Restriction Enzymes WHAT ARE RESTRICTION ENZYMES Restriction Enzymes









- Slides: 18

Restriction Enzymes

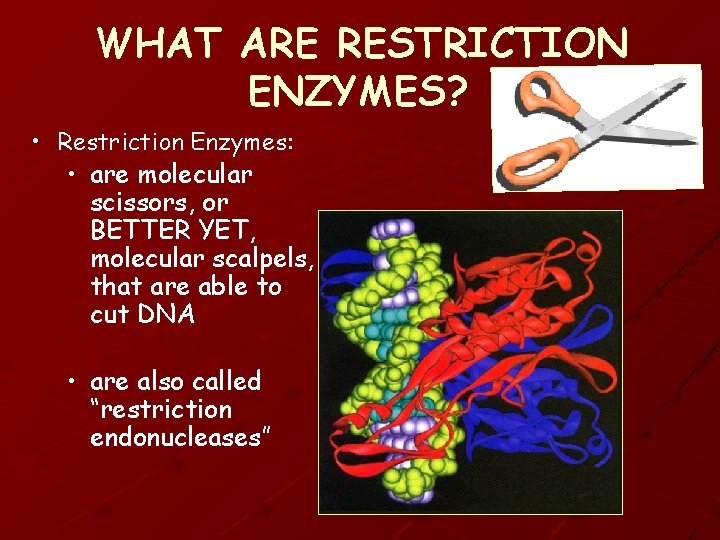
WHAT ARE RESTRICTION ENZYMES? • Restriction Enzymes: • are molecular scissors, or BETTER YET, molecular scalpels, that are able to cut DNA • are also called “restriction endonucleases”

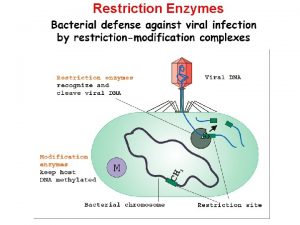
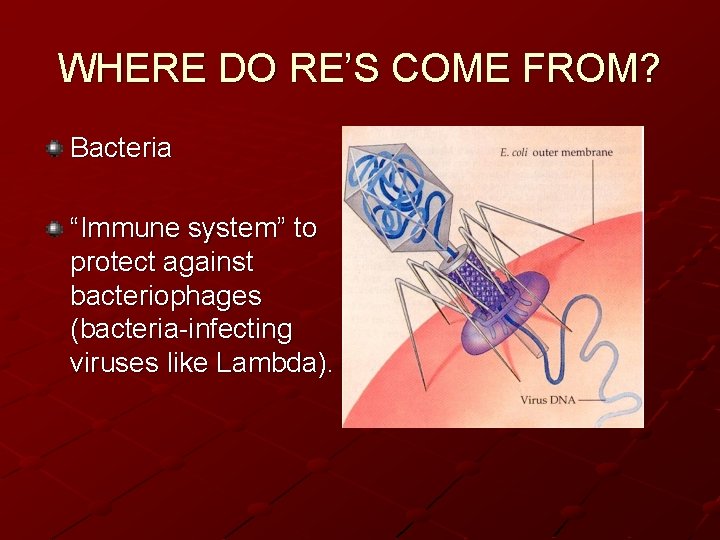
WHERE DO RE’S COME FROM? Bacteria “Immune system” to protect against bacteriophages (bacteria-infecting viruses like Lambda).

THOUGHT QUESTION Bacteria are prokaryotes. Prokaryotes do not have a nucleus. Both DNA and RE’s are in cytoplasm. Why isn’t bacterial DNA cut by RE’s?

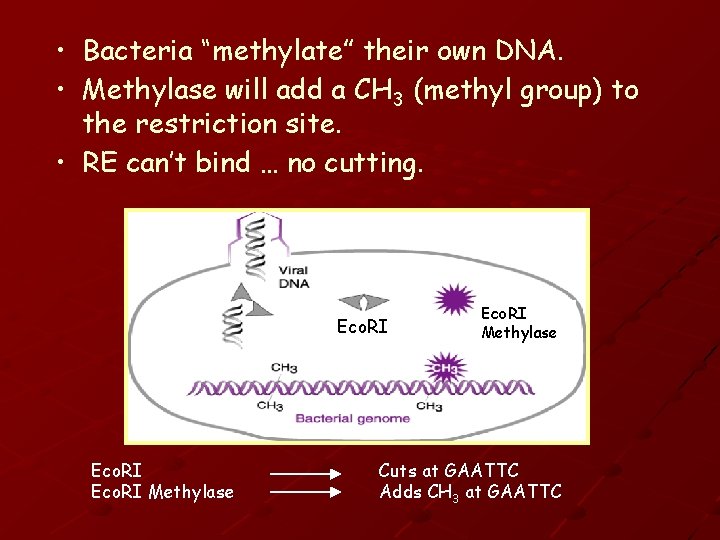
• Bacteria “methylate” their own DNA. • Methylase will add a CH 3 (methyl group) to the restriction site. • RE can’t bind … no cutting. Eco. RI Methylase Cuts at GAATTC Adds CH 3 at GAATTC

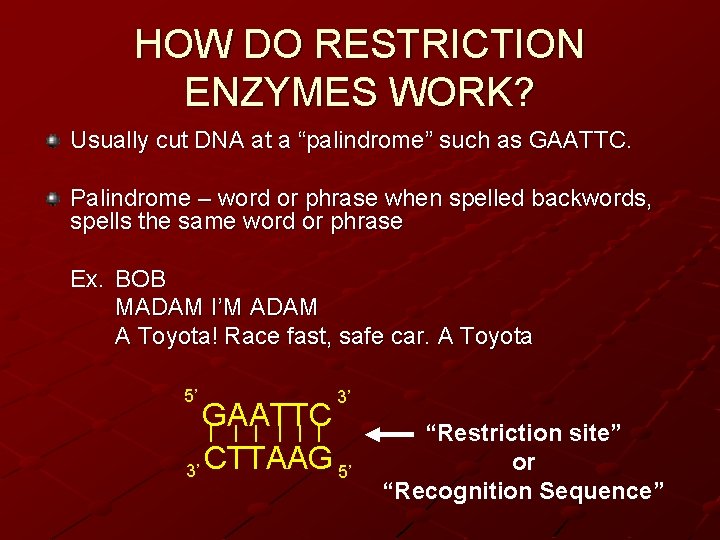
HOW DO RESTRICTION ENZYMES WORK? Usually cut DNA at a “palindrome” such as GAATTC. Palindrome – word or phrase when spelled backwords, spells the same word or phrase Ex. BOB MADAM I’M ADAM A Toyota! Race fast, safe car. A Toyota 5’ 3’ GAATTC | | | 3’ CTTAAG 5’ “Restriction site” or “Recognition Sequence”

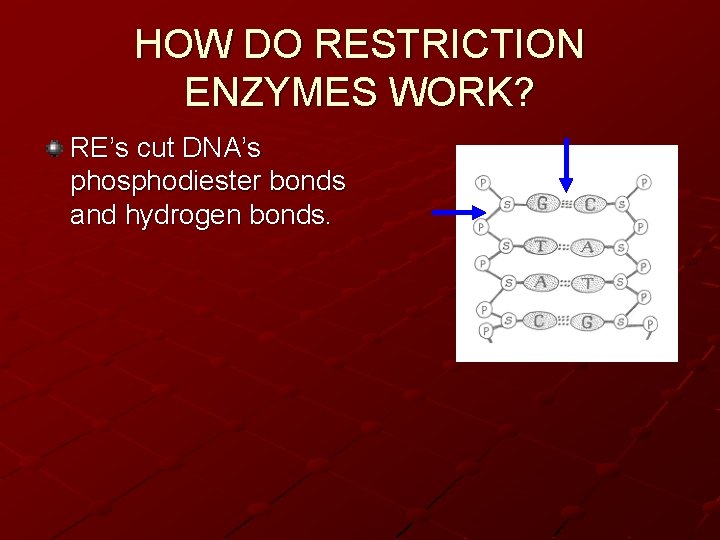
HOW DO RESTRICTION ENZYMES WORK? RE’s cut DNA’s phosphodiester bonds and hydrogen bonds.

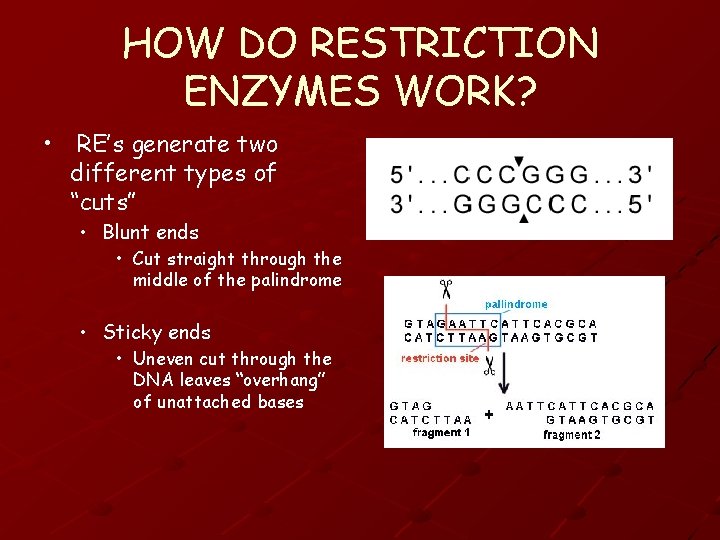
HOW DO RESTRICTION ENZYMES WORK? • RE’s generate two different types of “cuts” • Blunt ends • Cut straight through the middle of the palindrome • Sticky ends • Uneven cut through the DNA leaves “overhang” of unattached bases

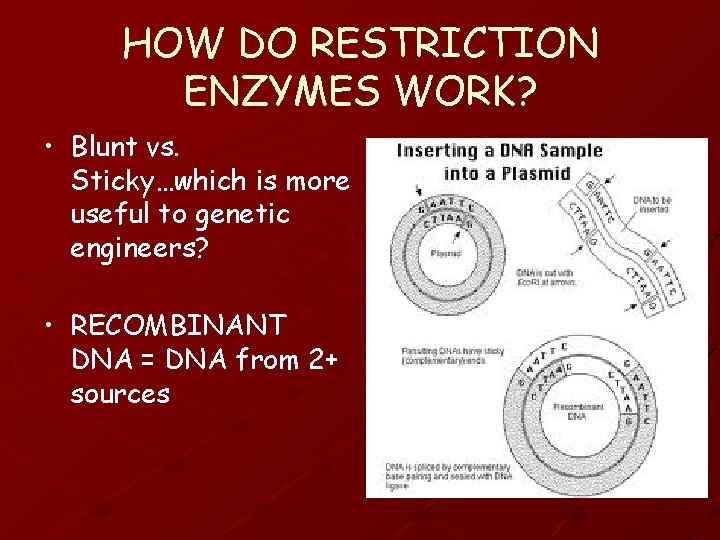
HOW DO RESTRICTION ENZYMES WORK? • Blunt vs. Sticky…which is more useful to genetic engineers? • RECOMBINANT DNA = DNA from 2+ sources

HOW IS DNA PASTED TOGETHER? Ligase – another enzyme which reconnects phosphodiester bonds.

In short… Enzymes BUILD or BREAK chemical bonds…

Enzymes - a brief reminder • Enzymes: • • Are proteins Usually end with “-ase” Speed up chemical reactions (catalysts) Have specific shapes that fit specific substrates Lower the activation energy of a reaction Can be used over, and over again Often require “cofactors” to work properly • metal ions - ex. iron, zinc, copper • organic molecules (COENZYMES) generally derived from vitamins

WHAT ARE RE’S USED FOR? • Genetic engineering – pasting together DNA from two different organisms. • Spider silk gene in goats • Glo-fish • GFP addition (to tag and track gene expression) • Golden Rice • Human Insulin

WHAT ELSE ARE RE’S USED FOR? Forensics – DNA Fingerprinting for crime scene investigation and paternity testing. Everyone’s DNA has a different sequence – even though only 0. 1% different. How frequently would Eco. RI cut DNA? 6 4 = once every 4096 bp Lambda (48, 514 bp) would expect about 12 Eco. RI sites

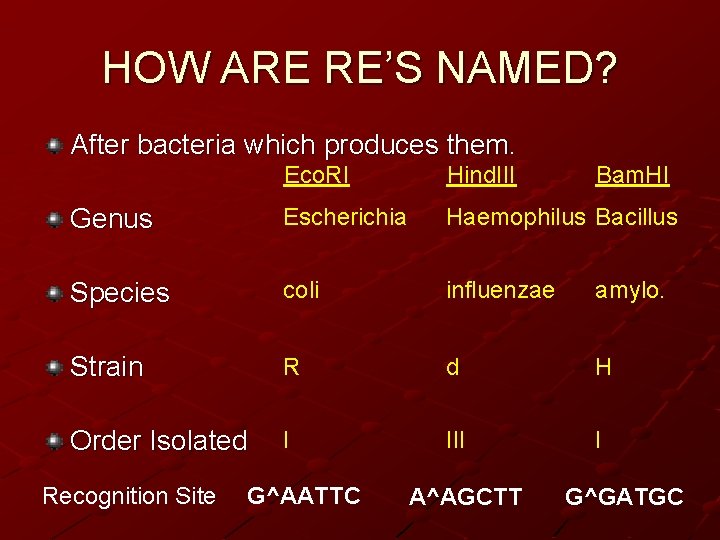
HOW ARE RE’S NAMED? • After bacteria which produces them. • Genus Recognition Site Eco. RI Escherichia • Species coli • Strain RY 13 • 1 st letter • 1 st two letters • 1 st letter • Order Isolated I G^AATTC

HOW ARE RE’S NAMED? After bacteria which produces them. Eco. RI Hind. III Genus Escherichia Haemophilus Bacillus Species coli influenzae amylo. Strain R d H Order Isolated I III I Recognition Site G^AATTC A^AGCTT Bam. HI G^GATGC

Quick review… • • • • 1. Monomer of DNA = 2. Three parts of the monomer = 3. A - T with ___ hydrogen bond 4. Which is bigger -- purines or pyrmidines? 5. What two bases are pyrimidines? 6. Charge on DNA = 7. DNA has a ____-wise and ____-hand twist. 8. Bond between sugars - phosphates = 9. Restriction enzymes are found only in… 10. Same forwards and backwards = 11. Bacteria add ____groups to their DNA to prevent cutting. 12. Overhangs of unpaired bases = 13. In Hind. III, the “in” comes from the _____ of bacteria. 14. Enzymes usually end in _____.

• 1. Monomer of DNA = Nucleotide • 2. Three parts of the monomer = Sugar (deoxyribose), phosphate, base • 3. A - T with 2 hydrogen bonds • 4. Which is bigger -- purines or pyrmidines? Purines • 5. What two bases are pyrimidines? Cytosine and thymine • 6. Charge on DNA = negative • 7. DNA has a Clock -wise and right -hand twist. • 8. Bond between sugars - phosphates = phosphodiester • 9. Restriction enzymes are found only in… bacteria • 10. Same forwards and backwards = palindrome • 11. Bacteria add CH 3 (methyl) groups to their DNA to prevent cutting. • 12. Overhangs of unpaired bases = sticky ends • 13. In Hind. III, the “in” comes from the species of bacteria. • 14. Enzymes usually end in -ase.
 Insidan region jh
Insidan region jh Example of type 3 restriction enzyme
Example of type 3 restriction enzyme Restriction enzyme
Restriction enzyme Restriction enzymes
Restriction enzymes Function of restriction enzymes
Function of restriction enzymes Restriction enzymes
Restriction enzymes Selective breeding definition biology
Selective breeding definition biology Restriction enzymes
Restriction enzymes Restriction enzymes
Restriction enzymes Calorie restriction
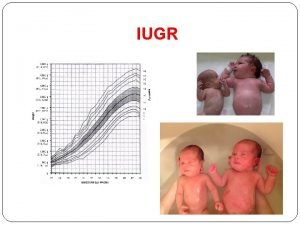
Calorie restriction Asymmetrical intrauterine growth restriction
Asymmetrical intrauterine growth restriction Restriction fragment length polymorphism
Restriction fragment length polymorphism Rflp analysis
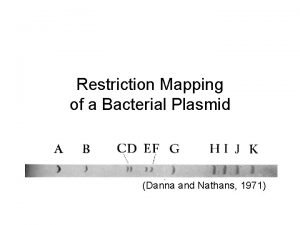
Rflp analysis Restriction mapping
Restriction mapping Asymmetrical and symmetrical iugr
Asymmetrical and symmetrical iugr Mechanism of restriction enzyme
Mechanism of restriction enzyme Student attribute restriction uiuc
Student attribute restriction uiuc Diffusion restriction
Diffusion restriction Restriction enzyme analysis of dna ap bio lab
Restriction enzyme analysis of dna ap bio lab