Mechanisms of Enzyme Action Transition TS State Intermediate



- Slides: 25

Mechanisms of Enzyme Action

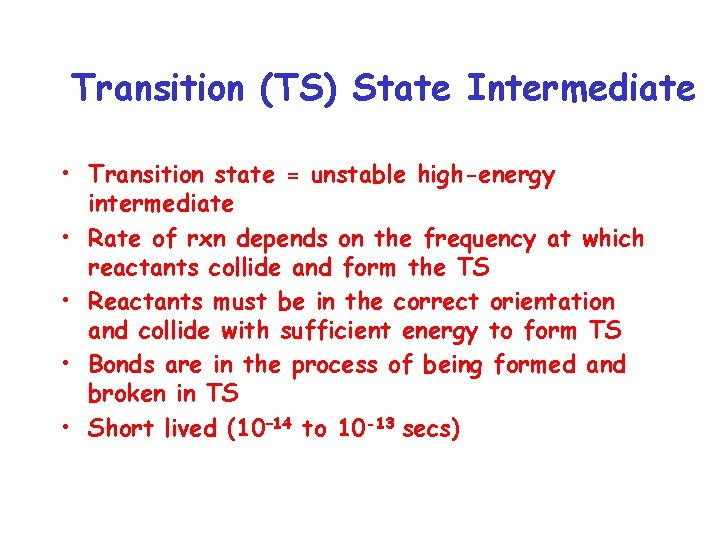
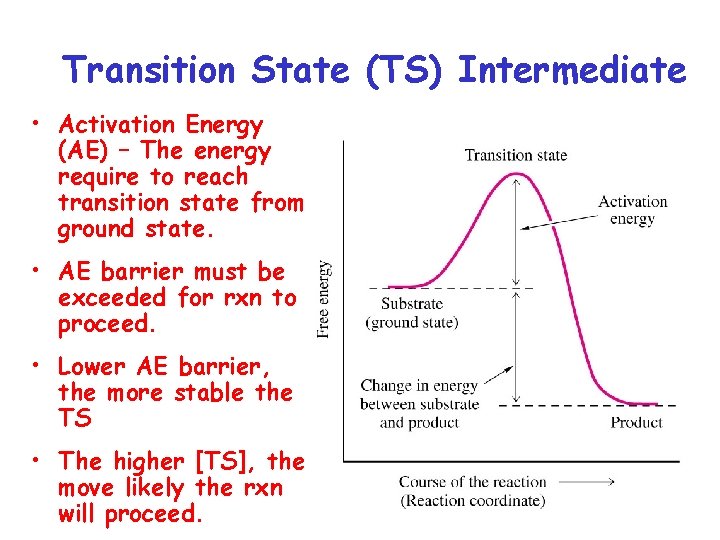
Transition (TS) State Intermediate • Transition state = unstable high-energy intermediate • Rate of rxn depends on the frequency at which reactants collide and form the TS • Reactants must be in the correct orientation and collide with sufficient energy to form TS • Bonds are in the process of being formed and broken in TS • Short lived (10– 14 to 10 -13 secs)

Transition State (TS) Intermediate • Activation Energy (AE) – The energy require to reach transition state from ground state. • AE barrier must be exceeded for rxn to proceed. • Lower AE barrier, the more stable the TS • The higher [TS], the move likely the rxn will proceed.

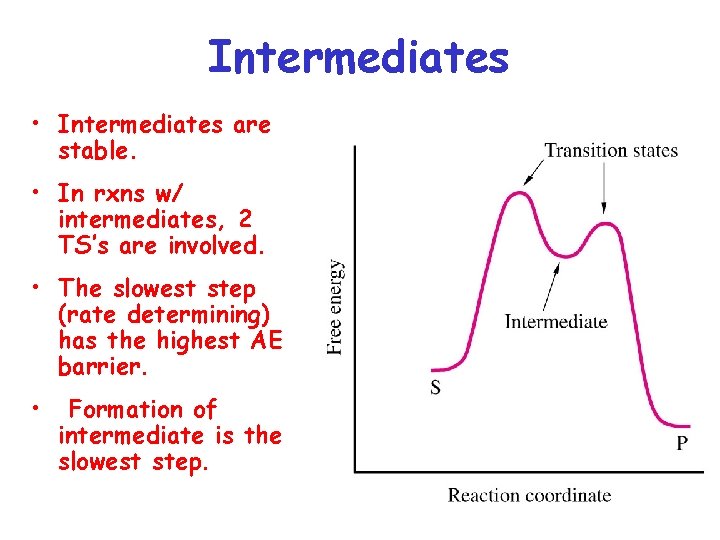
Intermediates • Intermediates are stable. • In rxns w/ intermediates, 2 TS’s are involved. • The slowest step (rate determining) has the highest AE barrier. • Formation of intermediate is the slowest step.

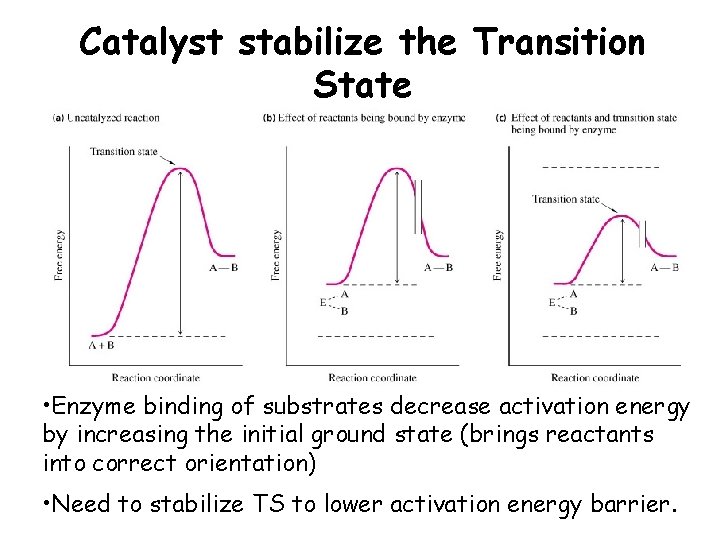
Catalyst stabilize the Transition State • Enzyme binding of substrates decrease activation energy by increasing the initial ground state (brings reactants into correct orientation) • Need to stabilize TS to lower activation energy barrier.

Common types of enzymatic mechanisms • Substitutions rxns • Bond cleavage rxns • Redox rxns

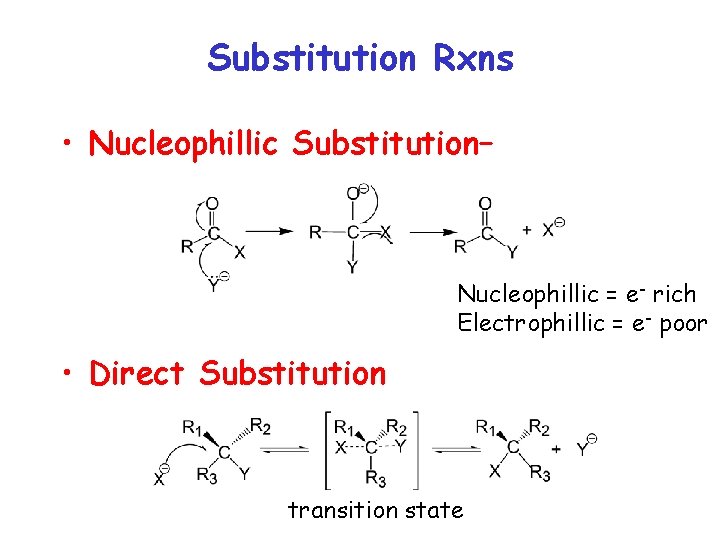
Substitution Rxns • Nucleophillic Substitution– Nucleophillic = e- rich Electrophillic = e- poor • Direct Substitution transition state

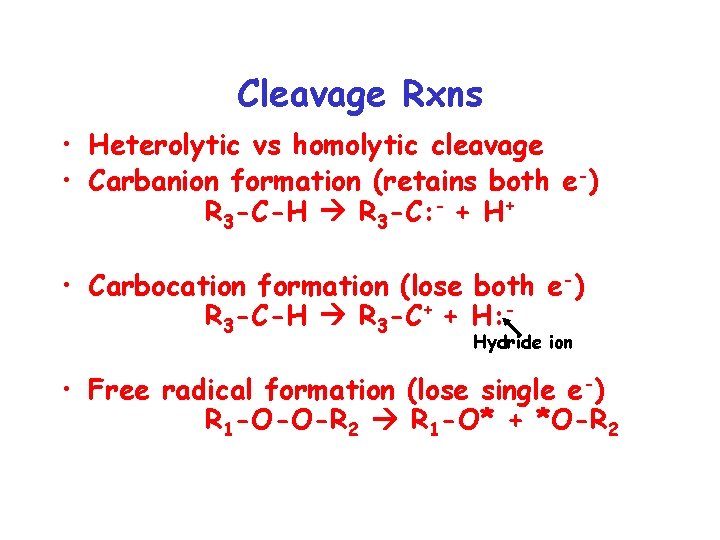
Cleavage Rxns • Heterolytic vs homolytic cleavage • Carbanion formation (retains both e-) R 3 -C-H R 3 -C: - + H+ • Carbocation formation (lose both e-) R 3 -C-H R 3 -C+ + H: Hydride ion • Free radical formation (lose single e-) R 1 -O-O-R 2 R 1 -O* + *O-R 2

Oxidation reduction (Redox) Rxns • Loose e- = oxidation (LEO) • Gain e- = reduction (GER) • Central to energy production • If something oxidized something must be reduced (reducing agent donates e- to oxidizing agent) • Oxidations = removal of hydrogen or addition of oxygen or removal of e • In biological systems reducing agent is usually a co-factor (NADH of NADPH)

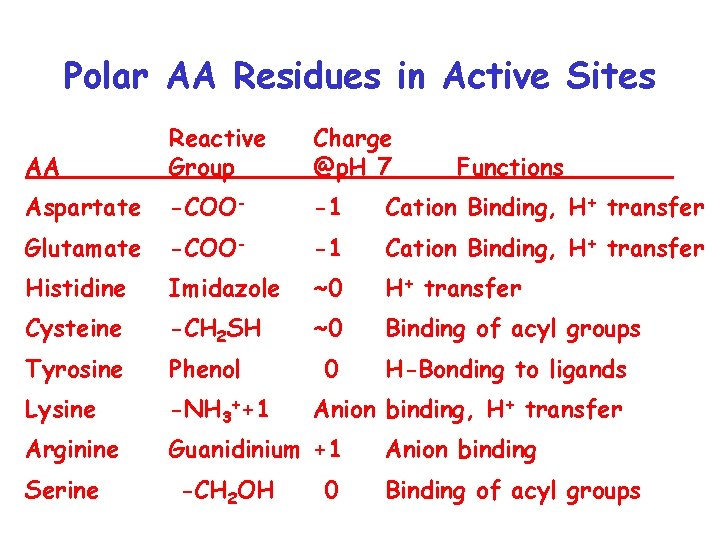
Polar AA Residues in Active Sites AA Reactive Group Charge @p. H 7 Aspartate -COO- -1 Cation Binding, H+ transfer Glutamate -COO- -1 Cation Binding, H+ transfer Histidine Imidazole ~0 H+ transfer Cysteine -CH 2 SH ~0 Binding of acyl groups Tyrosine Phenol 0 H-Bonding to ligands Lysine -NH 3++1 Arginine Guanidinium +1 Serine -CH 2 OH Functions Anion binding, H+ transfer 0 Anion binding Binding of acyl groups

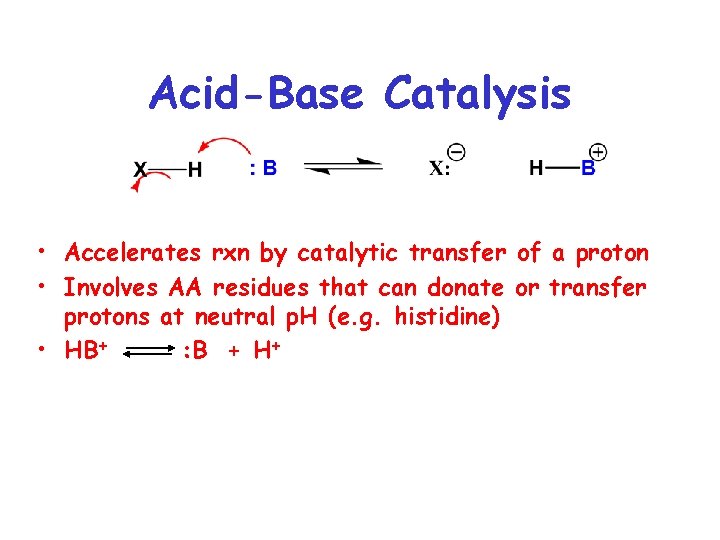
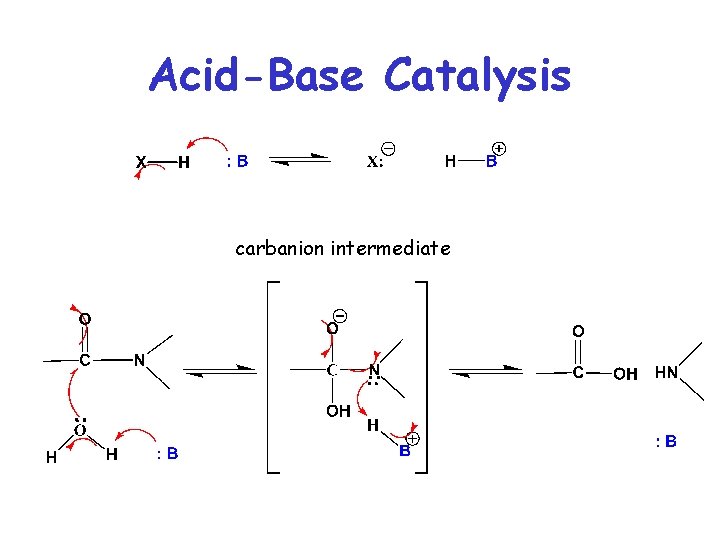
Acid-Base Catalysis • Accelerates rxn by catalytic transfer of a proton • Involves AA residues that can donate or transfer protons at neutral p. H (e. g. histidine) • HB+ : B + H+

Acid-Base Catalysis carbanion intermediate : :

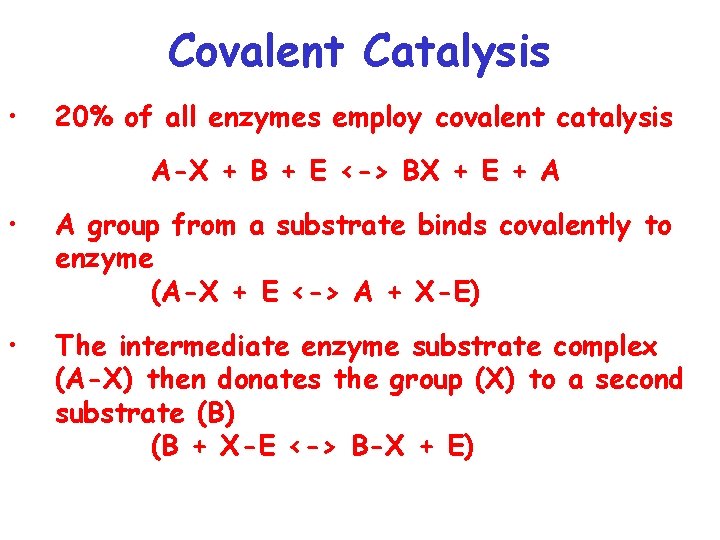
Covalent Catalysis • 20% of all enzymes employ covalent catalysis A-X + B + E <-> BX + E + A • A group from a substrate binds covalently to enzyme (A-X + E <-> A + X-E) • The intermediate enzyme substrate complex (A-X) then donates the group (X) to a second substrate (B) (B + X-E <-> B-X + E)

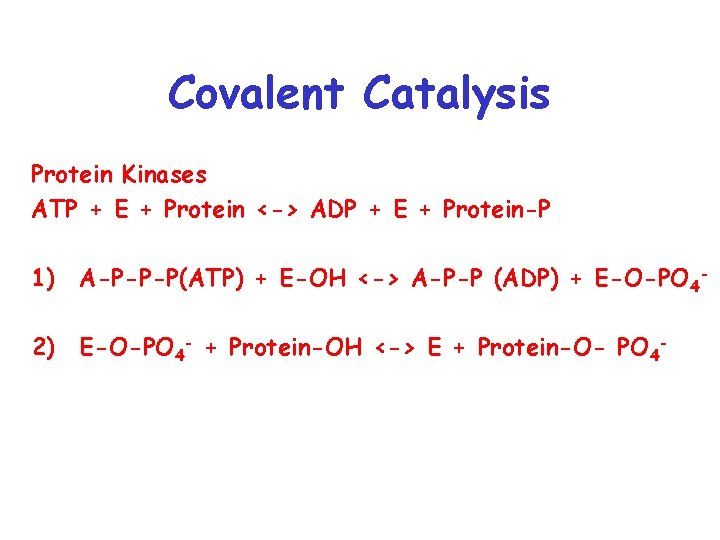
Covalent Catalysis Protein Kinases ATP + E + Protein <-> ADP + E + Protein-P 1) A-P-P-P(ATP) + E-OH <-> A-P-P (ADP) + E-O-PO 42) E-O-PO 4 - + Protein-OH <-> E + Protein-O- PO 4 -

Binding Modes of Enzymatic Catalysis • Proximity Effects • Transition State Stabilization

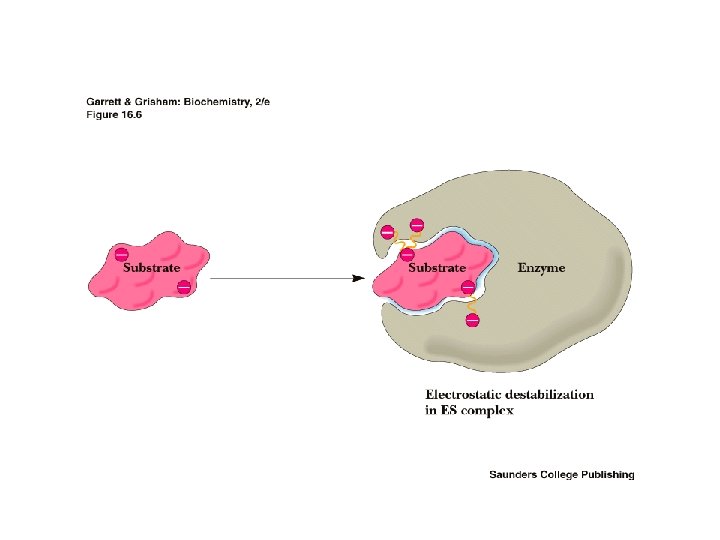
Proximity Effects • In multi-substrate reactions, collecting and correct positioning of substrates important • Increases effective concentration of substrates which favors formation of TS • Decreases activation energy by decreasing entropy • Increases rate of reaction > 10, 000 fold • E binding to S can not be too strong or conversion from ES to TS would require too much energy

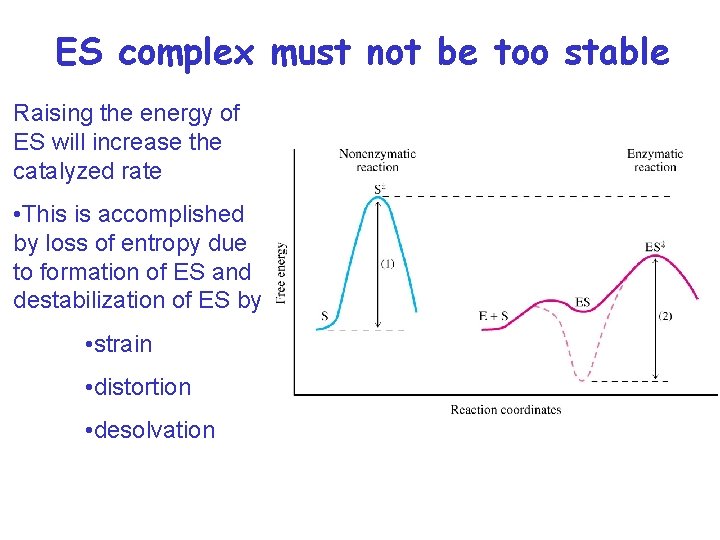
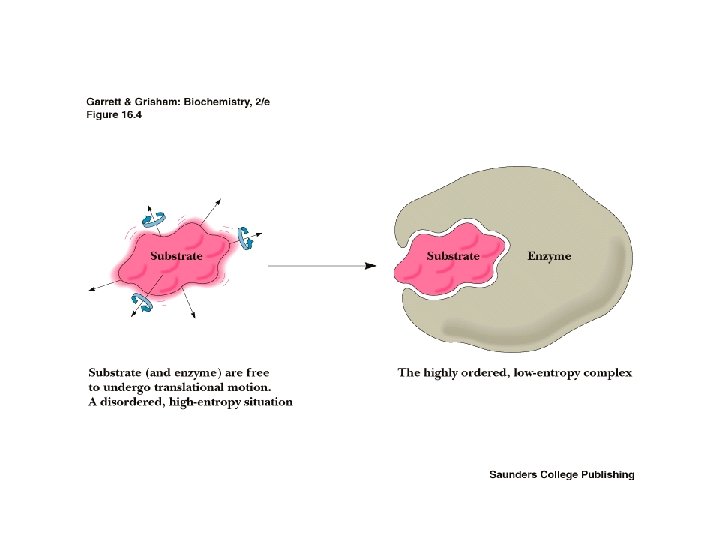
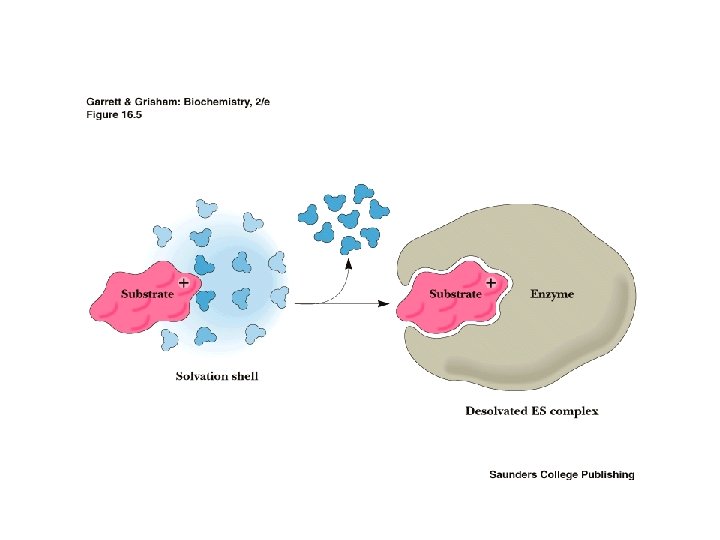
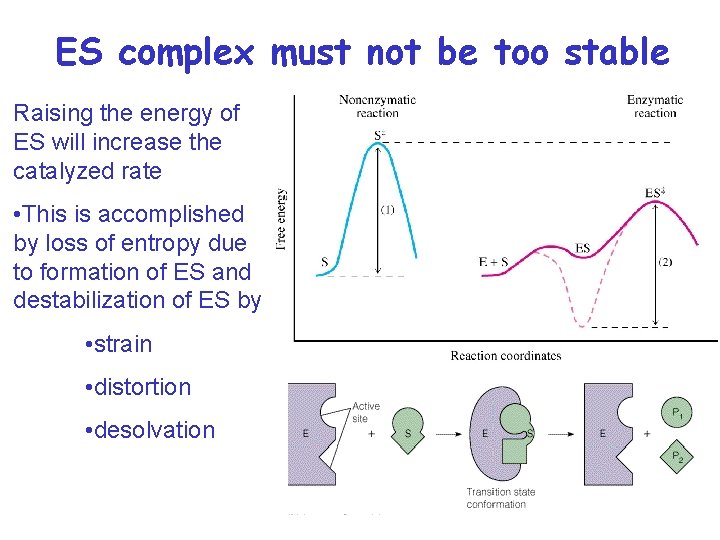
ES complex must not be too stable Raising the energy of ES will increase the catalyzed rate • This is accomplished by loss of entropy due to formation of ES and destabilization of ES by • strain • distortion • desolvation




ES complex must not be too stable Raising the energy of ES will increase the catalyzed rate • This is accomplished by loss of entropy due to formation of ES and destabilization of ES by • strain • distortion • desolvation

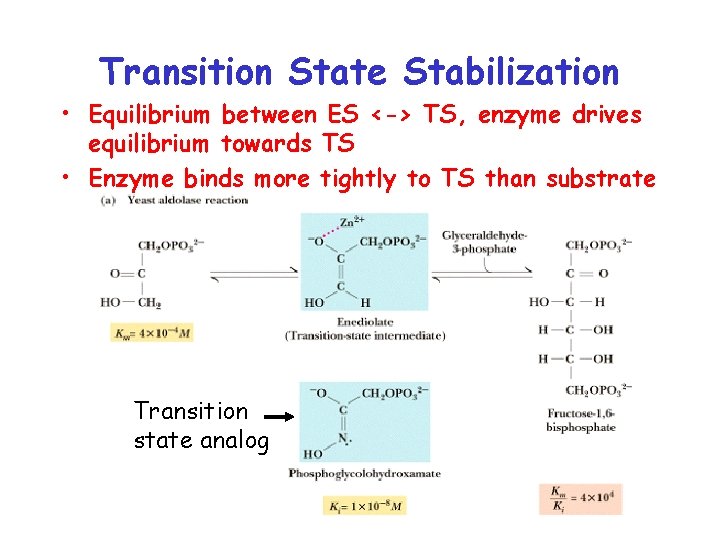
Transition State Stabilization • Equilibrium between ES <-> TS, enzyme drives equilibrium towards TS • Enzyme binds more tightly to TS than substrate Transition state analog

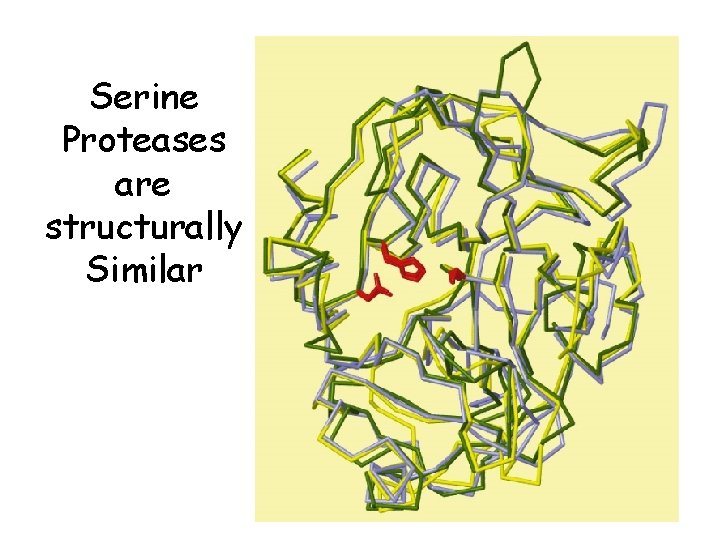
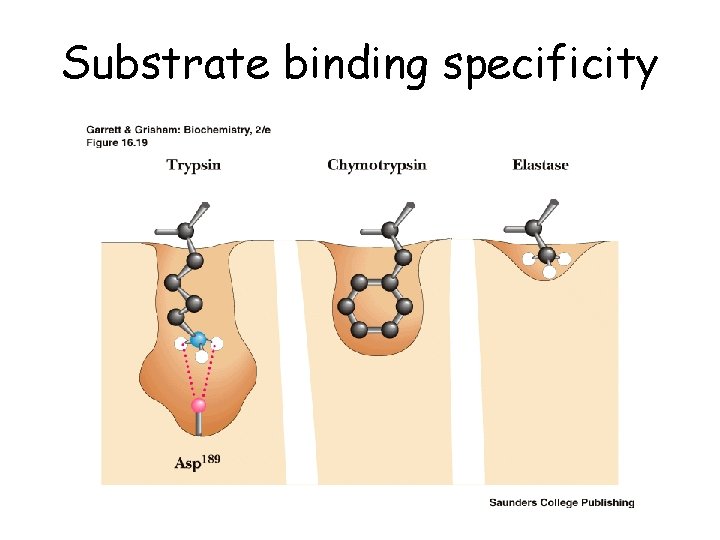
The Serine Proteases • • Trypsin, chymotrypsin, elastase, thrombin, subtilisin, plasmin, TPA All involve a serine in catalysis - thus the name Ser is part of a "catalytic triad" of Ser, His, Asp (show over head) Serine proteases are homologous, but locations of the three crucial residues differ somewhat Substrate specificity determined by binding pocket

Serine Proteases are structurally Similar

Substrate binding specificity
 Examples of transferases
Examples of transferases Transition state enzymes
Transition state enzymes State graph in software testing
State graph in software testing Aspirin action
Aspirin action Mechanism of action of asprin
Mechanism of action of asprin Traxler
Traxler Process state transition
Process state transition State transition testing adalah
State transition testing adalah What is state transition matrix
What is state transition matrix Mesi state transition diagram
Mesi state transition diagram What is state transition matrix
What is state transition matrix Linear system theory and design
Linear system theory and design Sctp state transition diagram
Sctp state transition diagram Transition state energy diagram
Transition state energy diagram Explain the process control block
Explain the process control block State transition table
State transition table Seven state process transition diagram
Seven state process transition diagram Dhcp state transition diagram
Dhcp state transition diagram State transition diagram
State transition diagram State transition diagram
State transition diagram State transition testing adalah
State transition testing adalah Syscall user dispatch
Syscall user dispatch State transition diagram пример
State transition diagram пример Transition state
Transition state Rearranged most stable carbocation is
Rearranged most stable carbocation is Diagonization
Diagonization