ENZYMES DR SHAMPA SARKAR ASSISTANT PROFESSOR DEPARTMENT OF





























- Slides: 76

ENZYMES DR. SHAMPA SARKAR ASSISTANT PROFESSOR DEPARTMENT OF ZOOLOGY NARASINHA DUTT COLLEGE

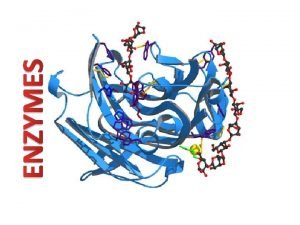
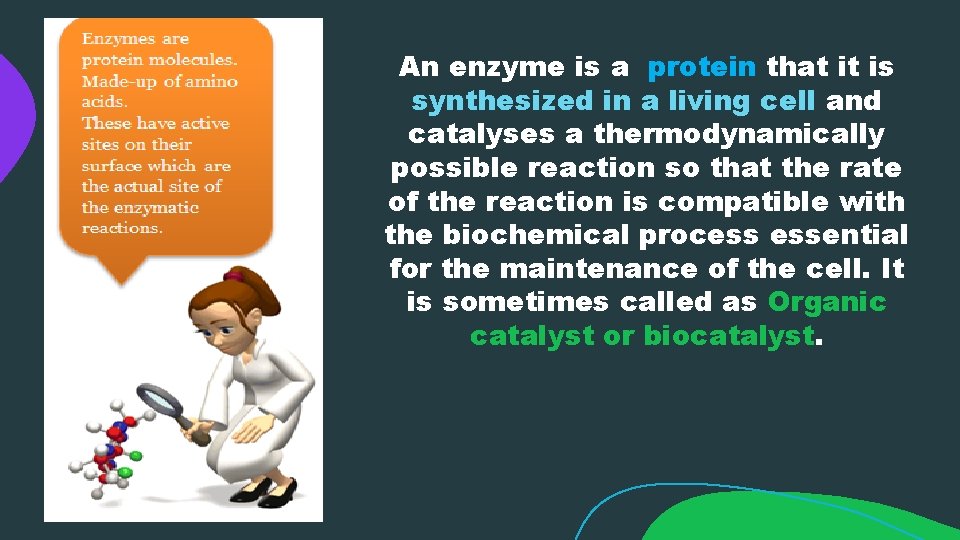
An enzyme is a protein that it is synthesized in a living cell and catalyses a thermodynamically possible reaction so that the rate of the reaction is compatible with the biochemical process essential for the maintenance of the cell. It is sometimes called as Organic catalyst or biocatalyst.

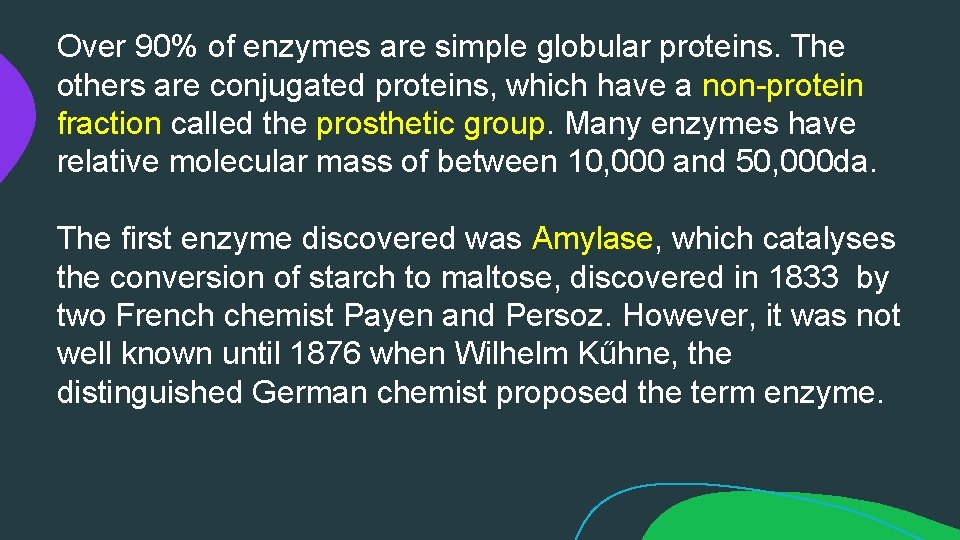
Over 90% of enzymes are simple globular proteins. The others are conjugated proteins, which have a non-protein fraction called the prosthetic group. Many enzymes have relative molecular mass of between 10, 000 and 50, 000 da. The first enzyme discovered was Amylase, which catalyses the conversion of starch to maltose, discovered in 1833 by two French chemist Payen and Persoz. However, it was not well known until 1876 when Wilhelm Kűhne, the distinguished German chemist proposed the term enzyme.

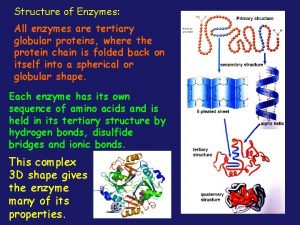
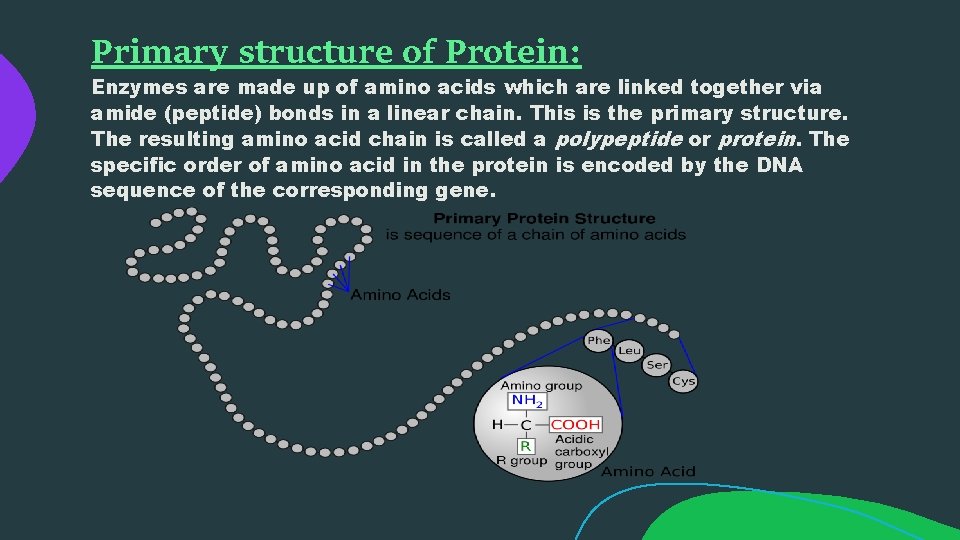
Primary structure of Protein: Enzymes are made up of amino acids which are linked together via amide (peptide) bonds in a linear chain. This is the primary structure. The resulting amino acid chain is called a polypeptide or protein. The specific order of amino acid in the protein is encoded by the DNA sequence of the corresponding gene.

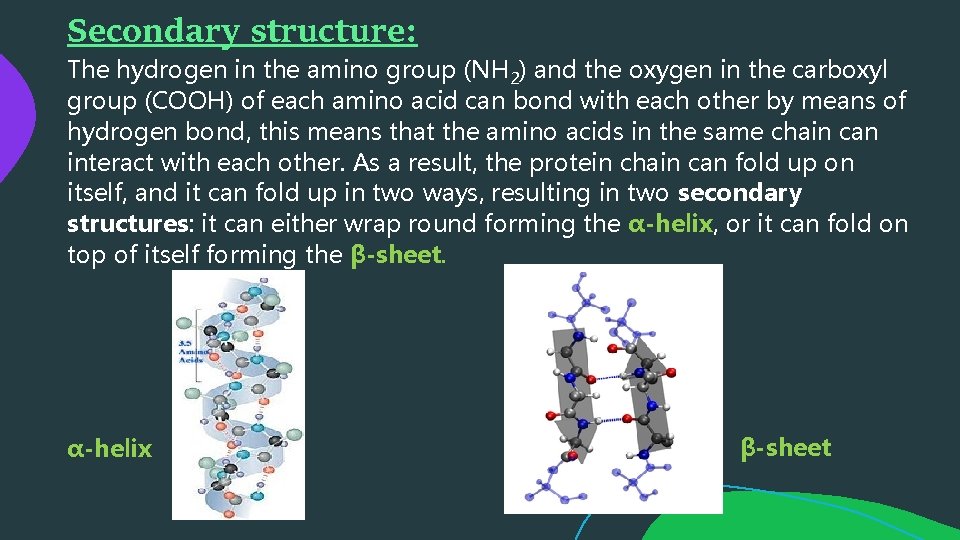
Secondary structure: The hydrogen in the amino group (NH 2) and the oxygen in the carboxyl group (COOH) of each amino acid can bond with each other by means of hydrogen bond, this means that the amino acids in the same chain can interact with each other. As a result, the protein chain can fold up on itself, and it can fold up in two ways, resulting in two secondary structures: it can either wrap round forming the α-helix, or it can fold on top of itself forming the β-sheet. α-helix β-sheet

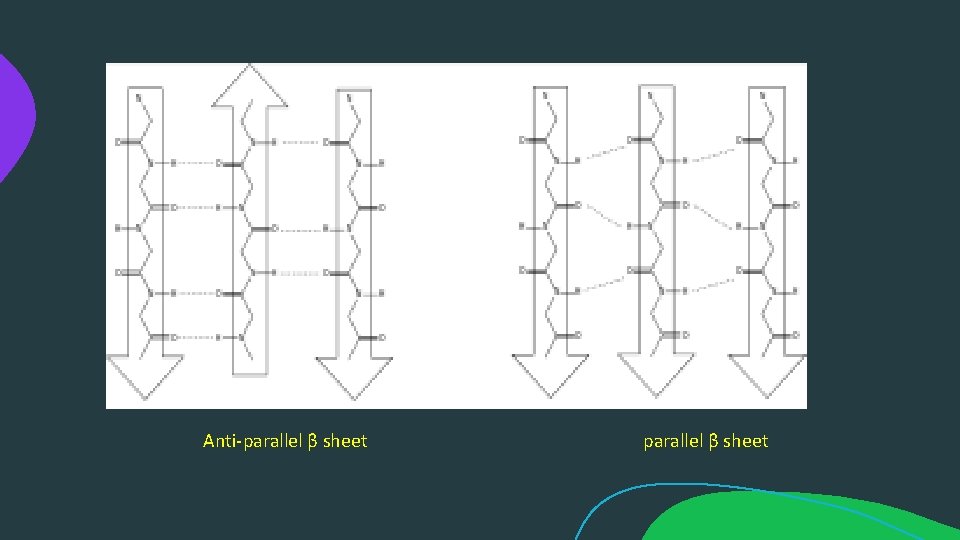
Anti-parallel β sheet parallel β sheet

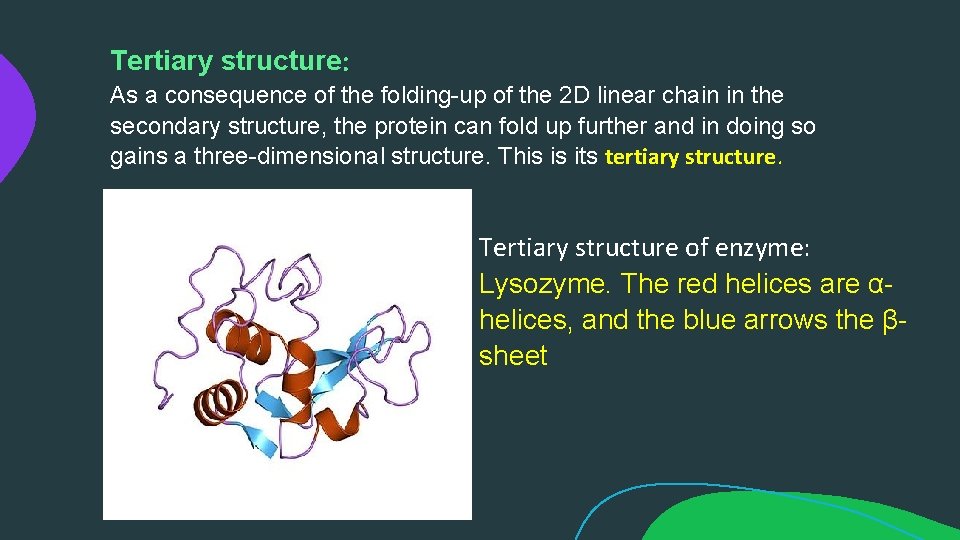
Tertiary structure: As a consequence of the folding-up of the 2 D linear chain in the secondary structure, the protein can fold up further and in doing so gains a three-dimensional structure. This is its tertiary structure. Tertiary structure of enzyme: Lysozyme. The red helices are αhelices, and the blue arrows the βsheet

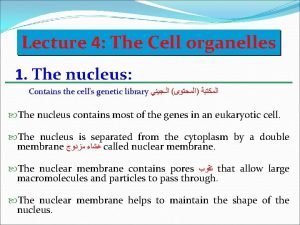
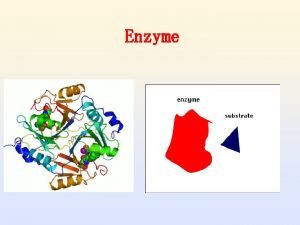
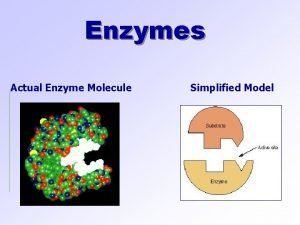
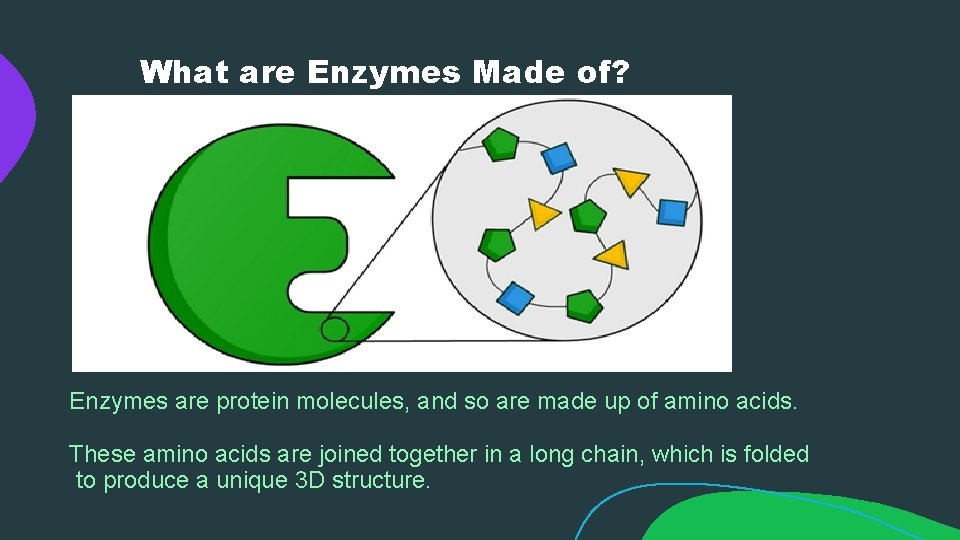
What are Enzymes Made of? Enzymes are protein molecules, and so are made up of amino acids. These amino acids are joined together in a long chain, which is folded to produce a unique 3 D structure.

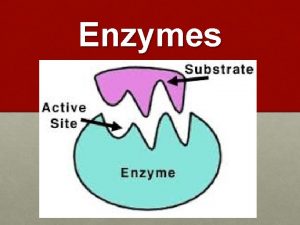
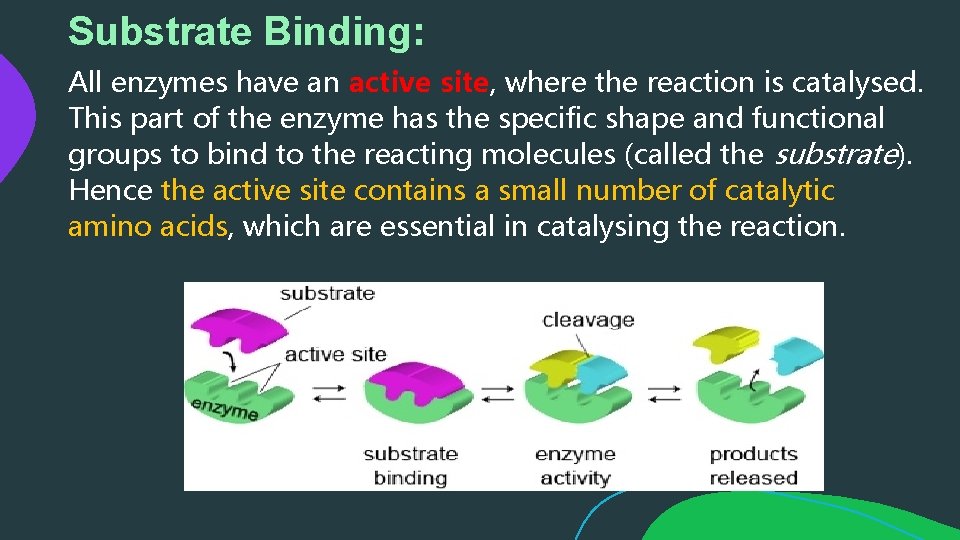
Substrate Binding: All enzymes have an active site, where the reaction is catalysed. This part of the enzyme has the specific shape and functional groups to bind to the reacting molecules (called the substrate). Hence the active site contains a small number of catalytic amino acids, which are essential in catalysing the reaction.


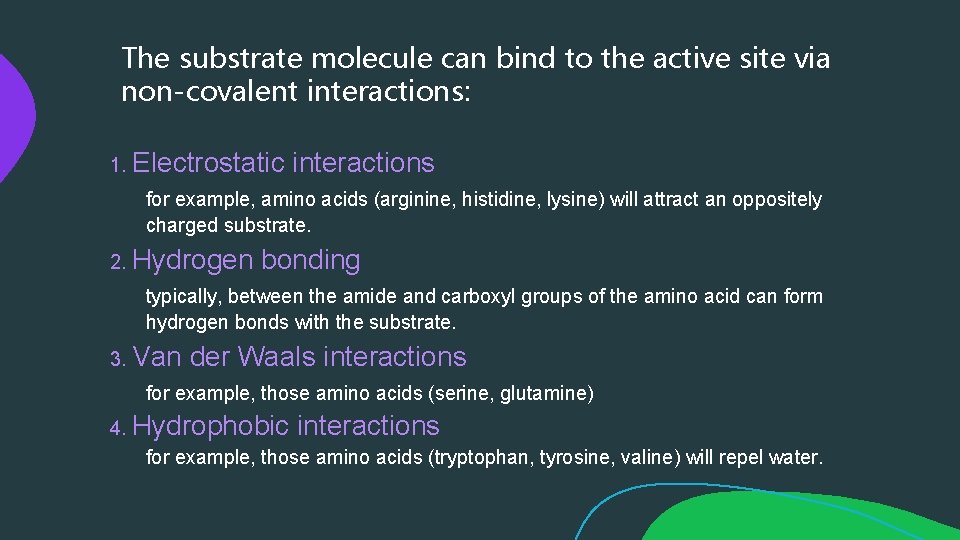
The substrate molecule can bind to the active site via non-covalent interactions: 1. Electrostatic interactions for example, amino acids (arginine, histidine, lysine) will attract an oppositely charged substrate. 2. Hydrogen bonding typically, between the amide and carboxyl groups of the amino acid can form hydrogen bonds with the substrate. 3. Van der Waals interactions for example, those amino acids (serine, glutamine) 4. Hydrophobic interactions for example, those amino acids (tryptophan, tyrosine, valine) will repel water.


Cofactor A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's activity as a catalyst, a substance that increases the rate of a chemical reaction. Cofactors can be considered "helper molecules" that assist in biochemical transformations. If the cofactor is removed from a complete enzyme (holoenzyme), the protein component (apoenzyme) no longer has catalytic activity. A cofactor that is firmly bound to the apoenzyme and cannot be removed without denaturing the latter is termed a prosthetic group; most such groups contain an atom of metal such as copper or iron. A cofactor that is bound loosely to the apoenzyme and can be readily separated from it is called a coenzyme. Coenzymes take part in the catalysed reaction, are modified during the reaction, and may require another enzyme-catalyzed reaction for restoration to their original state.


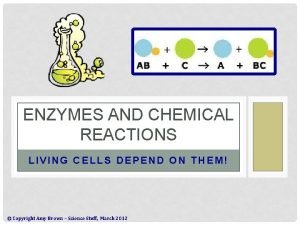
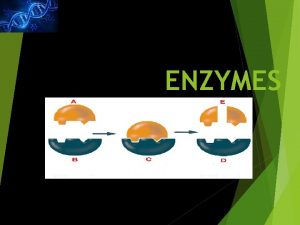
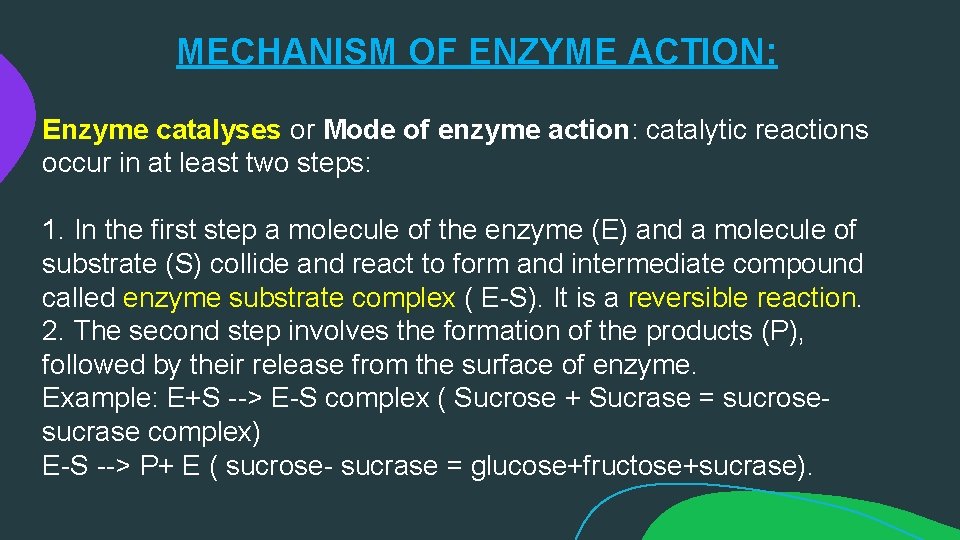
MECHANISM OF ENZYME ACTION: Enzyme catalyses or Mode of enzyme action: catalytic reactions occur in at least two steps: 1. In the first step a molecule of the enzyme (E) and a molecule of substrate (S) collide and react to form and intermediate compound called enzyme substrate complex ( E-S). It is a reversible reaction. 2. The second step involves the formation of the products (P), followed by their release from the surface of enzyme. Example: E+S --> E-S complex ( Sucrose + Sucrase = sucrosesucrase complex) E-S --> P+ E ( sucrose- sucrase = glucose+fructose+sucrase).


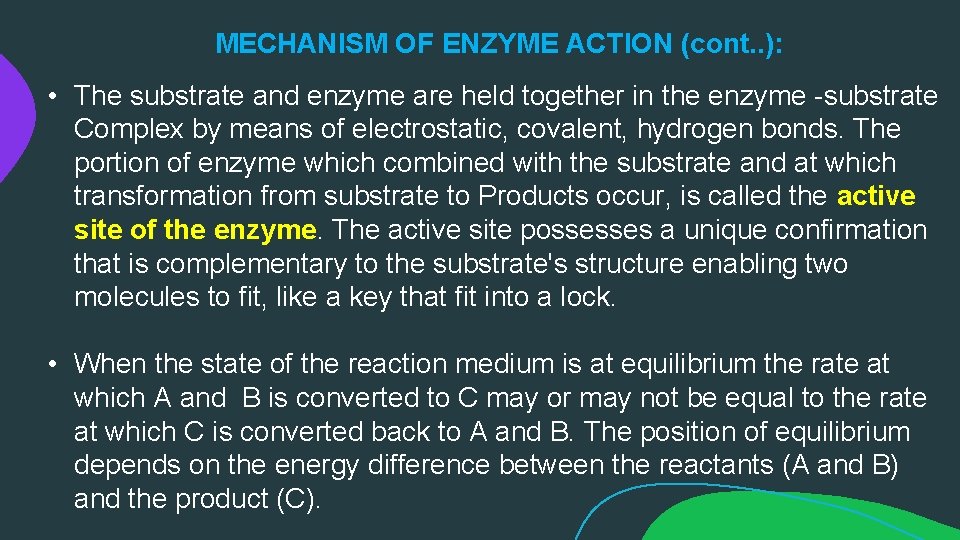
MECHANISM OF ENZYME ACTION (cont. . ): • The substrate and enzyme are held together in the enzyme -substrate Complex by means of electrostatic, covalent, hydrogen bonds. The portion of enzyme which combined with the substrate and at which transformation from substrate to Products occur, is called the active site of the enzyme. The active site possesses a unique confirmation that is complementary to the substrate's structure enabling two molecules to fit, like a key that fit into a lock. • When the state of the reaction medium is at equilibrium the rate at which A and B is converted to C may or may not be equal to the rate at which C is converted back to A and B. The position of equilibrium depends on the energy difference between the reactants (A and B) and the product (C).

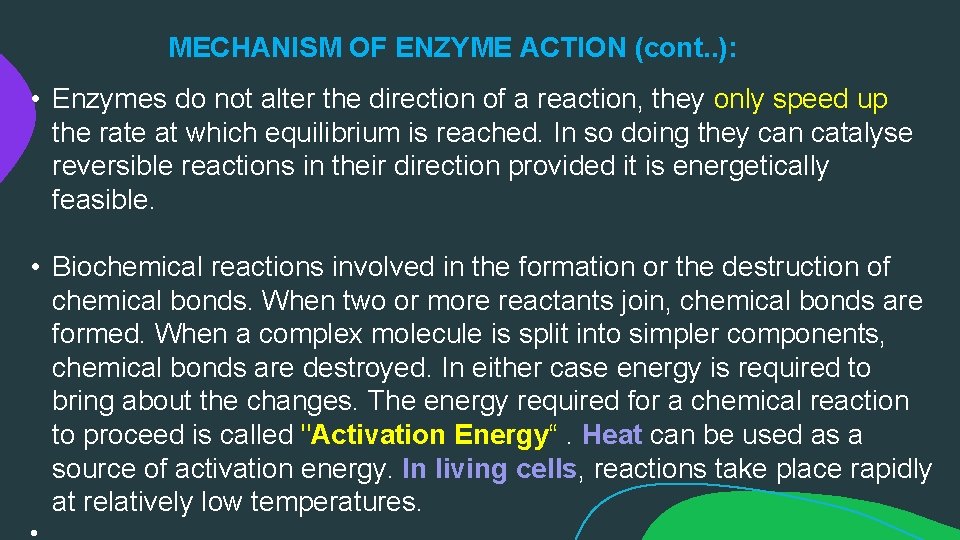
MECHANISM OF ENZYME ACTION (cont. . ): • Enzymes do not alter the direction of a reaction, they only speed up the rate at which equilibrium is reached. In so doing they can catalyse reversible reactions in their direction provided it is energetically feasible. • Biochemical reactions involved in the formation or the destruction of chemical bonds. When two or more reactants join, chemical bonds are formed. When a complex molecule is split into simpler components, chemical bonds are destroyed. In either case energy is required to bring about the changes. The energy required for a chemical reaction to proceed is called "Activation Energy“. Heat can be used as a source of activation energy. In living cells, reactions take place rapidly at relatively low temperatures.

The maximum number of molecules of substrate that can be converted to product by one enzyme molecule per minute is called as turnover number of that enzyme. A turnover number of 1000 is typical for enzymes.

Important characteristic features of enzyme 1. Catalytic power (ratio of enzyme catalysed rate of a reaction to the uncatalyzed rate) 2. Regulation ( control of enzymatic reaction) 3. Specificity ( selectivity of enzyme to their substrate)

Specificity of enzyme action: What is enzyme specificity? Specificity is the ability of an enzyme to choose exact substrate from a group of similar chemical molecules. The specificity is actually a molecular recognition mechanism and it operates through the structural and conformational complementary between enzyme and substrate. Enzymes show different degrees of specificity towards their substrate.

The specificity shown by enzymes are grouped into 6 categories. 1. Bond specificity 2. Group specificity 3. Substrate specificity 4. Stereo specificity(optical specificity) 5. Geometrical specificity 6. Cofactors specificity

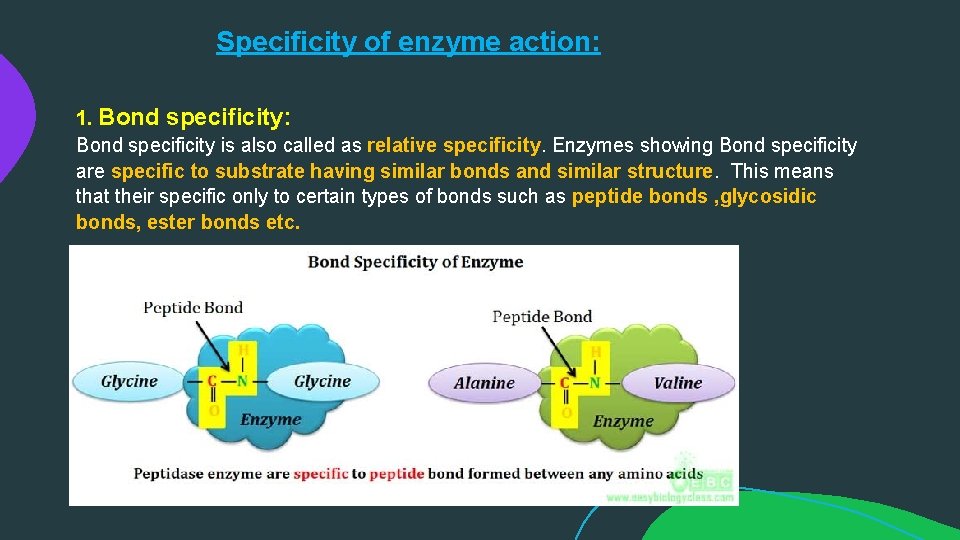
Specificity of enzyme action: 1. Bond specificity: Bond specificity is also called as relative specificity. Enzymes showing Bond specificity are specific to substrate having similar bonds and similar structure. This means that their specific only to certain types of bonds such as peptide bonds , glycosidic bonds, ester bonds etc.

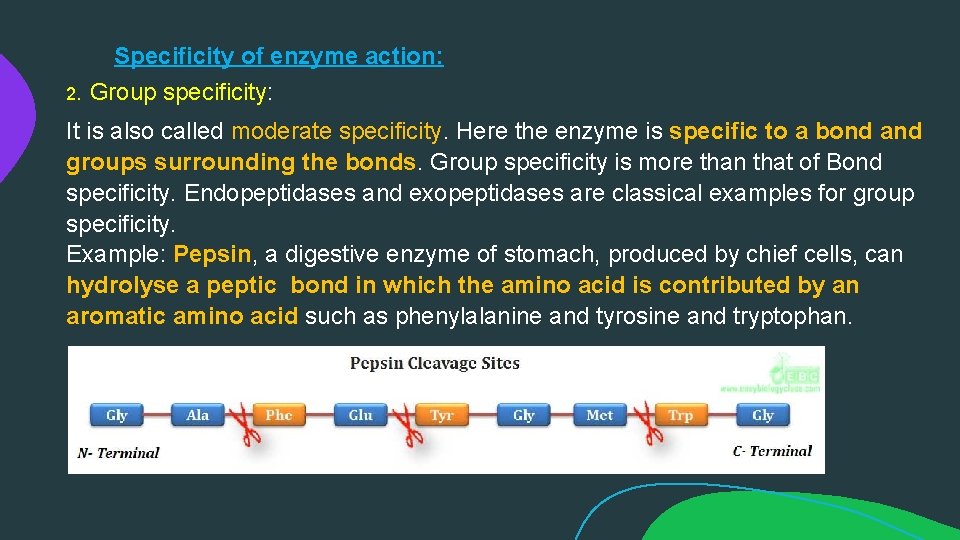
Specificity of enzyme action: 2. Group specificity: It is also called moderate specificity. Here the enzyme is specific to a bond and groups surrounding the bonds. Group specificity is more than that of Bond specificity. Endopeptidases and exopeptidases are classical examples for group specificity. Example: Pepsin, a digestive enzyme of stomach, produced by chief cells, can hydrolyse a peptic bond in which the amino acid is contributed by an aromatic amino acid such as phenylalanine and tyrosine and tryptophan.

Specificity of enzyme action: 3. Substrate specificity: Substrate specificity is also called as absolute specificity since here the specificity is very high. Enzymes showing substrate specificity are specific only to one substrate and one reaction. Example: enzyme lactase can only hydrolyse the β 1 -4 glycosidic bond of lactose to yield galactose and glucose. Similarly, Maltase can only act on the α 1 -4 Glycosidic linkage of two glucose molecules in maltose.


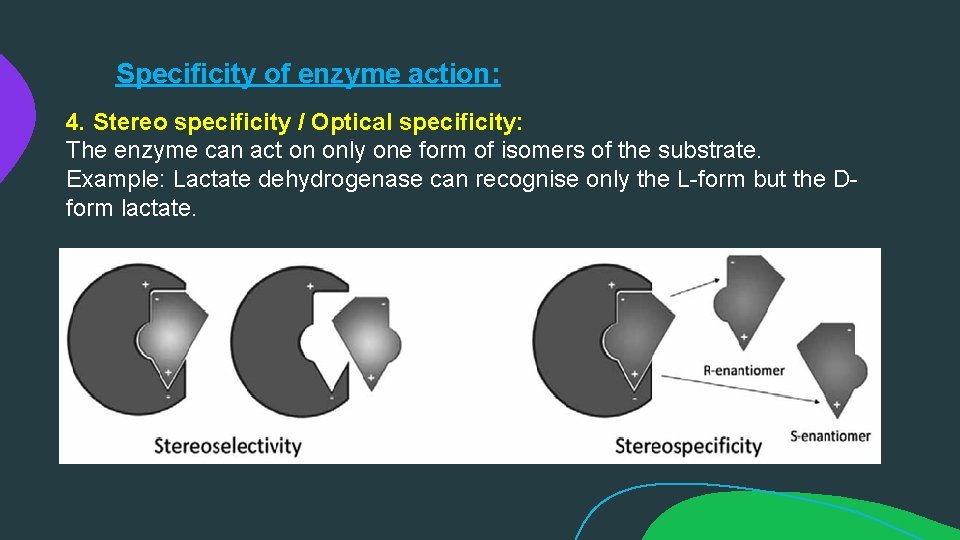
Specificity of enzyme action: 4. Stereo specificity / Optical specificity: The enzyme can act on only one form of isomers of the substrate. Example: Lactate dehydrogenase can recognise only the L-form but the Dform lactate.


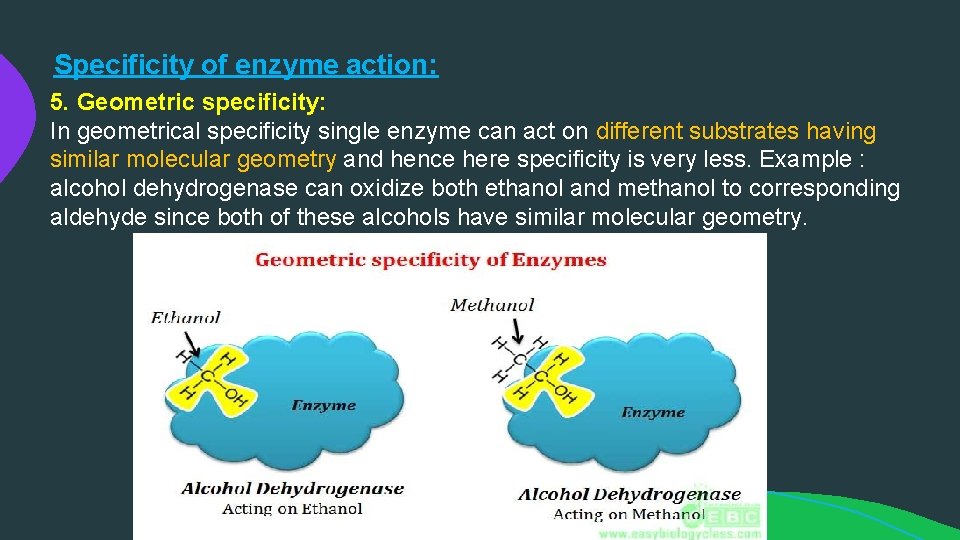
Specificity of enzyme action: 5. Geometric specificity: In geometrical specificity single enzyme can act on different substrates having similar molecular geometry and hence here specificity is very less. Example : alcohol dehydrogenase can oxidize both ethanol and methanol to corresponding aldehyde since both of these alcohols have similar molecular geometry.

6. Co-factor specificity: Co-factors are non-protein part of enzyme required for the functioning of some enzymes. Enzyme which requires cofactors for their activity , shows cofactors specificity. Only correct combination of substrate and cofactor allows enzyme reaction. In the absence of specific cofactor, the enzyme will be inactive even if there are plenty of substrates.

Enzyme nomenclature The International Union of Biochemistry and Molecular Biology (IUB in 1961) have developed a nomenclature for enzymes. Enzymes are identified by EC (Enzyme Commission) numbers. These are also valuable for relating the information to other databases. They were divided into 6 major classes according to the type of reaction catalysed and a seventh, the translocases, was added in 2018.

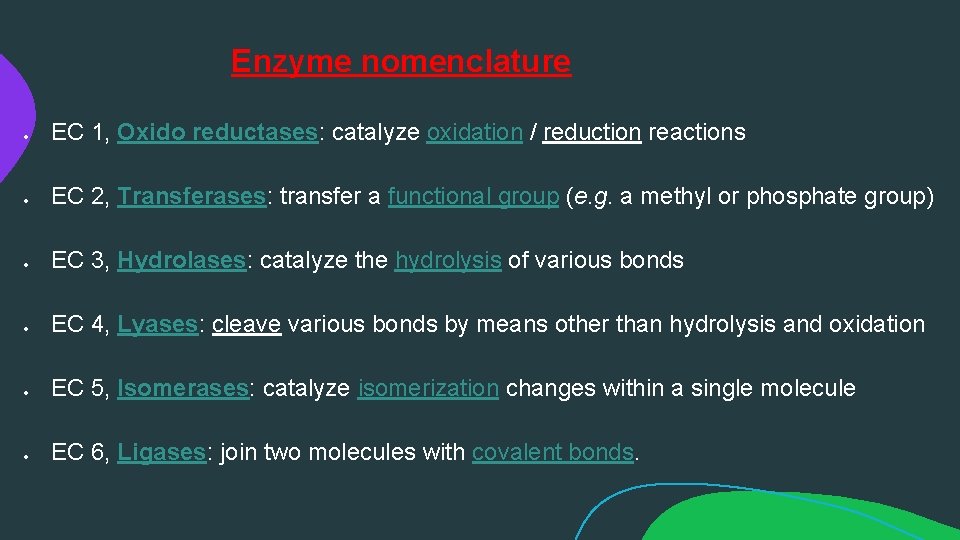
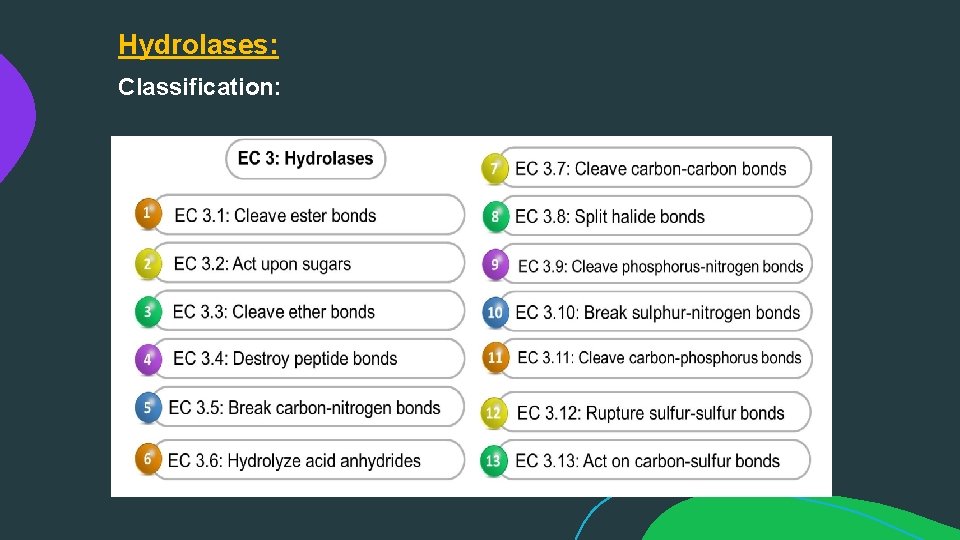
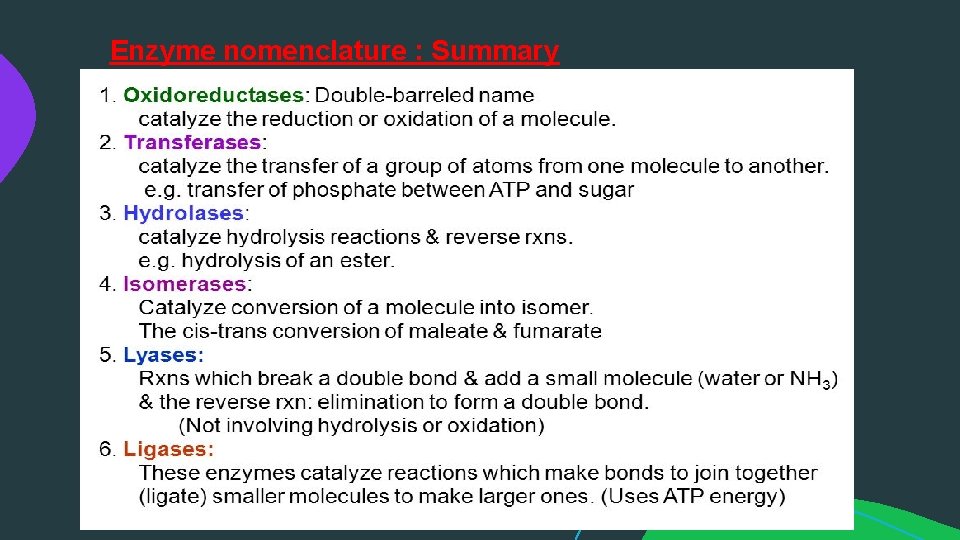
Enzyme nomenclature EC 1, Oxido reductases: catalyze oxidation / reduction reactions EC 2, Transferases: transfer a functional group (e. g. a methyl or phosphate group) EC 3, Hydrolases: catalyze the hydrolysis of various bonds EC 4, Lyases: cleave various bonds by means other than hydrolysis and oxidation EC 5, Isomerases: catalyze isomerization changes within a single molecule EC 6, Ligases: join two molecules with covalent bonds.

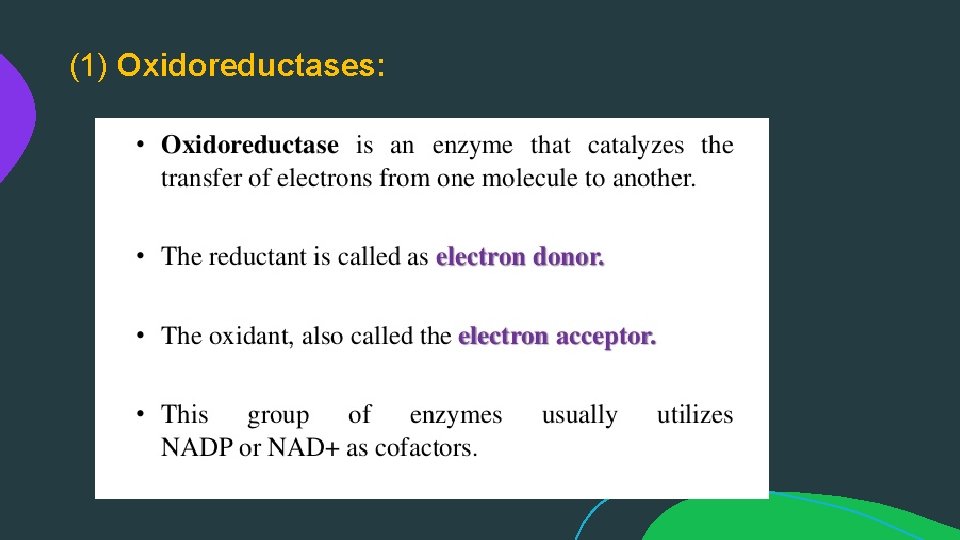
(1) Oxidoreductases:

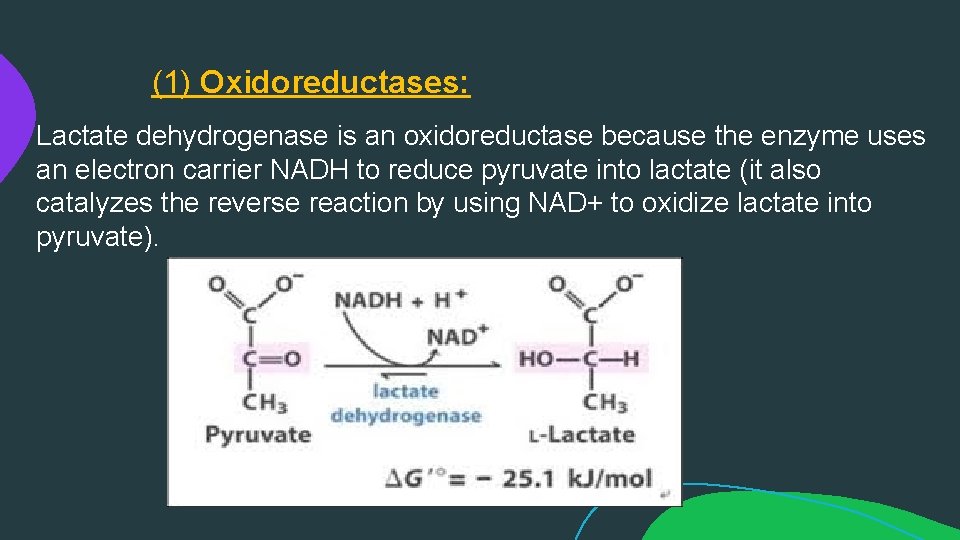
(1) Oxidoreductases: Lactate dehydrogenase is an oxidoreductase because the enzyme uses an electron carrier NADH to reduce pyruvate into lactate (it also catalyzes the reverse reaction by using NAD+ to oxidize lactate into pyruvate).


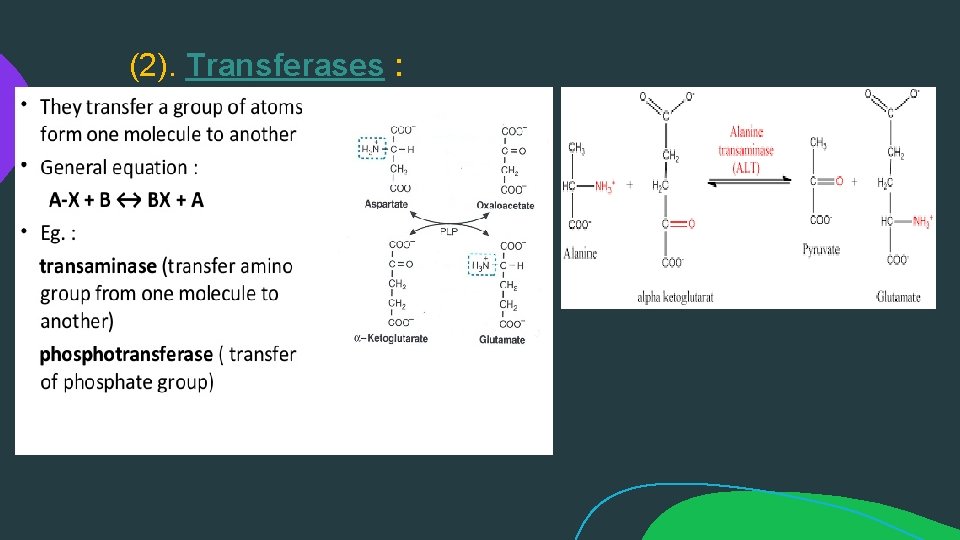
(2). Transferases :

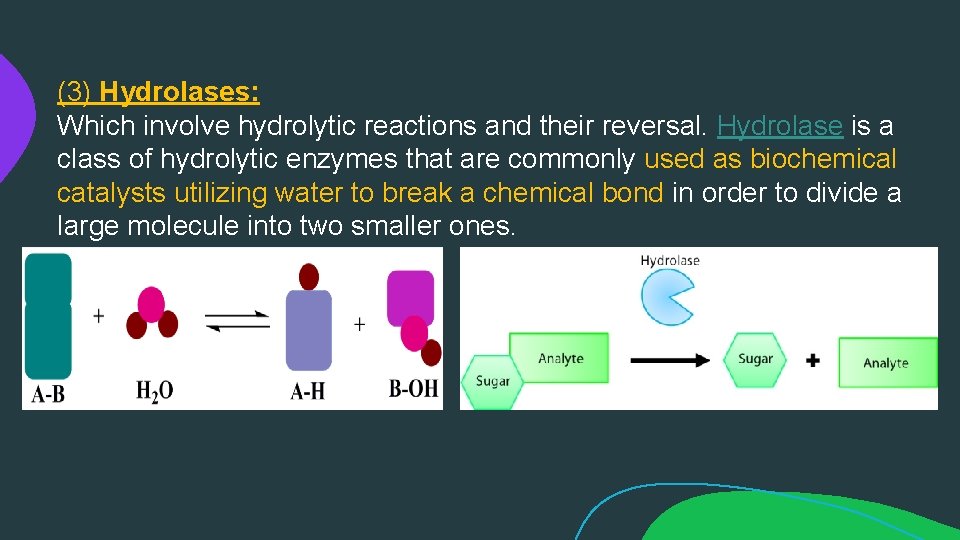
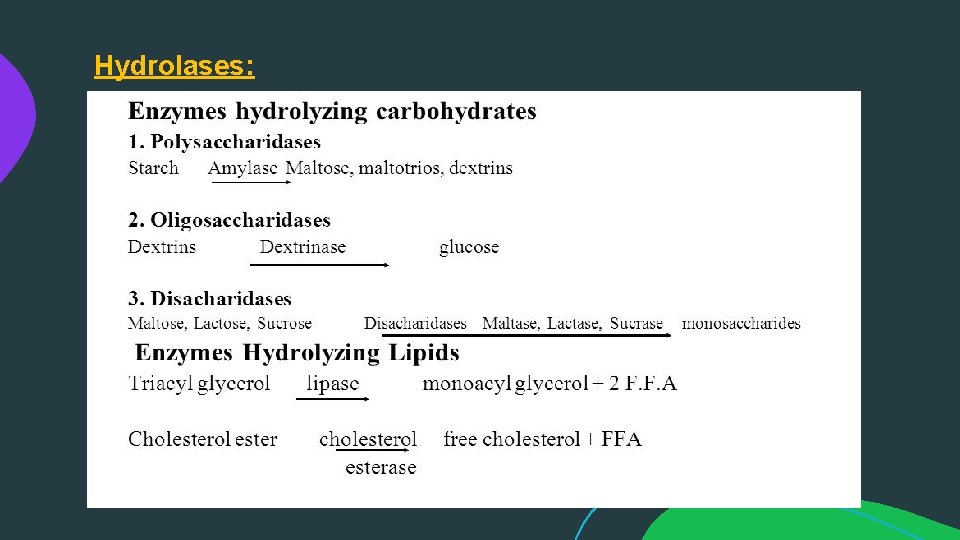
(3) Hydrolases: Which involve hydrolytic reactions and their reversal. Hydrolase is a class of hydrolytic enzymes that are commonly used as biochemical catalysts utilizing water to break a chemical bond in order to divide a large molecule into two smaller ones.

Hydrolases: Classification:

Hydrolases:

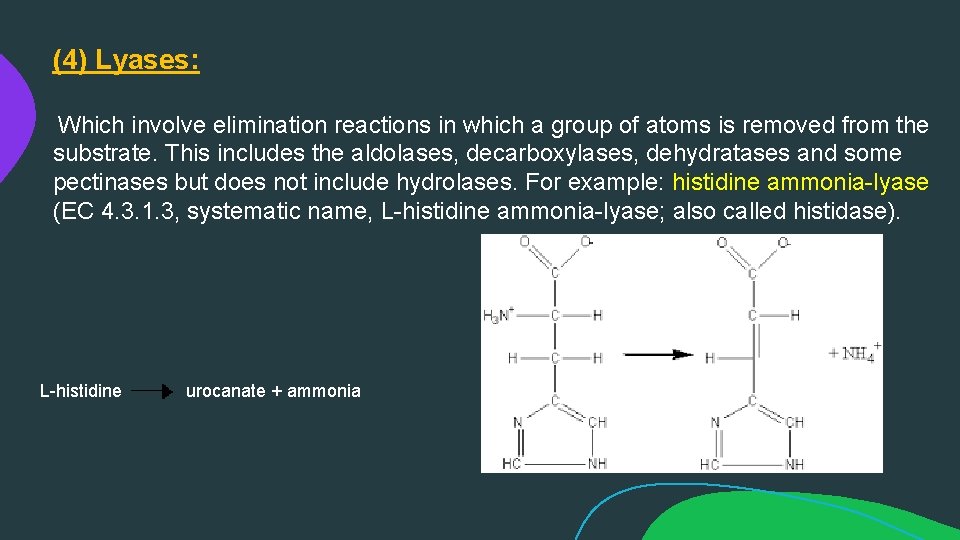
(4) Lyases: Which involve elimination reactions in which a group of atoms is removed from the substrate. This includes the aldolases, decarboxylases, dehydratases and some pectinases but does not include hydrolases. For example: histidine ammonia-lyase (EC 4. 3. 1. 3, systematic name, L-histidine ammonia-lyase; also called histidase). L-histidine urocanate + ammonia

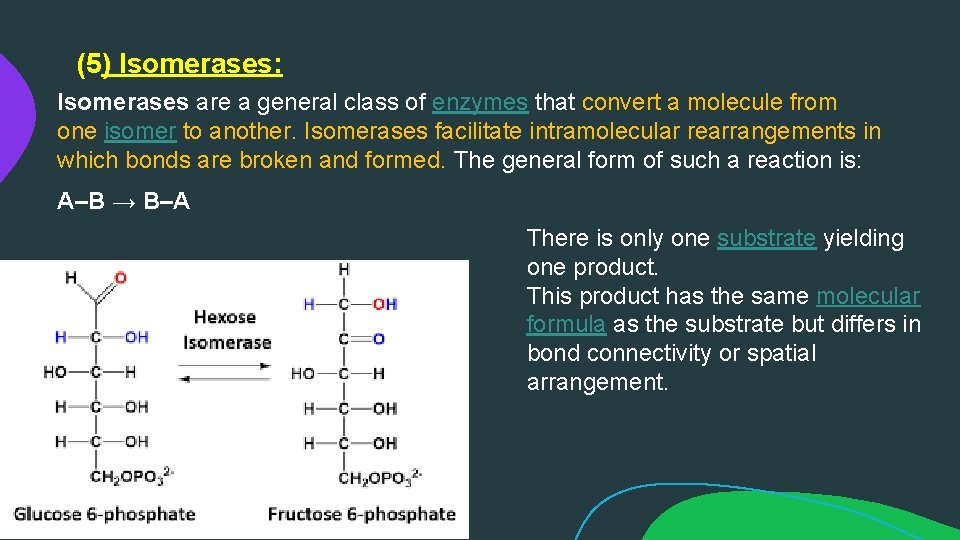
(5) Isomerases: Isomerases are a general class of enzymes that convert a molecule from one isomer to another. Isomerases facilitate intramolecular rearrangements in which bonds are broken and formed. The general form of such a reaction is: A–B → B–A There is only one substrate yielding one product. This product has the same molecular formula as the substrate but differs in bond connectivity or spatial arrangement.

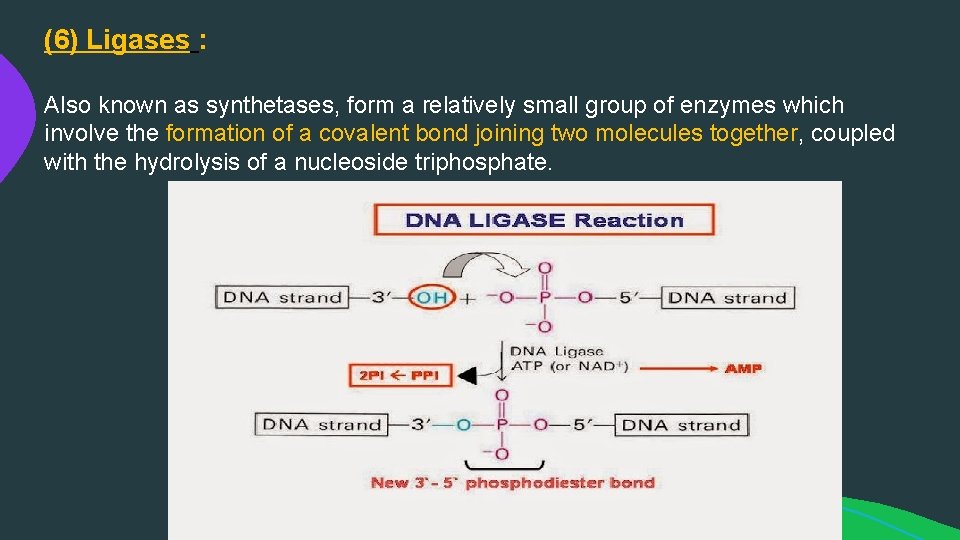
(6) Ligases : Also known as synthetases, form a relatively small group of enzymes which involve the formation of a covalent bond joining two molecules together, coupled with the hydrolysis of a nucleoside triphosphate.

Enzyme nomenclature : Summary

Isozymes: § Isozymes (also known as isoenzymes or more generally as multiple forms of enzymes) are enzymes that differ in amino acid sequence but catalyze the same chemical reaction. § Isozymes (or isoenzymes) are isoforms (closely related variants) of enzymes. In many cases, they are coded for by homologous genes that have diverged over time.

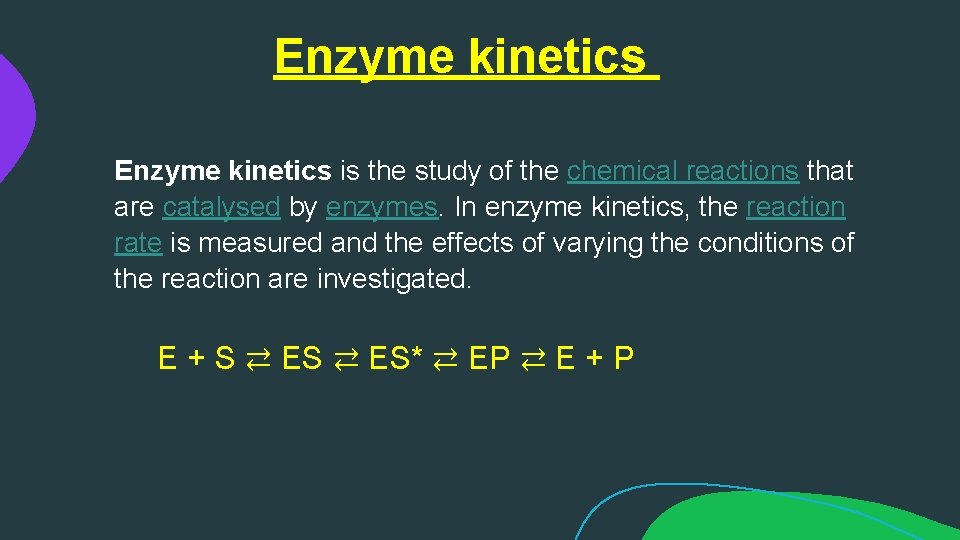
Enzyme kinetics is the study of the chemical reactions that are catalysed by enzymes. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. E + S ⇄ ES* ⇄ EP ⇄ E + P




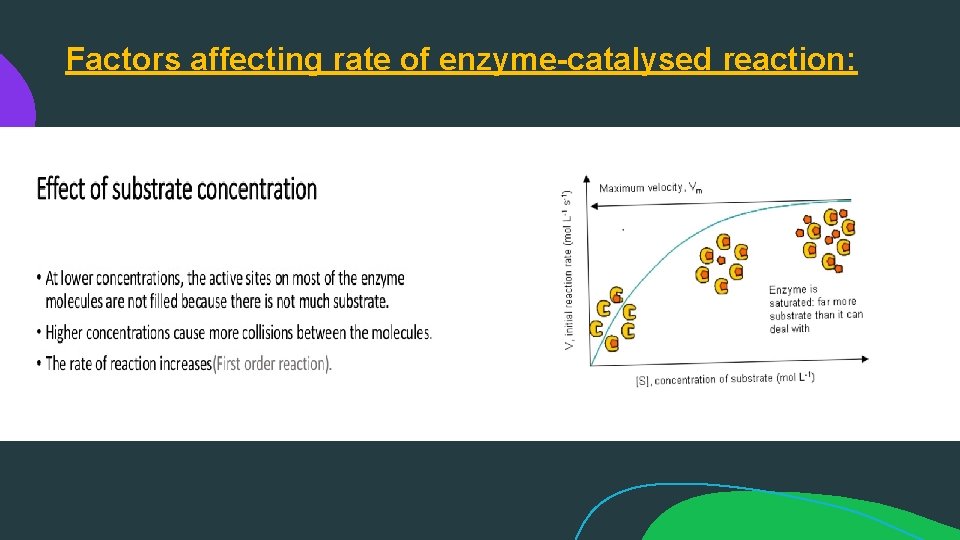
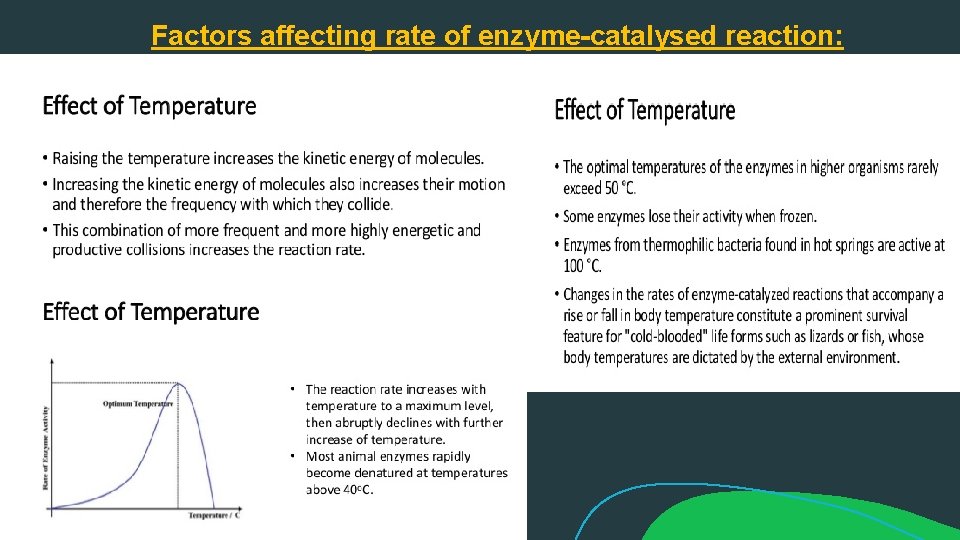
Factors affecting rate of enzyme-catalysed reaction:

Factors affecting rate of enzyme-catalysed reaction:

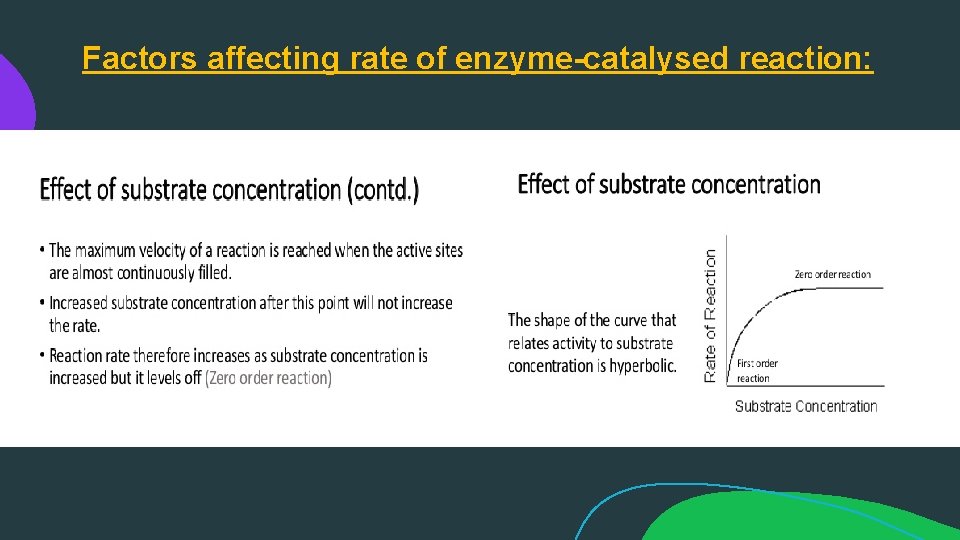
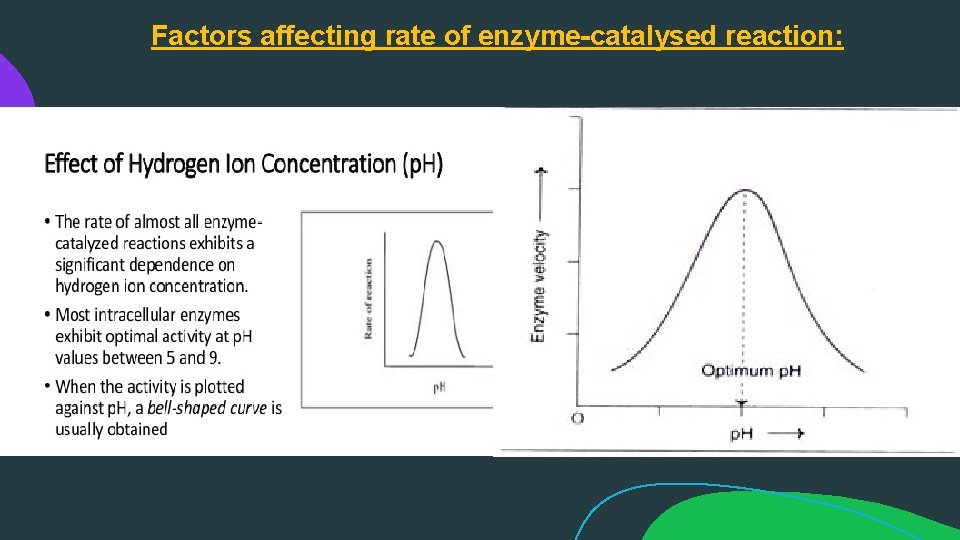
Factors affecting rate of enzyme-catalysed reaction:

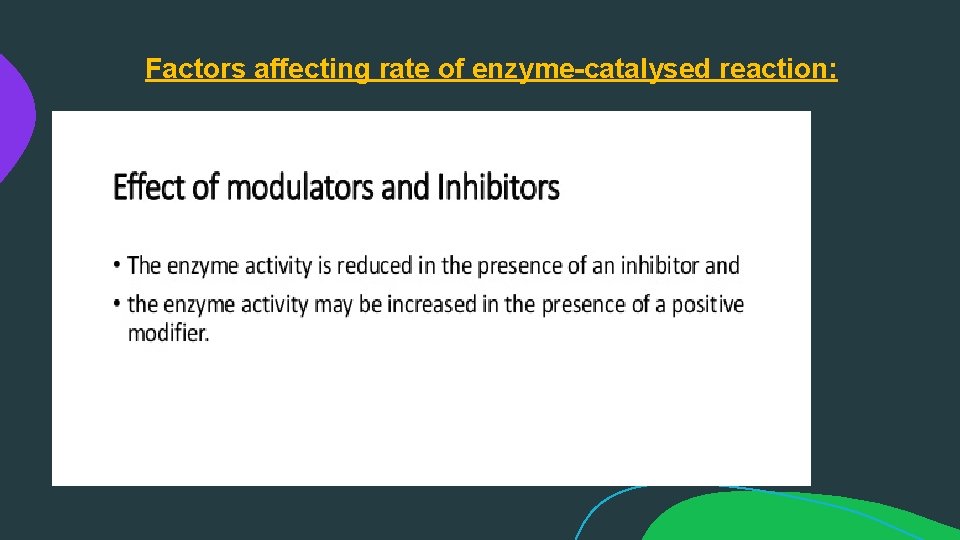
Factors affecting rate of enzyme-catalysed reaction:

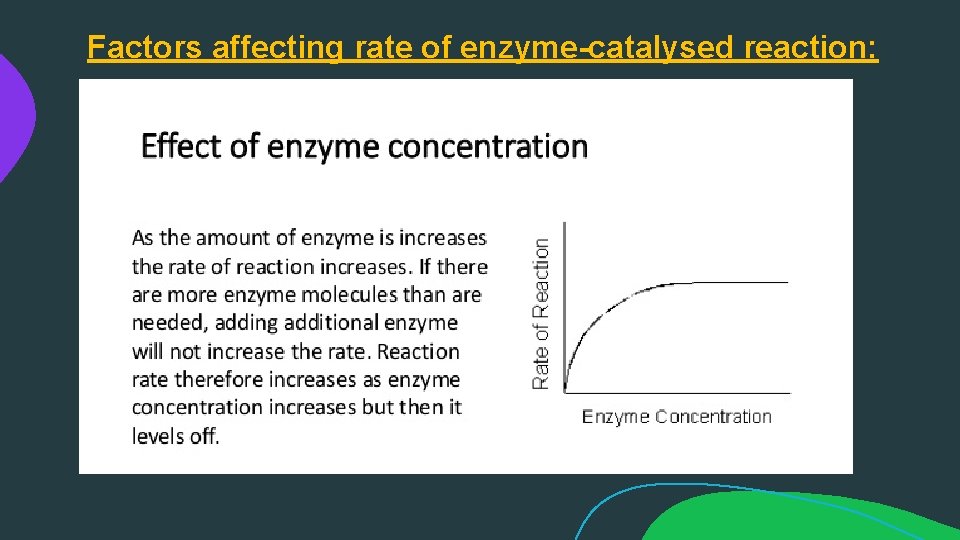
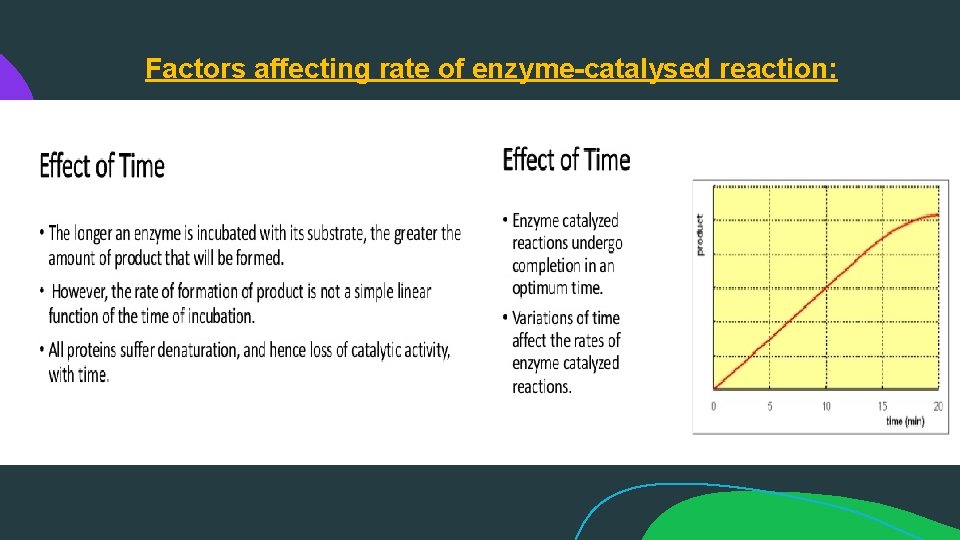
Factors affecting rate of enzyme-catalysed reaction:

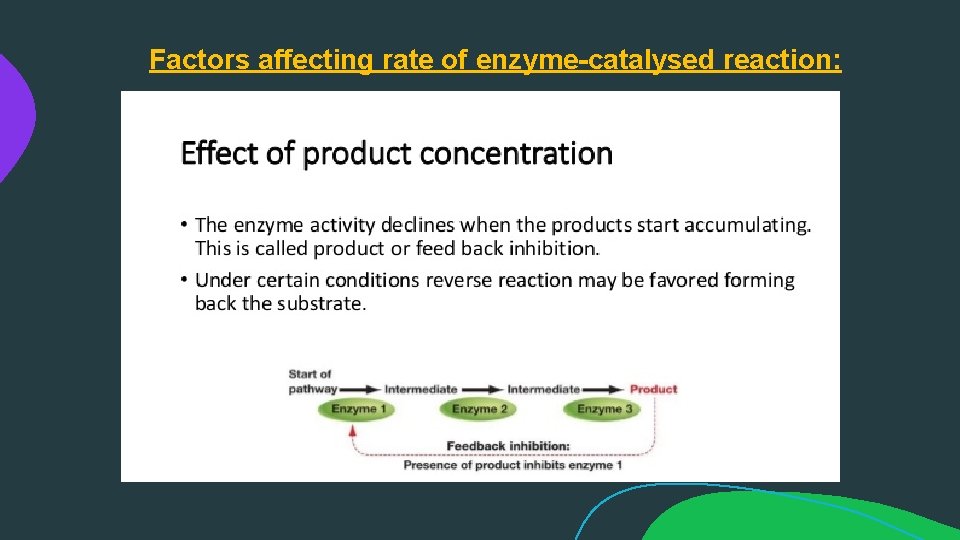
Factors affecting rate of enzyme-catalysed reaction:

Factors affecting rate of enzyme-catalysed reaction:

Factors affecting rate of enzyme-catalysed reaction:

The Michaelis–Menten equation is the rate equation for a one-substrate enzyme-catalyzed reaction. This equation relates the initial reaction rate (v 0), the maximum reaction rate (Vmax), and the initial substrate concentration [S] through the Michaelis constant KM—a measure of the substrate-binding affinity.







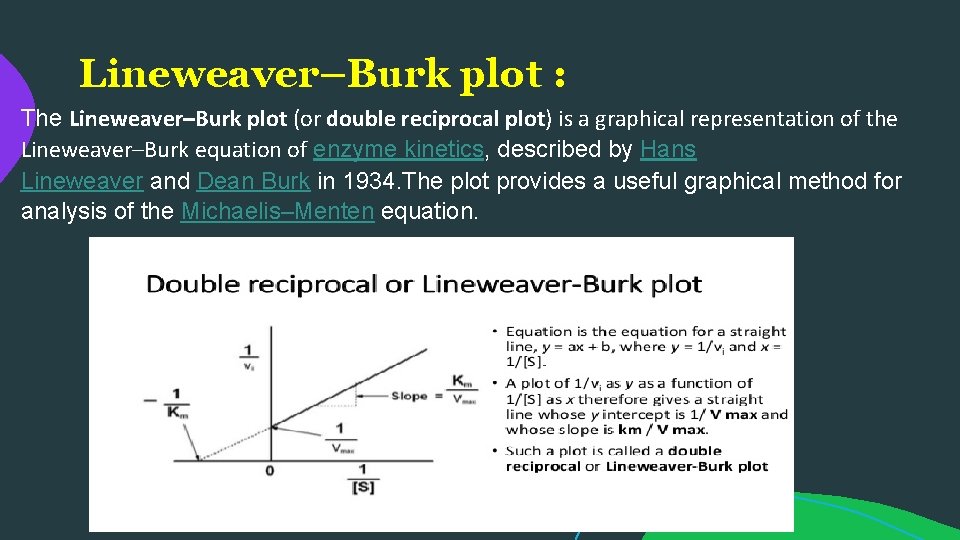
Lineweaver–Burk plot : The Lineweaver–Burk plot (or double reciprocal plot) is a graphical representation of the Lineweaver–Burk equation of enzyme kinetics, described by Hans Lineweaver and Dean Burk in 1934. The plot provides a useful graphical method for analysis of the Michaelis–Menten equation.

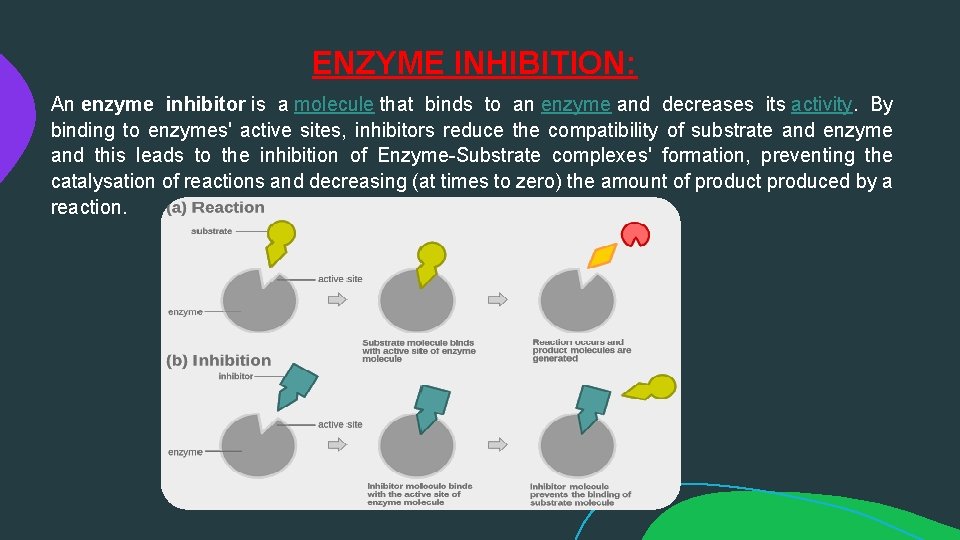
ENZYME INHIBITION: An enzyme inhibitor is a molecule that binds to an enzyme and decreases its activity. By binding to enzymes' active sites, inhibitors reduce the compatibility of substrate and enzyme and this leads to the inhibition of Enzyme-Substrate complexes' formation, preventing the catalysation of reactions and decreasing (at times to zero) the amount of product produced by a reaction.





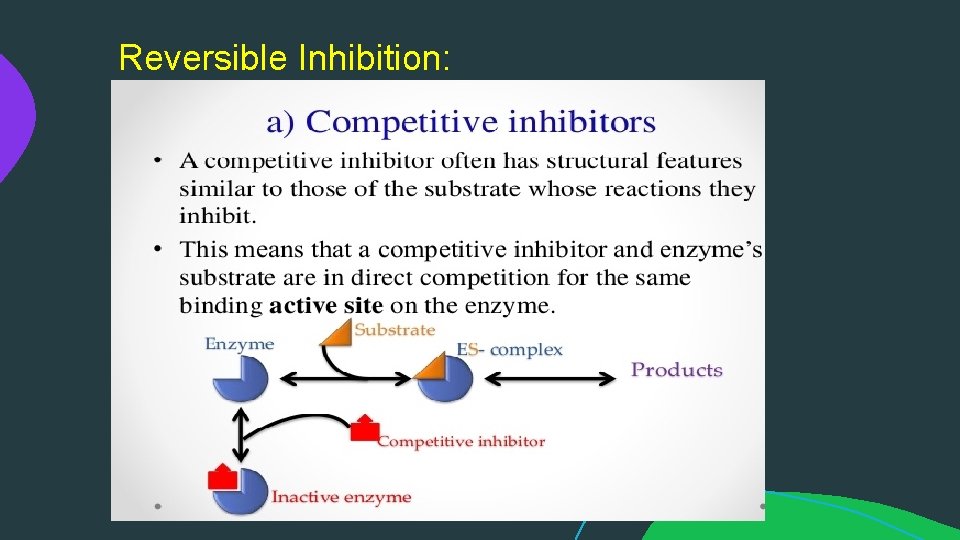
Reversible Inhibition:

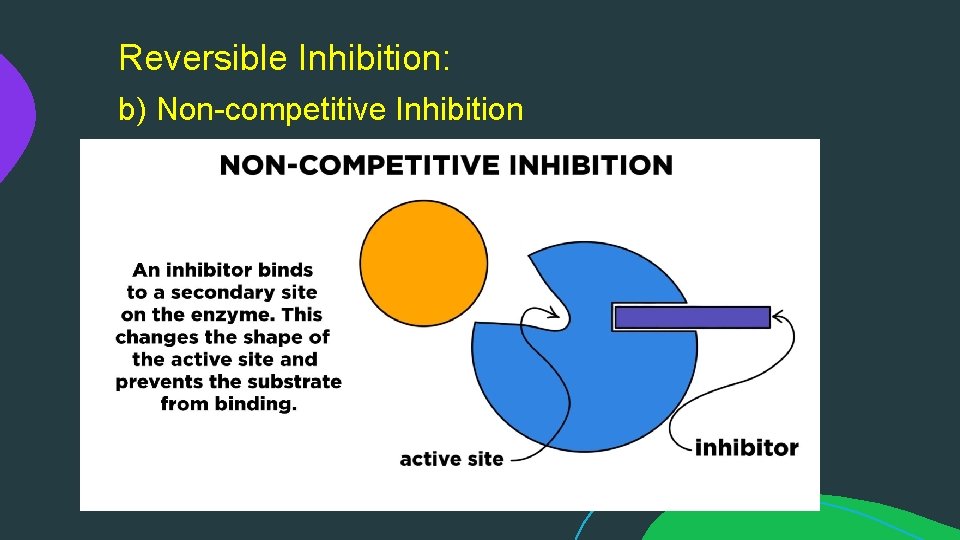
Reversible Inhibition: b) Non-competitive Inhibition




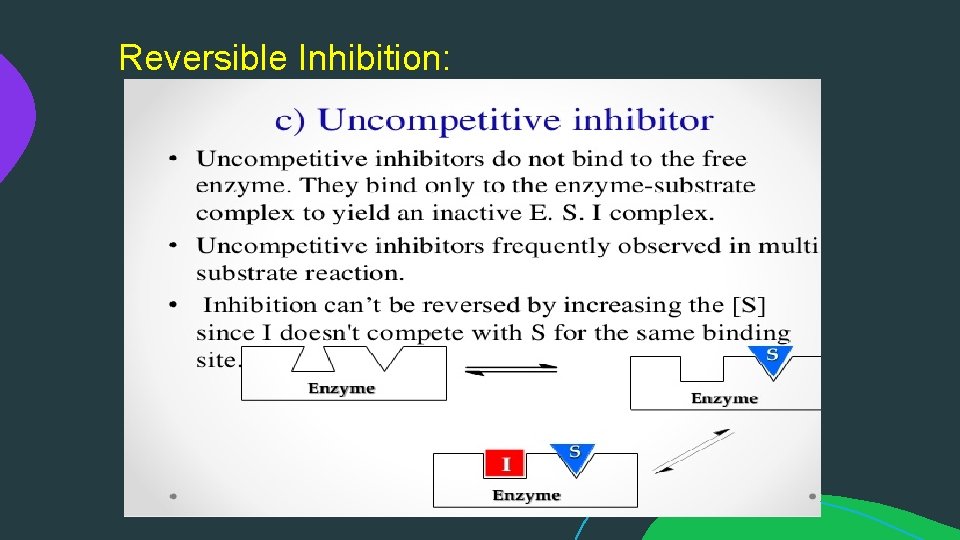
Reversible Inhibition:

THANK YOU
 Promotion from assistant to associate professor
Promotion from assistant to associate professor Cuhk salary scale 2020
Cuhk salary scale 2020 Purnamrita sarkar
Purnamrita sarkar Complement of a set definition
Complement of a set definition Purnamrita sarkar
Purnamrita sarkar Purnamrita sarkar
Purnamrita sarkar Taher haveliwala
Taher haveliwala Dr anirban sarkar
Dr anirban sarkar 370hr dielectric constant
370hr dielectric constant Alcohol enzymes market
Alcohol enzymes market Chemical nature of enzymes
Chemical nature of enzymes Brush border enzymes
Brush border enzymes Parietal layer and visceral layer
Parietal layer and visceral layer Most enzymes are *
Most enzymes are * Examples of enzymes
Examples of enzymes Buprenorphine and elevated liver enzymes
Buprenorphine and elevated liver enzymes Non functional plasma enzymes
Non functional plasma enzymes Enzymes guided notes
Enzymes guided notes Ap biology enzymes
Ap biology enzymes Restriction enzymes
Restriction enzymes Enzymes a
Enzymes a Enzyme foldable instructions
Enzyme foldable instructions What are 3 characteristics of enzymes
What are 3 characteristics of enzymes Enzymes
Enzymes Enzymes and their characteristics
Enzymes and their characteristics Lysosomal enzymes
Lysosomal enzymes Optimal temperature for enzymes
Optimal temperature for enzymes Enzymes in biological washing powder
Enzymes in biological washing powder Krebs cycle atp yield
Krebs cycle atp yield Biology-roots.com
Biology-roots.com Pepsin and renin
Pepsin and renin Bacteriophage characteristics
Bacteriophage characteristics Immobilized enzymes
Immobilized enzymes What are enzymes made of
What are enzymes made of Section 2-4 chemical reactions and enzymes
Section 2-4 chemical reactions and enzymes Immobilized enzymes
Immobilized enzymes Enzymes
Enzymes Nomenclature of enzymes
Nomenclature of enzymes How enzymes work mcgraw hill
How enzymes work mcgraw hill Section 2-4 chemical reactions and enzymes
Section 2-4 chemical reactions and enzymes Enzymes of staphylococcus aureus
Enzymes of staphylococcus aureus Examples of enzymes
Examples of enzymes Enzymes speed up chemical reactions by ____
Enzymes speed up chemical reactions by ____ The principal enzyme involved in dna replication is
The principal enzyme involved in dna replication is Ctfr
Ctfr Restriction enzymes
Restriction enzymes Elevated liver enzymes causes
Elevated liver enzymes causes Four properties of enzymes
Four properties of enzymes Transition state
Transition state What is enzymes
What is enzymes Small intestine enzymes
Small intestine enzymes Why are enzymes so important?
Why are enzymes so important? Non functional plasma enzymes
Non functional plasma enzymes Enzymes in small intestine
Enzymes in small intestine Lipid digestion enzymes
Lipid digestion enzymes Lumis enzymes
Lumis enzymes 3 characteristics of enzymes
3 characteristics of enzymes Enzymes used in recombinant dna technology ppt
Enzymes used in recombinant dna technology ppt Enzyme molecule
Enzyme molecule Factors affecting enzyme activity slideshare
Factors affecting enzyme activity slideshare Enzymes of staphylococcus aureus
Enzymes of staphylococcus aureus What are enzymes in food ingredients
What are enzymes in food ingredients Enzymes of glycogenesis
Enzymes of glycogenesis Enzymes
Enzymes All enzymes are globular proteins
All enzymes are globular proteins Homotrophic effects for allosteric enzymes involve
Homotrophic effects for allosteric enzymes involve What is a prosthetic group in biochemistry
What is a prosthetic group in biochemistry 5 examples of palindromic dna sequences
5 examples of palindromic dna sequences What is immobilisation of enzymes
What is immobilisation of enzymes What is enzyme
What is enzyme Othlil theory of enzymes with examples
Othlil theory of enzymes with examples Ligases enzymes examples
Ligases enzymes examples Restriction enzymes
Restriction enzymes Not all enzymes are proteins
Not all enzymes are proteins Tca cycle
Tca cycle Chapter 9 cellular respiration harvesting chemical energy
Chapter 9 cellular respiration harvesting chemical energy Factors that affect enzymes
Factors that affect enzymes