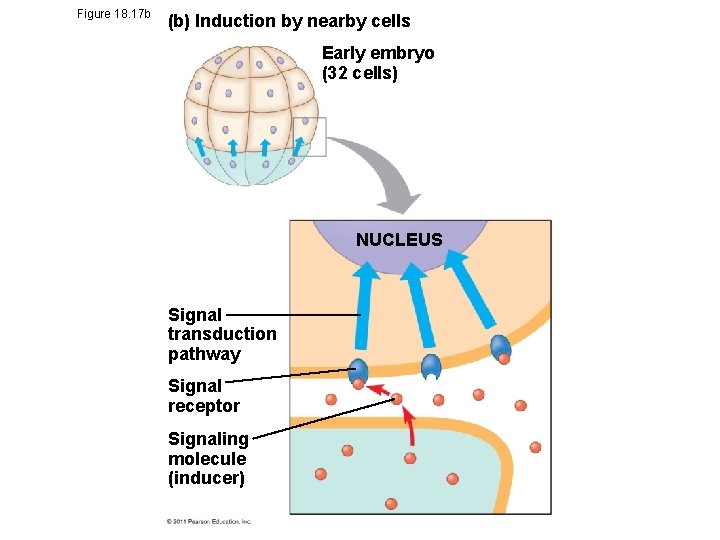
• In the process called induction, signal molecules from embryonic cells cause transcriptional changes in nearby target cells • Thus, interactions between cells induce differentiation of specialized cell types © 2011 Pearson Education, Inc.

Figure 18. 17 b (b) Induction by nearby cells Early embryo (32 cells) NUCLEUS Signal transduction pathway Signal receptor Signaling molecule (inducer)

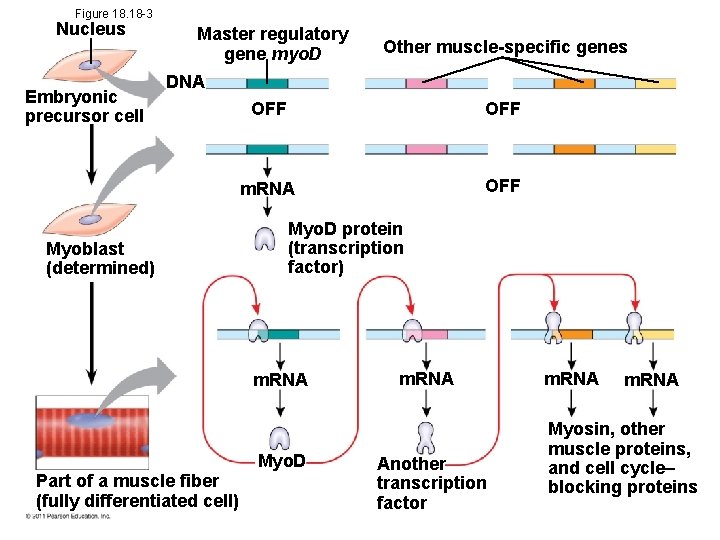
Sequential Regulation of Gene Expression During Cellular Differentiation • Determination commits a cell to its final fate • Determination precedes differentiation • Cell differentiation is marked by the production of tissue-specific proteins – Ex- muscle cell- regulatory gene commits cell to being muscle - makes myo D transcription factor to bind to enhancers and stimulates expression © 2011 Pearson Education, Inc.

Figure 18. 18 -3 Nucleus Embryonic precursor cell Master regulatory gene myo. D Other muscle-specific genes DNA Myoblast (determined) OFF m. RNA OFF Myo. D protein (transcription factor) m. RNA Part of a muscle fiber (fully differentiated cell) Myo. D m. RNA Another transcription factor m. RNA Myosin, other muscle proteins, and cell cycle– blocking proteins

Pattern Formation: Setting Up the Body Plan • Pattern formation is the development of a spatial organization of tissues and organs – In animals, pattern formation begins with the establishment of the major axes – Levels of morphogens establish an embryo’s axes and other features • Positional information, the molecular cues that control pattern formation, tells a cell its location relative to the body axes and to neighboring cells – Studied extensively with Drosophila- used mutants © 2011 Pearson Education, Inc.

Concept 18. 5: Cancer results from genetic changes that affect cell cycle control • The gene regulation systems that go wrong during cancer are the very same systems involved in embryonic development © 2011 Pearson Education, Inc.

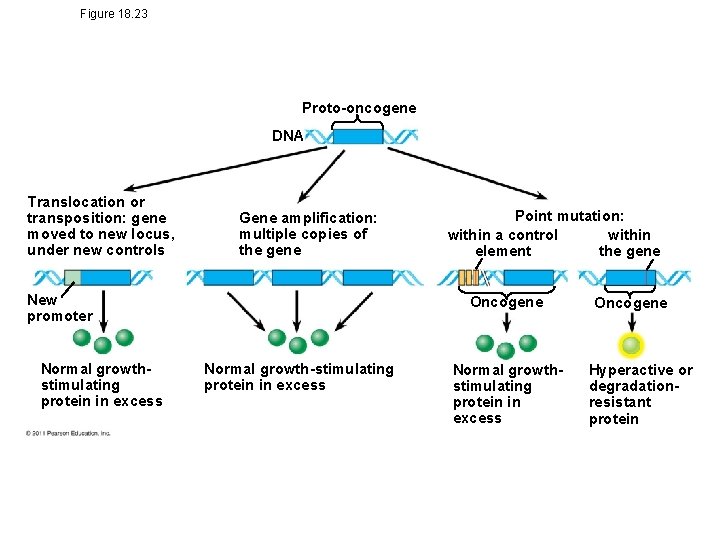
Types of Genes Associated with Cancer • Cancer can be caused by mutations to genes that regulate cell growth and division – Oncogenes are cancer-causing genes – Proto-oncogenes are the corresponding normal cellular genes responsible for normal cell growth and division – Conversion of a proto-oncogene to an oncogene can lead to abnormal stimulation of the cell cycle • Tumor viruses can cause cancer in animals including humans © 2011 Pearson Education, Inc.

Figure 18. 23 Proto-oncogene DNA Translocation or transposition: gene moved to new locus, under new controls Gene amplification: multiple copies of the gene New promoter Normal growthstimulating protein in excess Point mutation: within a control within element the gene Oncogene Normal growth-stimulating protein in excess Normal growthstimulating protein in excess Oncogene Hyperactive or degradationresistant protein

• Proto-oncogenes can be converted to oncogenes by – Movement of DNA within the genome: if it ends up near an active promoter, transcription may increase – Amplification of a proto-oncogene: increases the number of copies of the gene – Point mutations in the proto-oncogene or its control elements: cause an increase in gene expression © 2011 Pearson Education, Inc.

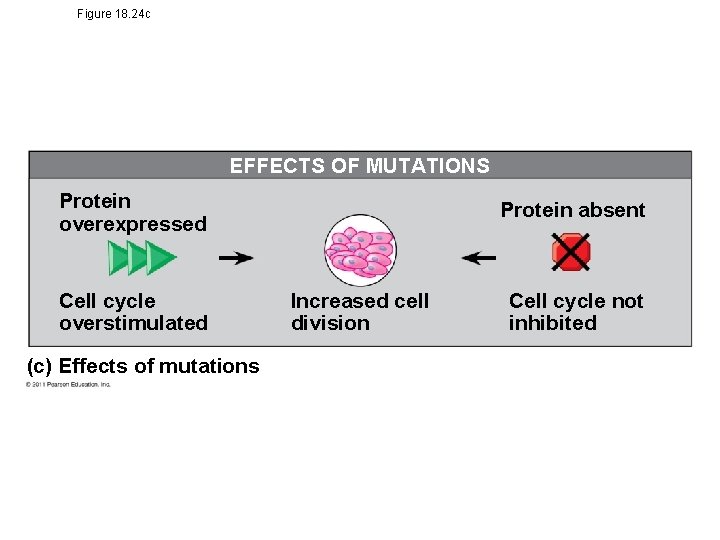
Tumor-Suppressor Genes • Tumor-suppressor genes help prevent uncontrolled cell growth • Mutations that decrease protein products of tumorsuppressor genes may contribute to cancer onset • Tumor-suppressor proteins – Repair damaged DNA – Control cell adhesion – Inhibit the cell cycle in the cell-signaling pathway © 2011 Pearson Education, Inc.

Figure 18. 24 c EFFECTS OF MUTATIONS Protein overexpressed Cell cycle overstimulated (c) Effects of mutations Protein absent Increased cell division Cell cycle not inhibited

Figure 18. UN 04 Transcription Chromatin modification • Genes in highly compacted chromatin are generally not transcribed. • Histone acetylation seems to loosen chromatin structure, enhancing transcription. • DNA methylation generally reduces transcription. • Regulation of transcription initiation: DNA control elements in enhancers bind specific transcription factors. Bending of the DNA enables activators to contact proteins at the promoter, initiating transcription. • Coordinate regulation: Enhancer for liver-specific genes lens-specific genes Chromatin modification Transcription RNA processing m. RNA degradation Translation Protein processing and degradation m. RNA degradation • Each m. RNA has a characteristic life span, determined in part by sequences in the 5 and 3 UTRs. RNA processing • Alternative RNA splicing: Primary RNA transcript m. RNA or Translation • Initiation of translation can be controlled via regulation of initiation factors. Protein processing and degradation • Protein processing and degradation by proteasomes are subject to regulation.