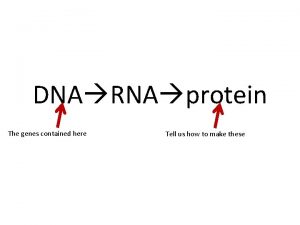
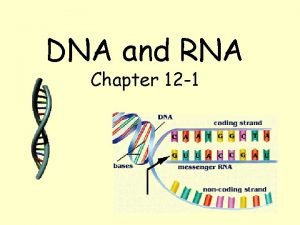
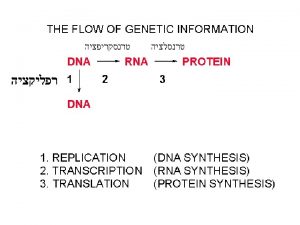
DNA RNA and Protein Synthesis Molecular structure of












- Slides: 96

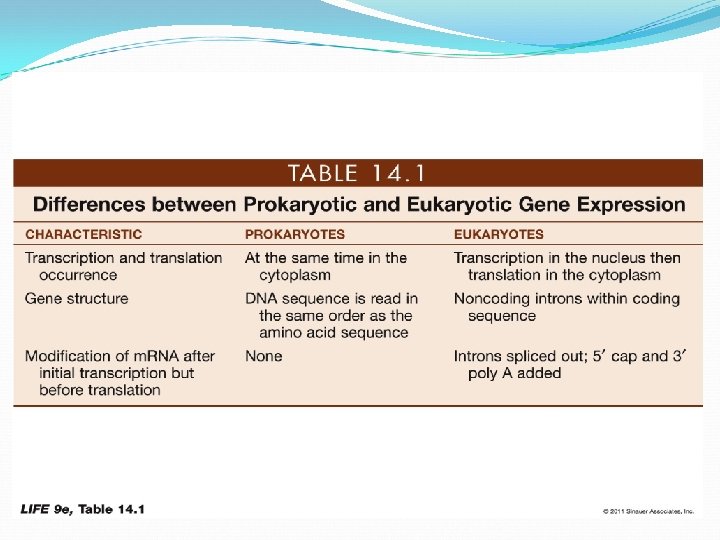
DNA, RNA, and Protein Synthesis

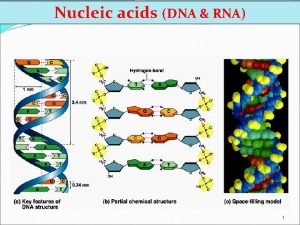
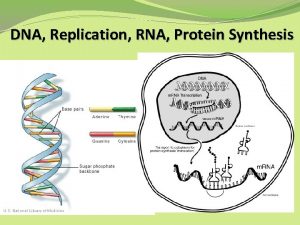
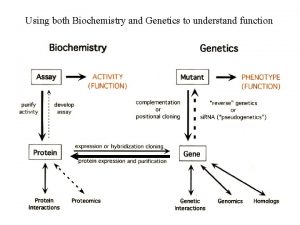
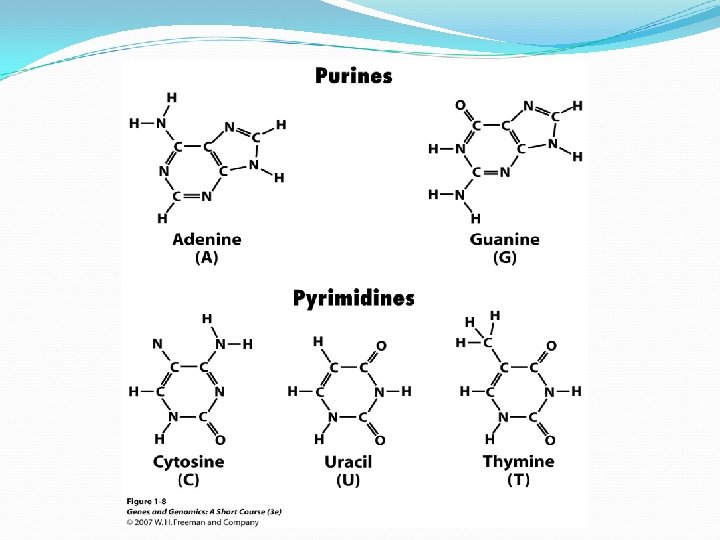
Molecular structure of DNA Chromosomes contain both nucleic acid and protein. There are two types of nucleic acid, DNA and RNA. Nucleic acid is a polymer of nucleotides, each comprised of a ribose sugar, a phosphate group and a nitrogen containing base. There are two types of bases, Purines and Pyrimidines.

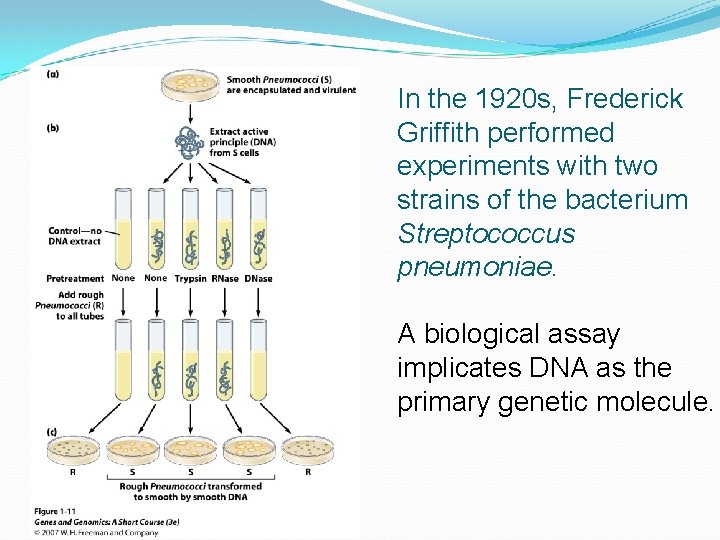
In the 1920 s, Frederick Griffith performed experiments with two strains of the bacterium Streptococcus pneumoniae. A biological assay implicates DNA as the primary genetic molecule.

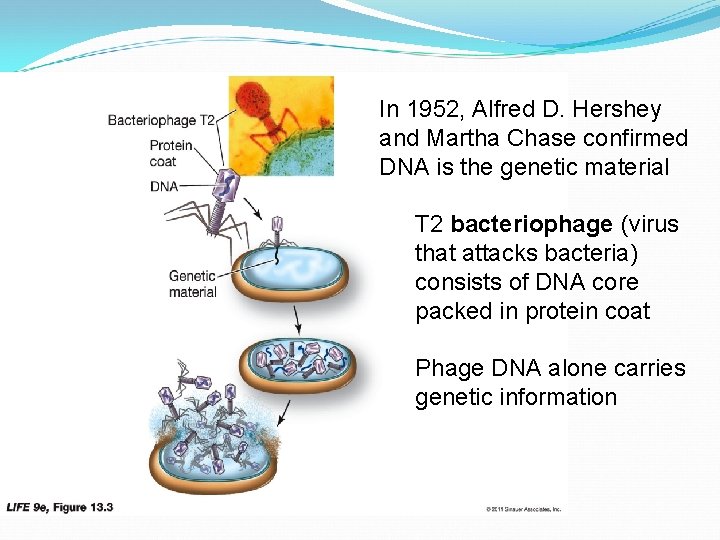
In 1952, Alfred D. Hershey and Martha Chase confirmed DNA is the genetic material T 2 bacteriophage (virus that attacks bacteria) consists of DNA core packed in protein coat Phage DNA alone carries genetic information




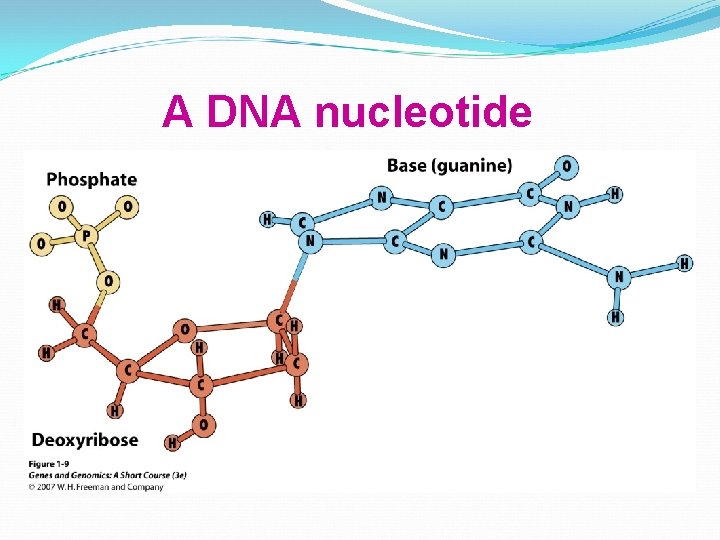
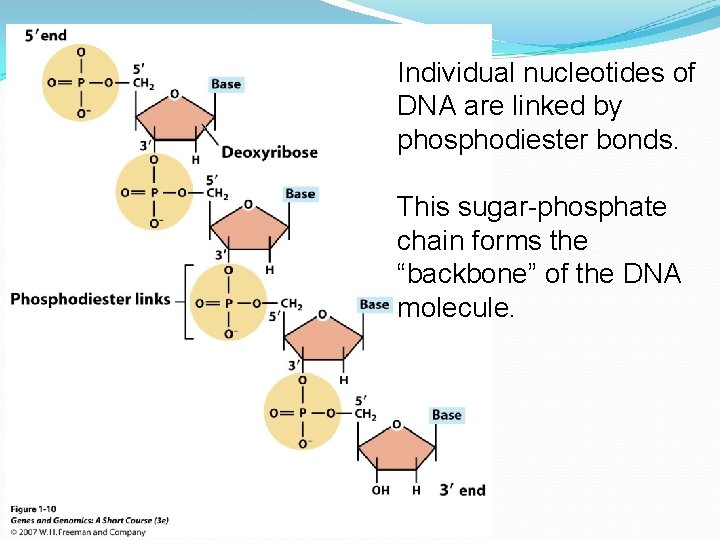
A DNA nucleotide

Individual nucleotides of DNA are linked by phosphodiester bonds. This sugar-phosphate chain forms the “backbone” of the DNA molecule.


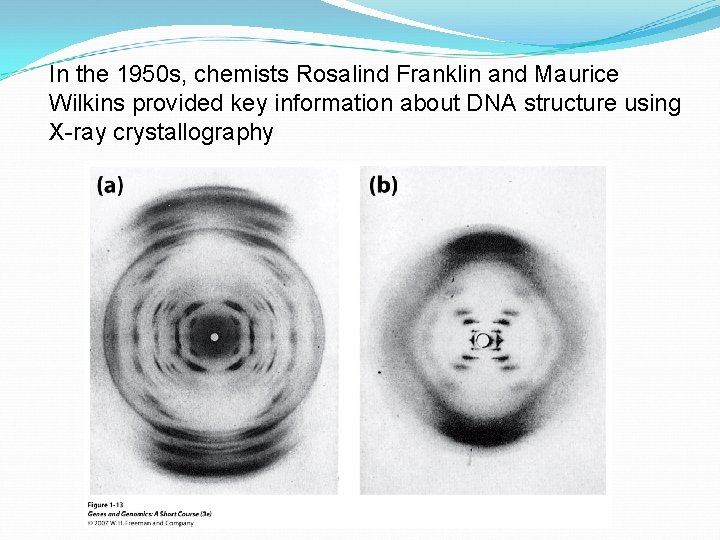
In the 1950 s, chemists Rosalind Franklin and Maurice Wilkins provided key information about DNA structure using X-ray crystallography

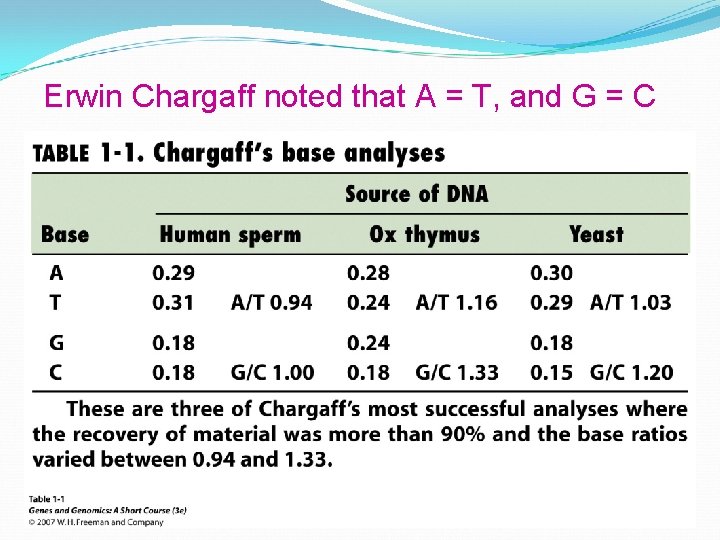
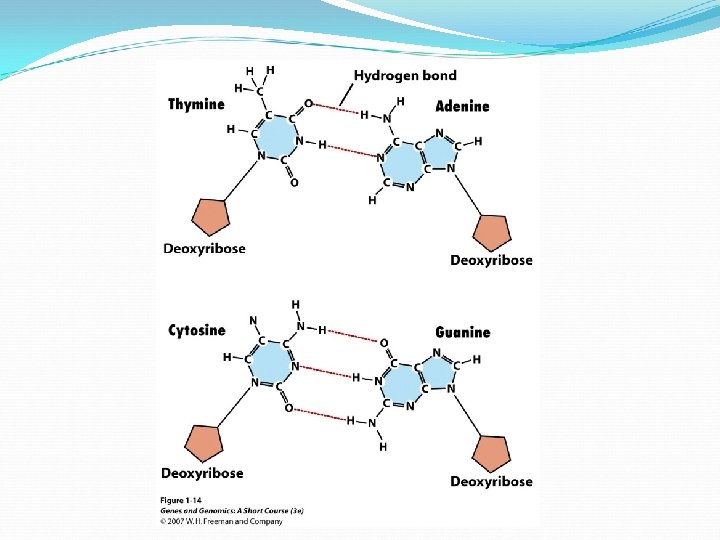
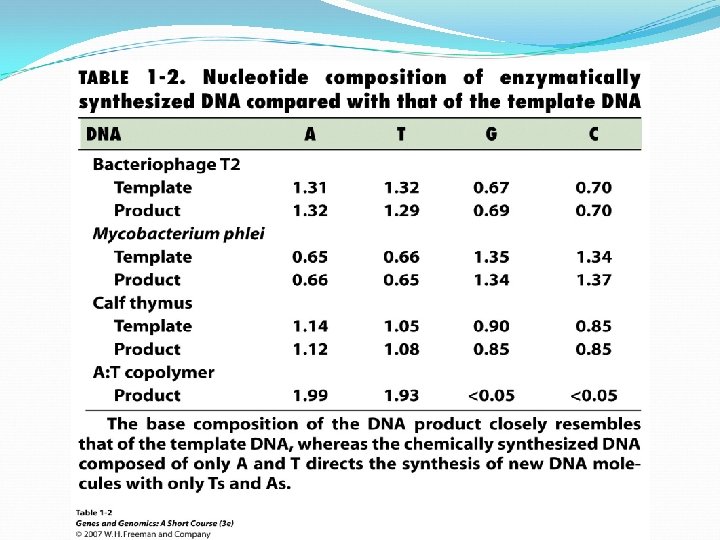
Erwin Chargaff noted that A = T, and G = C


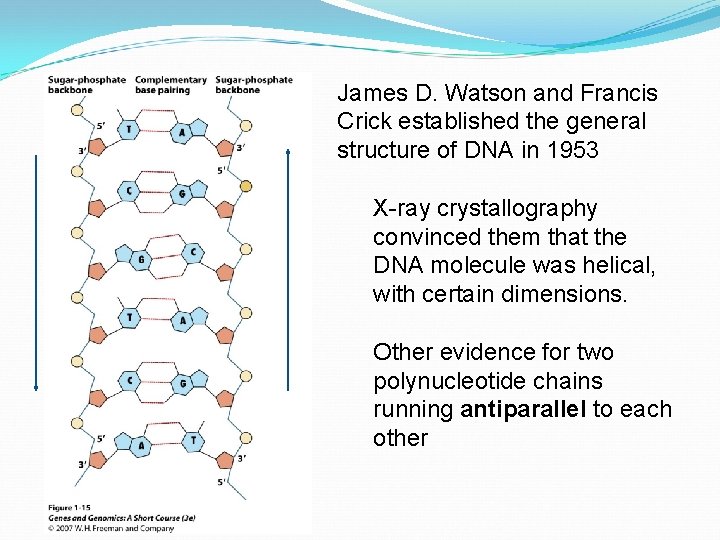
James D. Watson and Francis Crick established the general structure of DNA in 1953 X-ray crystallography convinced them that the DNA molecule was helical, with certain dimensions. Other evidence for two polynucleotide chains running antiparallel to each other

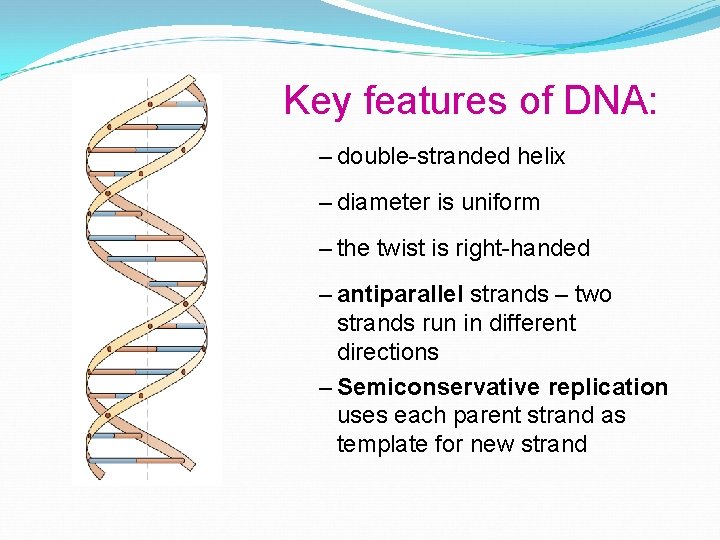
Key features of DNA: – double-stranded helix – diameter is uniform – the twist is right-handed – antiparallel strands – two strands run in different directions – Semiconservative replication uses each parent strand as template for new strand

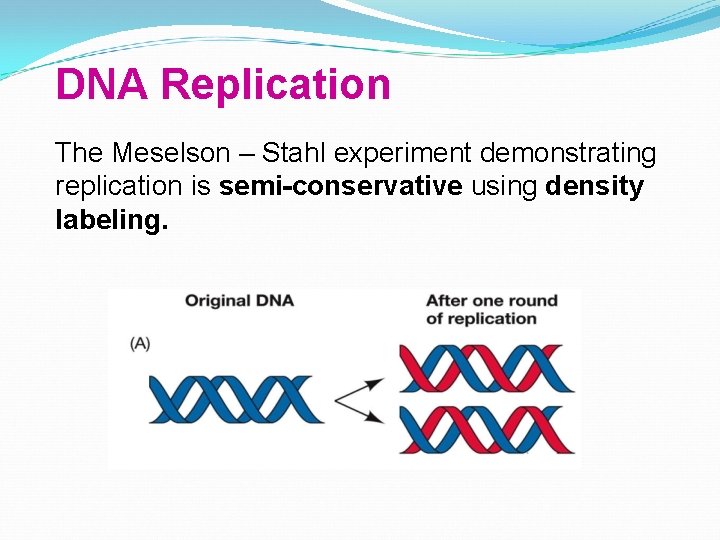
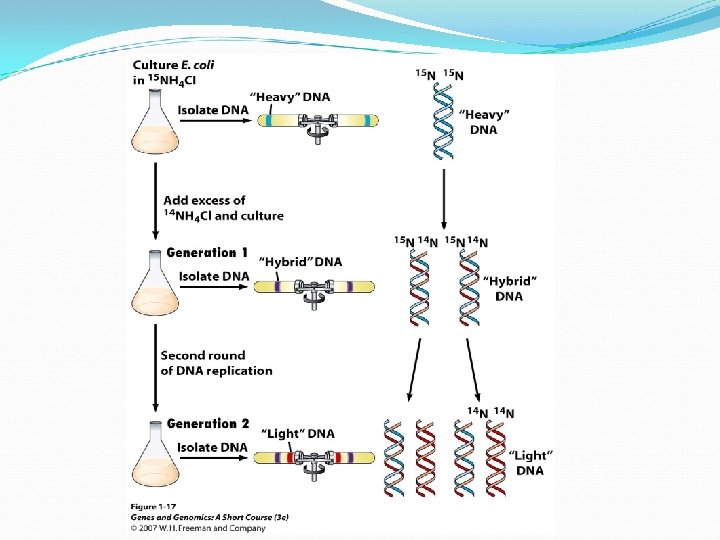
DNA Replication The Meselson – Stahl experiment demonstrating replication is semi-conservative using density labeling.



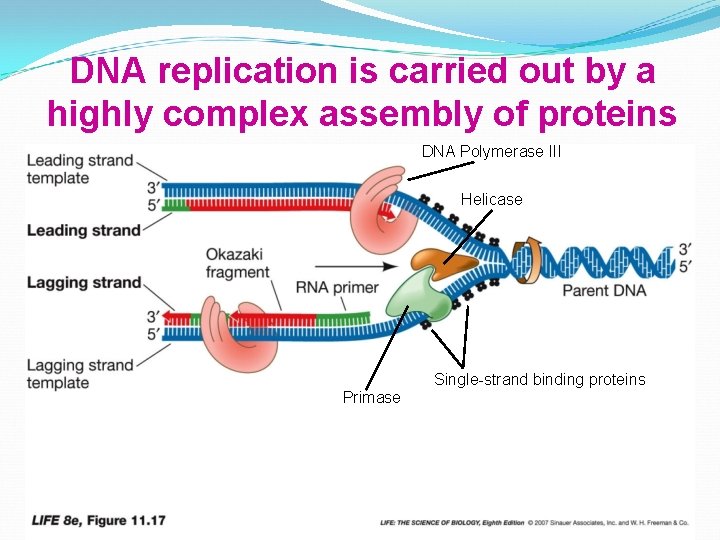
DNA replication is carried out by a highly complex assembly of proteins DNA Polymerase III Helicase Single-strand binding proteins Primase

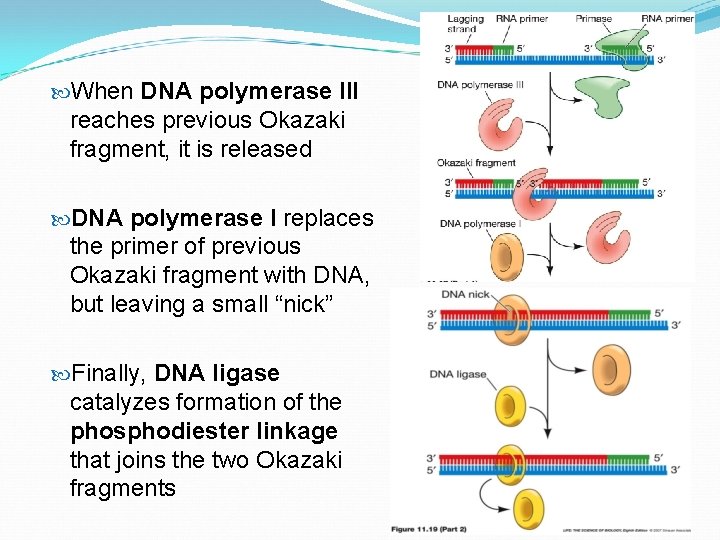
When DNA polymerase III reaches previous Okazaki fragment, it is released DNA polymerase I replaces the primer of previous Okazaki fragment with DNA, but leaving a small “nick” Finally, DNA ligase catalyzes formation of the phosphodiester linkage that joins the two Okazaki fragments

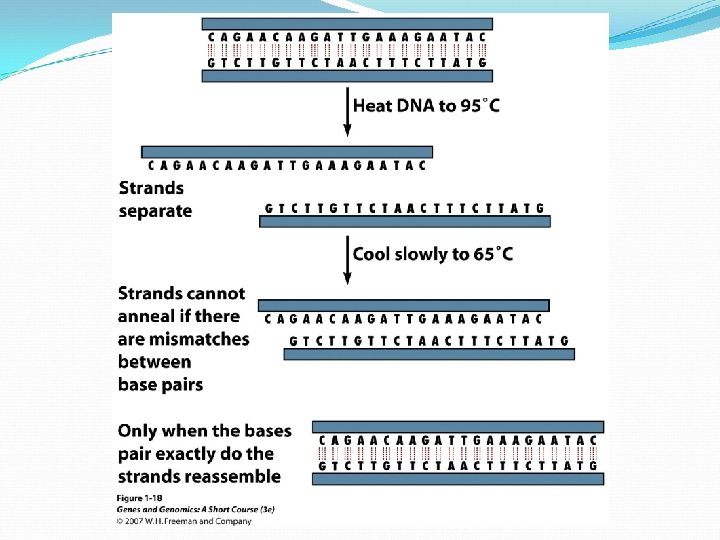
Denature and Renature of DNA double helix is stabilized by numerous hydrogen bonds between complimentary base pairs. DNA is denatured or melt if exposed to high temperature or extreme of p. H. The melting temperature (Tm) is the temperature at which half of the DNA molecules in a sample have been denatured. Double helices with an access of G: C base pairs are more stable and have higher Tm than helices in which A: T base pairs predominate.


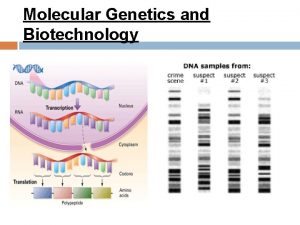
Annealing or hybridization is an essential and powerful tool Labeled DNA fragment can be used as a probe to find its complement, even in a whole genome. The gene of interest can be identified and isolate from DNA library. Specific primers can be designed to amplify the gene of interest in polymerase chain reaction. By observing the extent of annealing between DNA strands in solution, researchers can determine the degree of similarity between DNA molecules from different species.

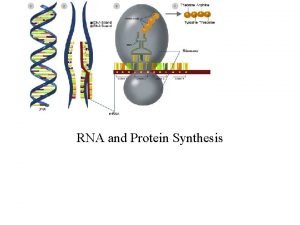
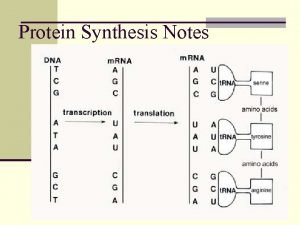
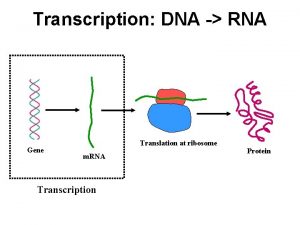
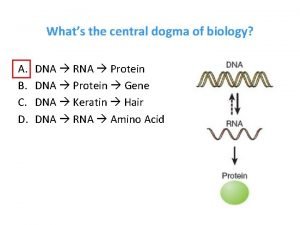
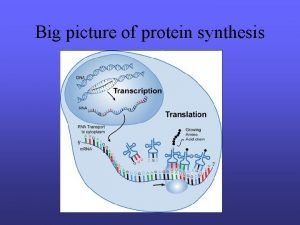
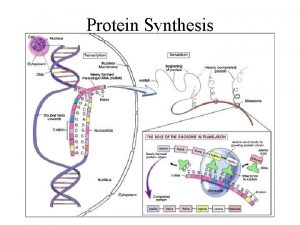
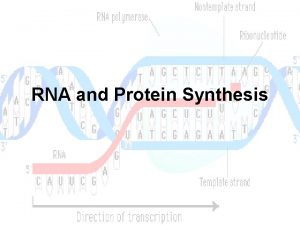
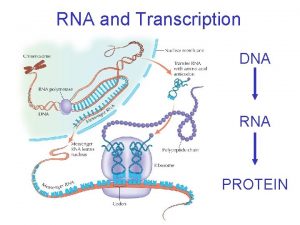
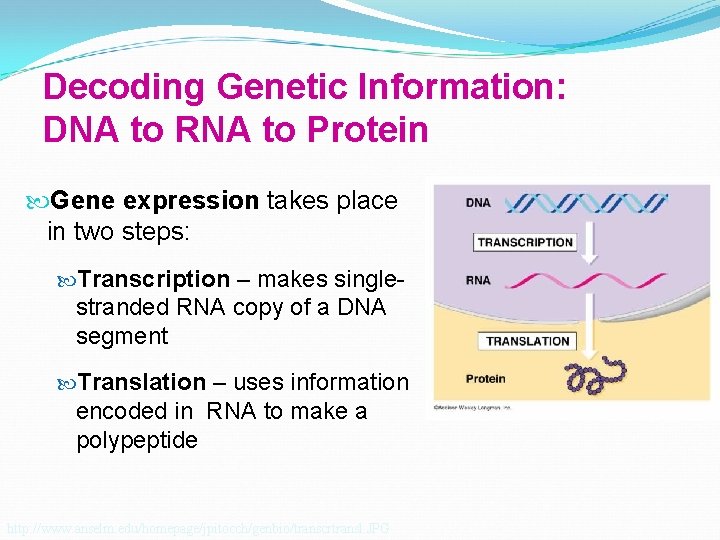
Decoding Genetic Information: DNA to RNA to Protein Gene expression takes place in two steps: Transcription – makes single- stranded RNA copy of a DNA segment Translation – uses information encoded in RNA to make a polypeptide http: //www. anselm. edu/homepage/jpitocch/genbio/transcrtransl. JPG

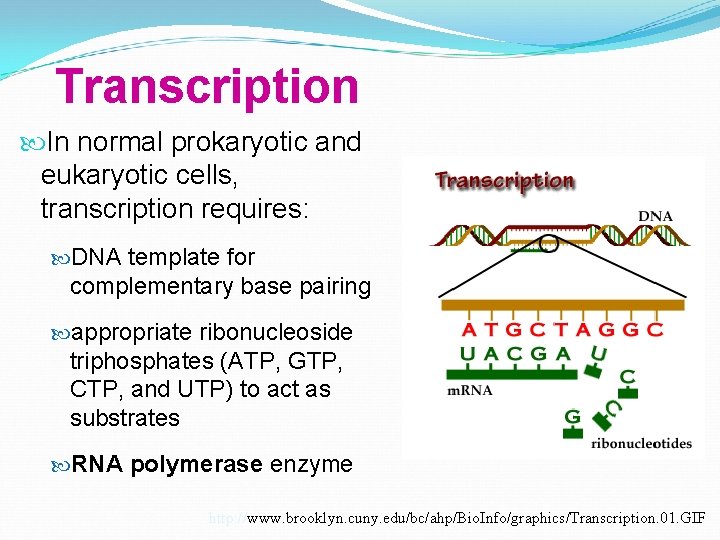
Transcription In normal prokaryotic and eukaryotic cells, transcription requires: DNA template for complementary base pairing appropriate ribonucleoside triphosphates (ATP, GTP, CTP, and UTP) to act as substrates RNA polymerase enzyme http: //www. brooklyn. cuny. edu/bc/ahp/Bio. Info/graphics/Transcription. 01. GIF


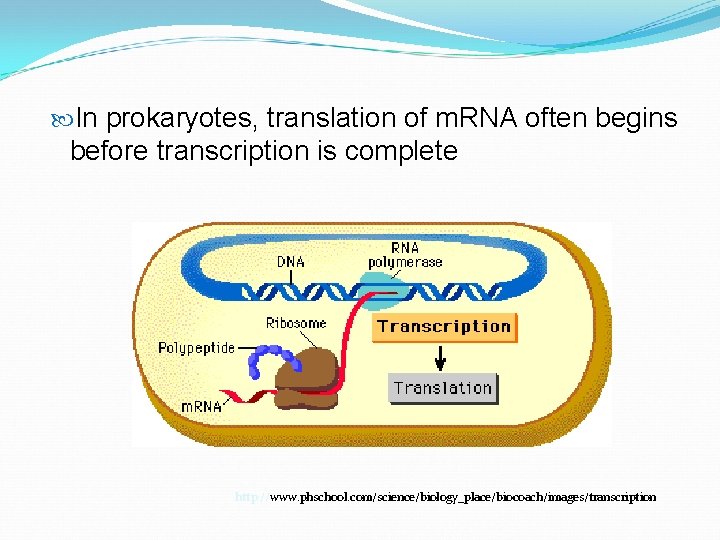
In prokaryotes, translation of m. RNA often begins before transcription is complete http: //www. phschool. com/science/biology_place/biocoach/images/transcription

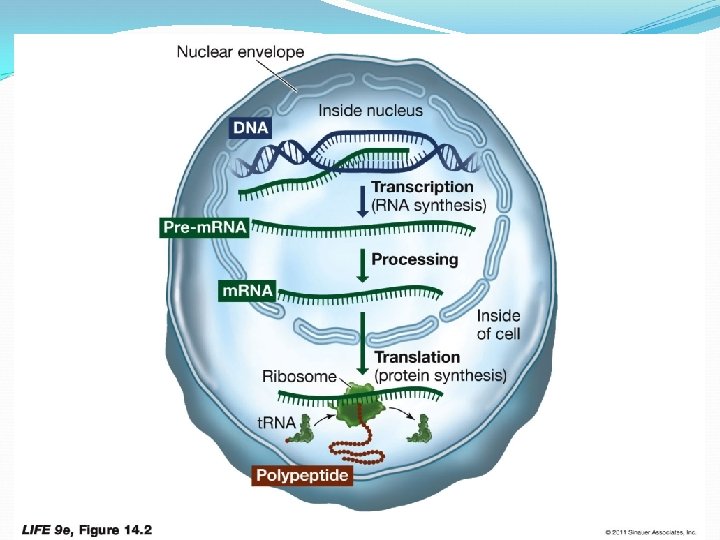
Figure 14. 2 From Gene to Protein

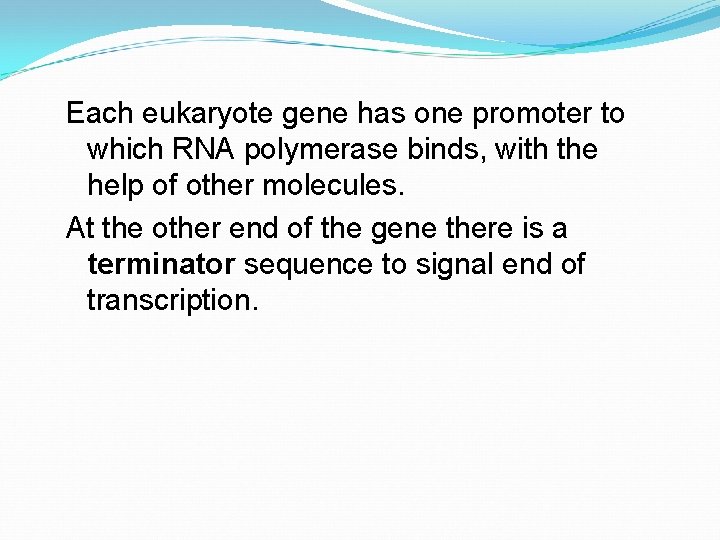
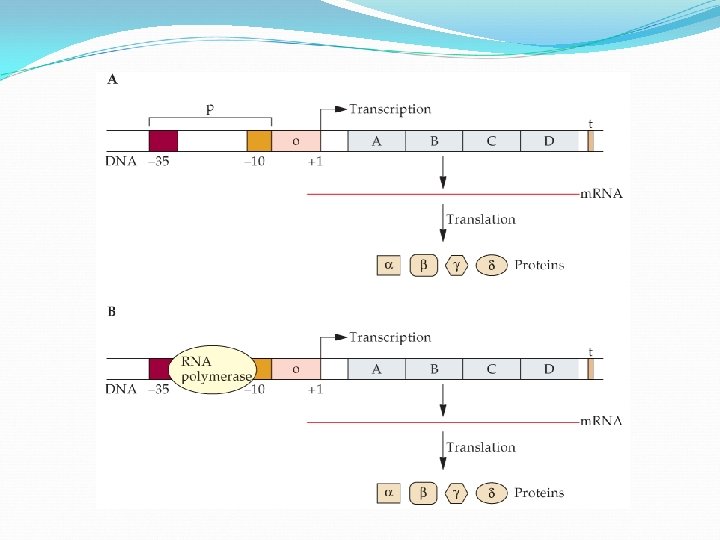
Each eukaryote gene has one promoter to which RNA polymerase binds, with the help of other molecules. At the other end of the gene there is a terminator sequence to signal end of transcription.

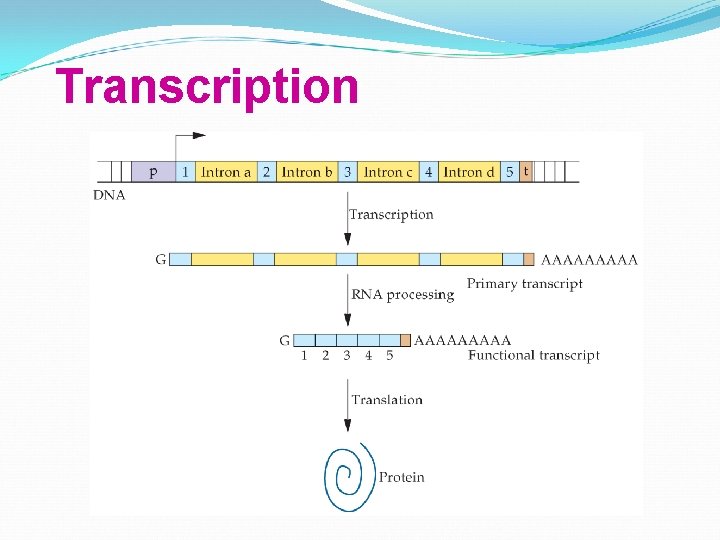
Transcription

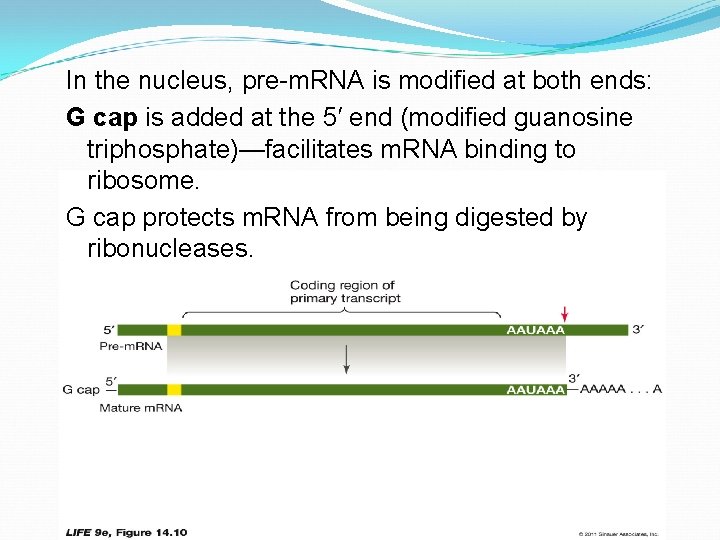
In the nucleus, pre-m. RNA is modified at both ends: G cap is added at the 5′ end (modified guanosine triphosphate)—facilitates m. RNA binding to ribosome. G cap protects m. RNA from being digested by ribonucleases.

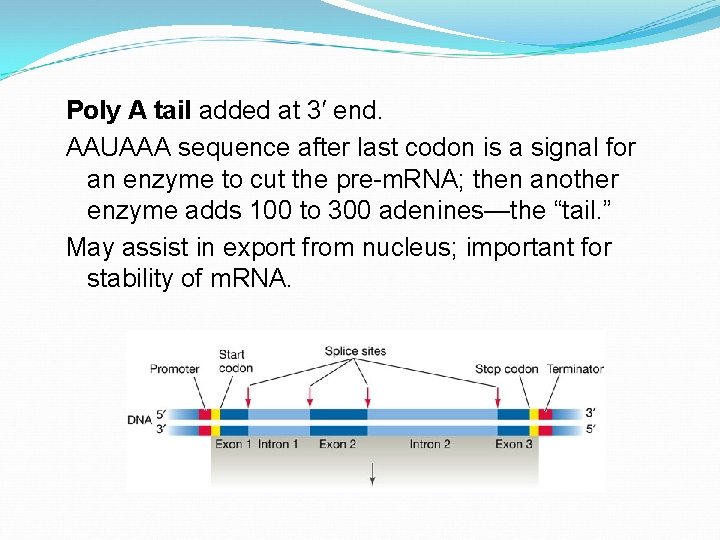
Poly A tail added at 3′ end. AAUAAA sequence after last codon is a signal for an enzyme to cut the pre-m. RNA; then another enzyme adds 100 to 300 adenines—the “tail. ” May assist in export from nucleus; important for stability of m. RNA.

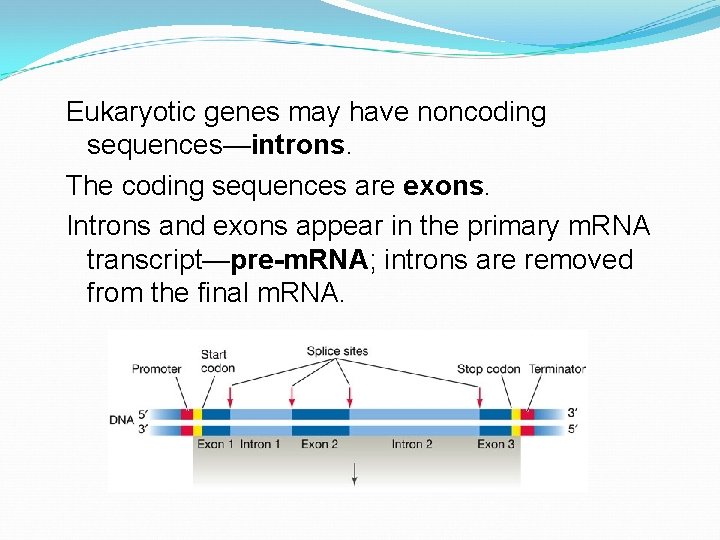
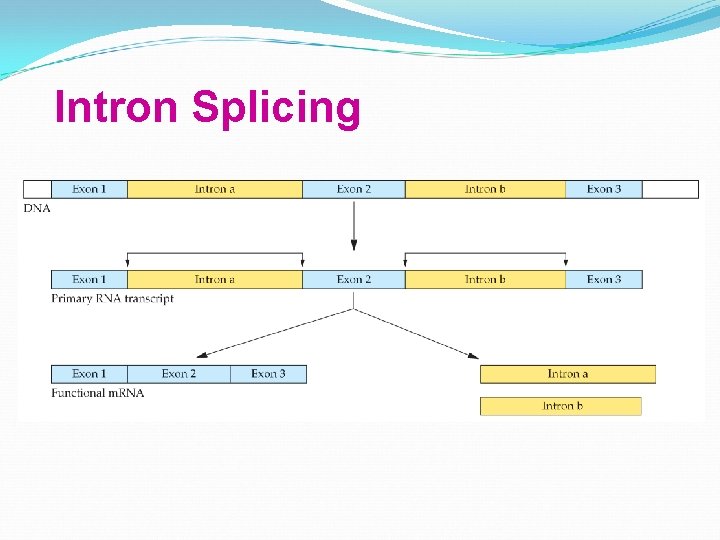
Eukaryotic genes may have noncoding sequences—introns. The coding sequences are exons. Introns and exons appear in the primary m. RNA transcript—pre-m. RNA; introns are removed from the final m. RNA.

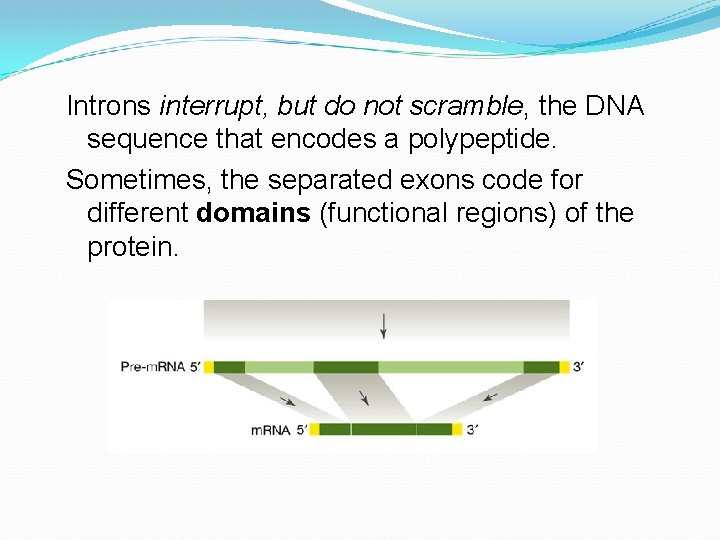
Introns interrupt, but do not scramble, the DNA sequence that encodes a polypeptide. Sometimes, the separated exons code for different domains (functional regions) of the protein.

Intron Splicing

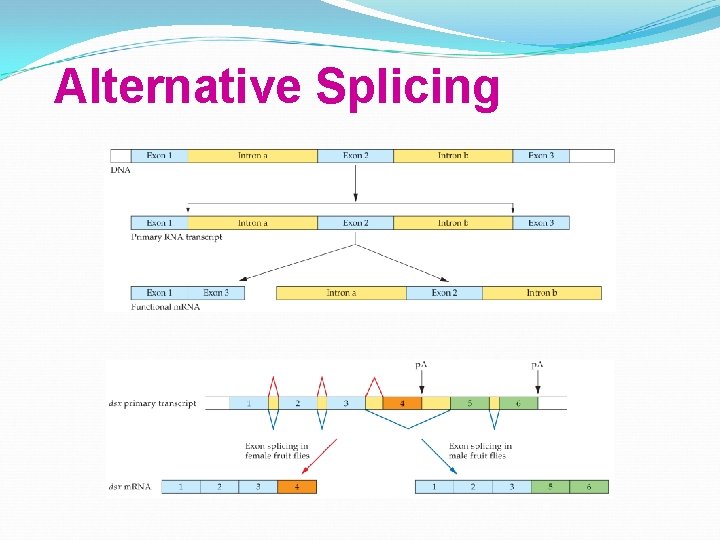
Alternative Splicing

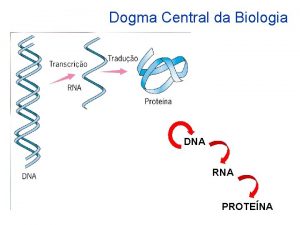
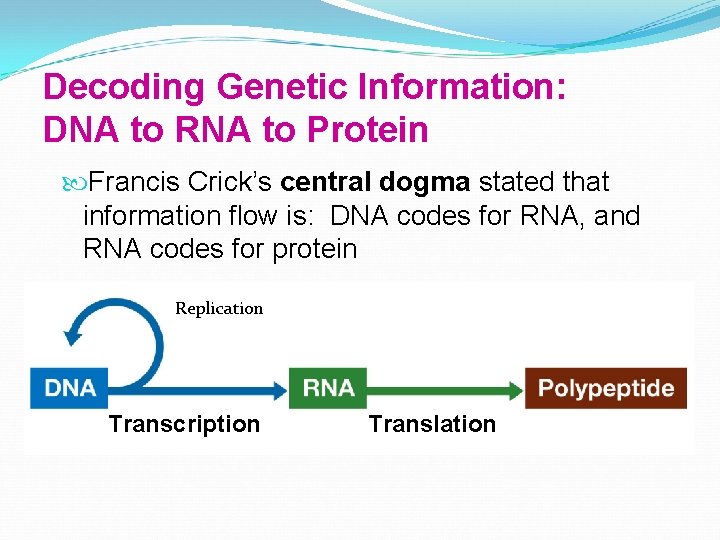
Decoding Genetic Information: DNA to RNA to Protein Francis Crick’s central dogma stated that information flow is: DNA codes for RNA, and RNA codes for protein Replication Transcription Translation

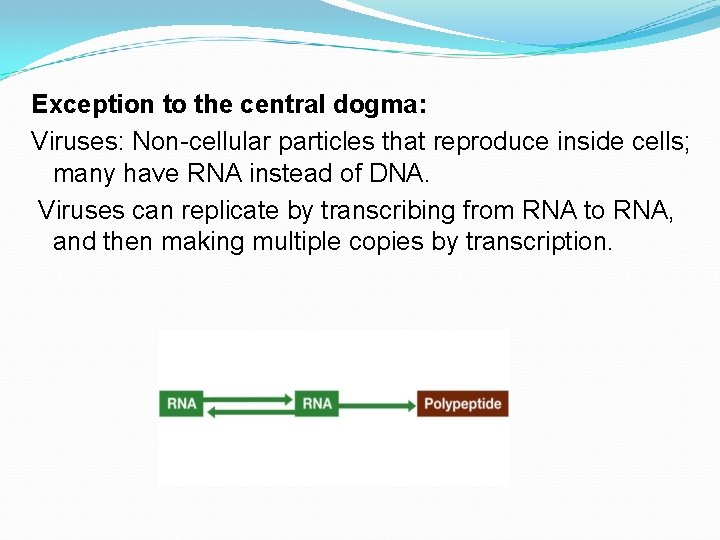
Exception to the central dogma: Viruses: Non-cellular particles that reproduce inside cells; many have RNA instead of DNA. Viruses can replicate by transcribing from RNA to RNA, and then making multiple copies by transcription.

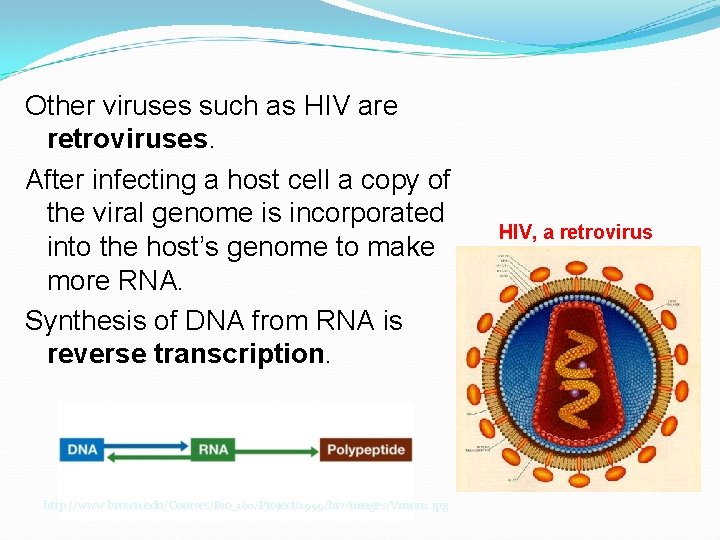
Other viruses such as HIV are retroviruses. After infecting a host cell a copy of the viral genome is incorporated into the host’s genome to make more RNA. Synthesis of DNA from RNA is reverse transcription. http: //www. brown. edu/Courses/Bio_160/Projects 1999/hiv/images/Virion 2. jpg HIV, a retrovirus

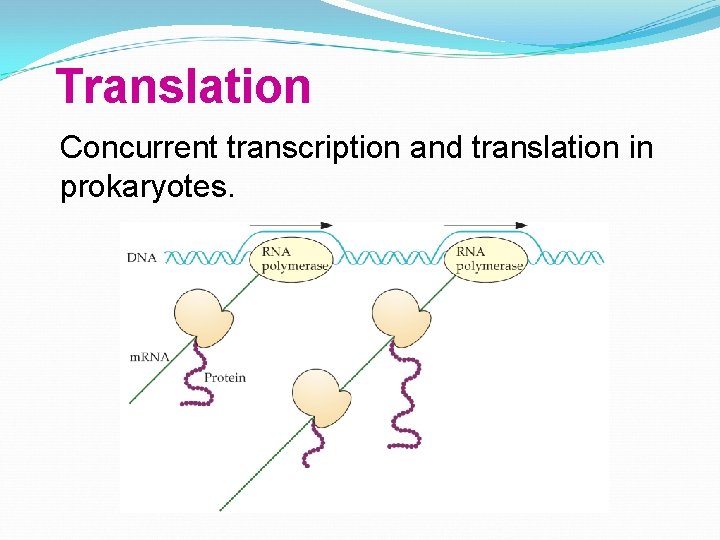
Translation Concurrent transcription and translation in prokaryotes.

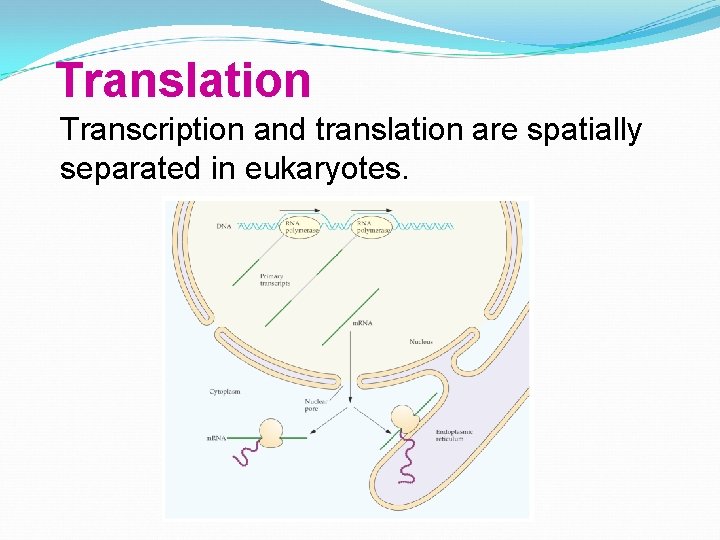
Translation Transcription and translation are spatially separated in eukaryotes.

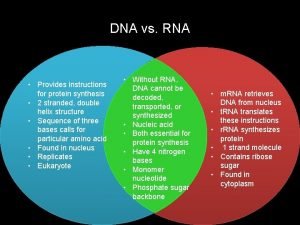
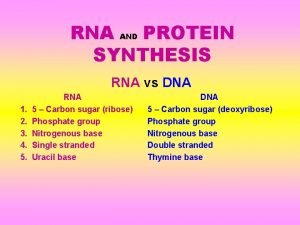
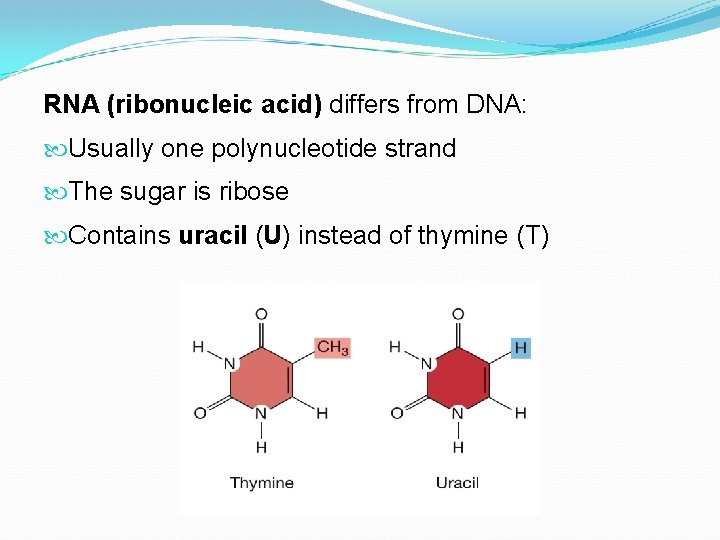
RNA (ribonucleic acid) differs from DNA: Usually one polynucleotide strand The sugar is ribose Contains uracil (U) instead of thymine (T)

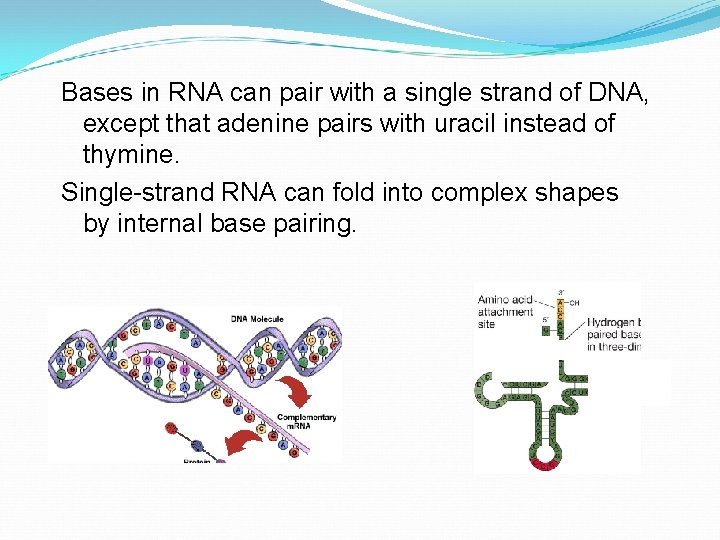
Bases in RNA can pair with a single strand of DNA, except that adenine pairs with uracil instead of thymine. Single-strand RNA can fold into complex shapes by internal base pairing.

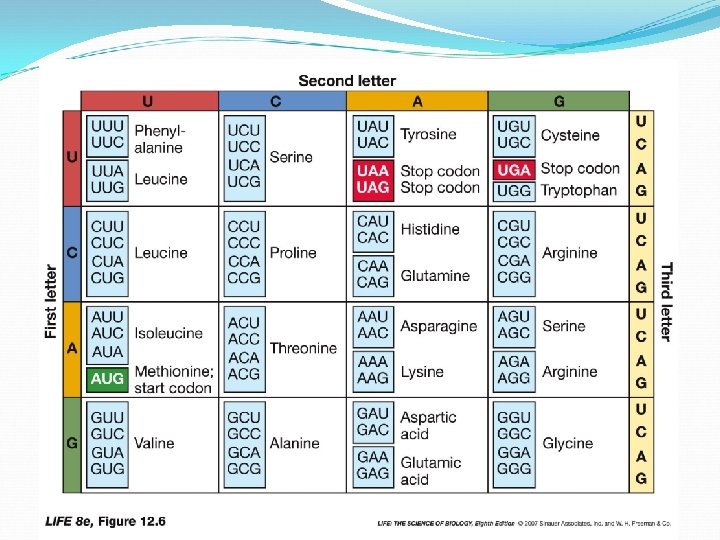
Three kinds of RNA in protein synthesis: Messenger RNA (m. RNA)—carries copy of a DNA sequence to site of protein synthesis at the ribosome Transfer RNA (t. RNA)—carries amino acids for polypeptide assembly Ribosomal RNA (r. RNA)—catalyzes peptide bonds and provides structure

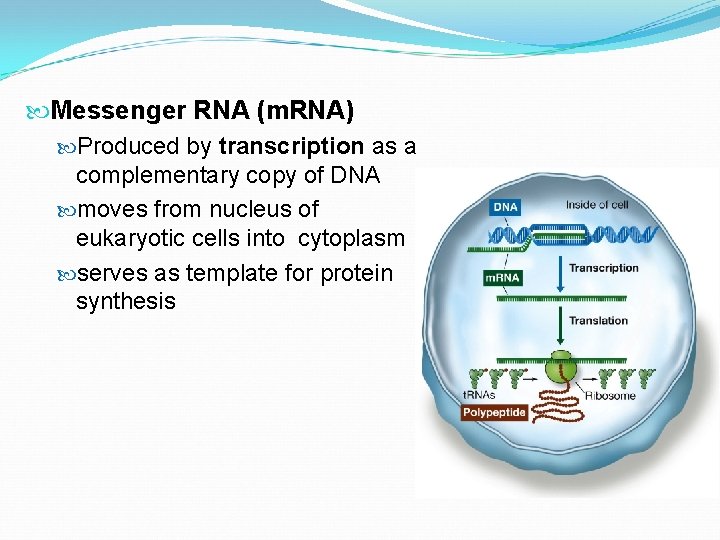
Messenger RNA (m. RNA) Produced by transcription as a complementary copy of DNA moves from nucleus of eukaryotic cells into cytoplasm serves as template for protein synthesis

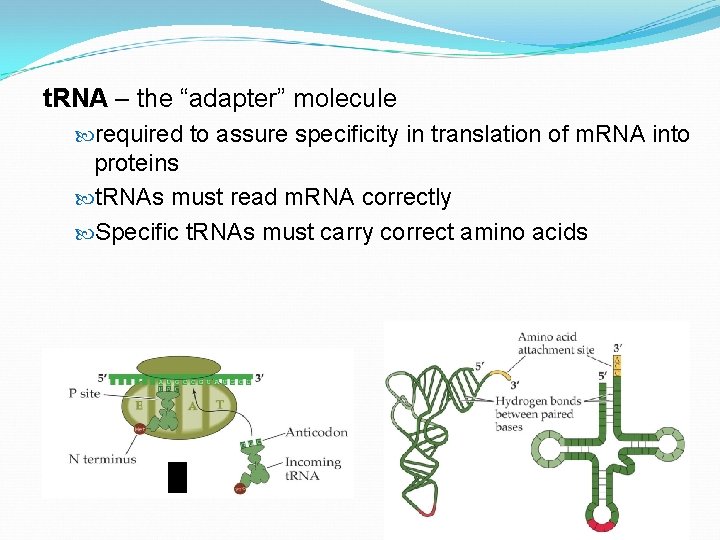
t. RNA – the “adapter” molecule required to assure specificity in translation of m. RNA into proteins t. RNAs must read m. RNA correctly Specific t. RNAs must carry correct amino acids

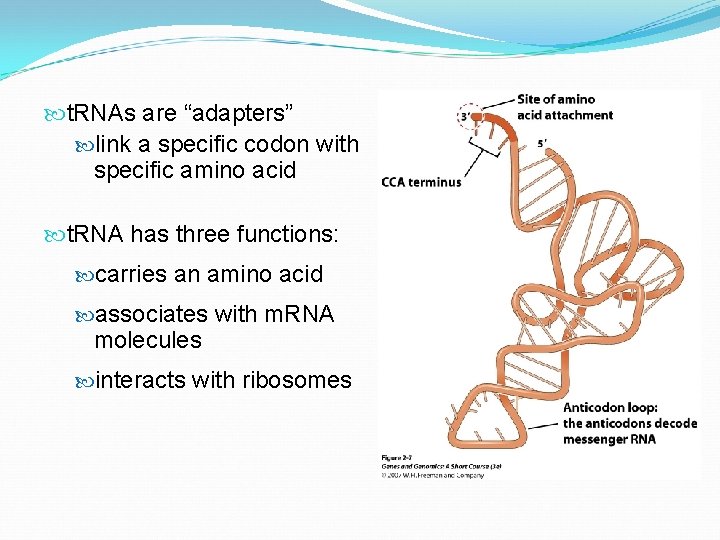
t. RNAs are “adapters” link a specific codon with specific amino acid t. RNA has three functions: carries an amino acid associates with m. RNA molecules interacts with ribosomes

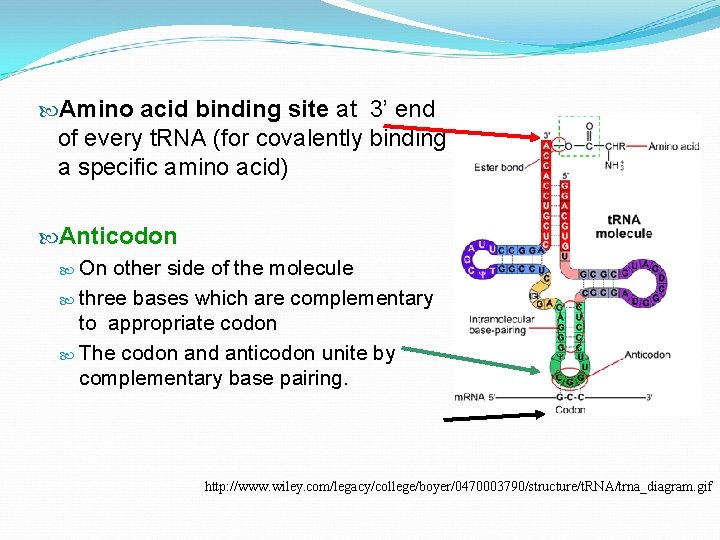
Amino acid binding site at 3’ end of every t. RNA (for covalently binding a specific amino acid) Anticodon On other side of the molecule three bases which are complementary to appropriate codon The codon and anticodon unite by complementary base pairing. http: //www. wiley. com/legacy/college/boyer/0470003790/structure/t. RNA/trna_diagram. gif

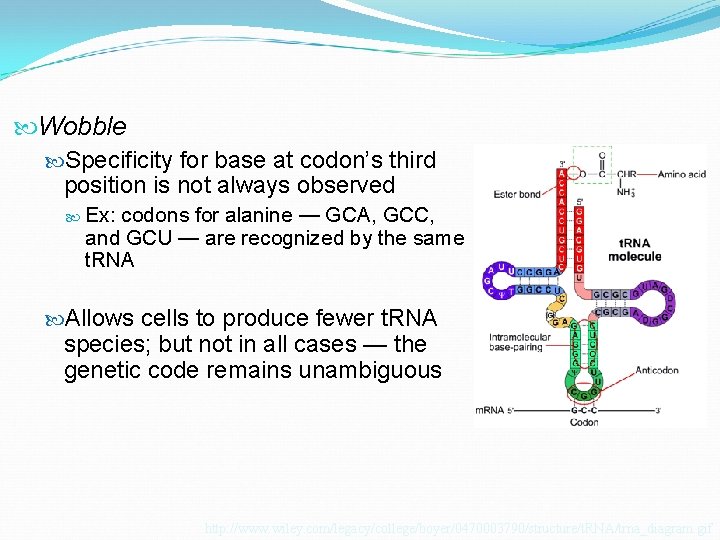
Wobble Specificity for base at codon’s third position is not always observed Ex: codons for alanine — GCA, GCC, and GCU — are recognized by the same t. RNA Allows cells to produce fewer t. RNA species; but not in all cases — the genetic code remains unambiguous http: //www. wiley. com/legacy/college/boyer/0470003790/structure/t. RNA/trna_diagram. gif

Aminoacyl-t. RNA synthetases Attaches amino acids to t. RNAs Their three-part active sites bind: a specific amino acid ATP a specific t. RNA, charged with a high-energy bond High-energy bond provides energy for making a peptide bond


Translation also occurs in three steps: Initiation Elongation Termination http: //srv 2. lycoming. edu/~newman/courses/bio 43704/ribosome 1. jpg

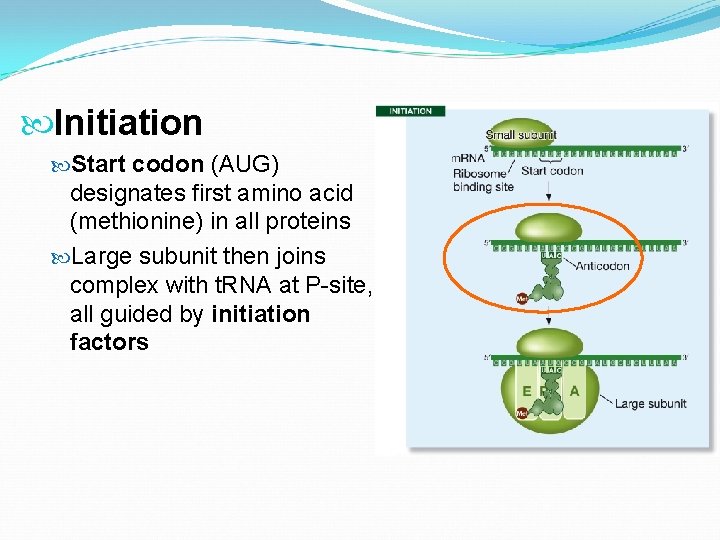
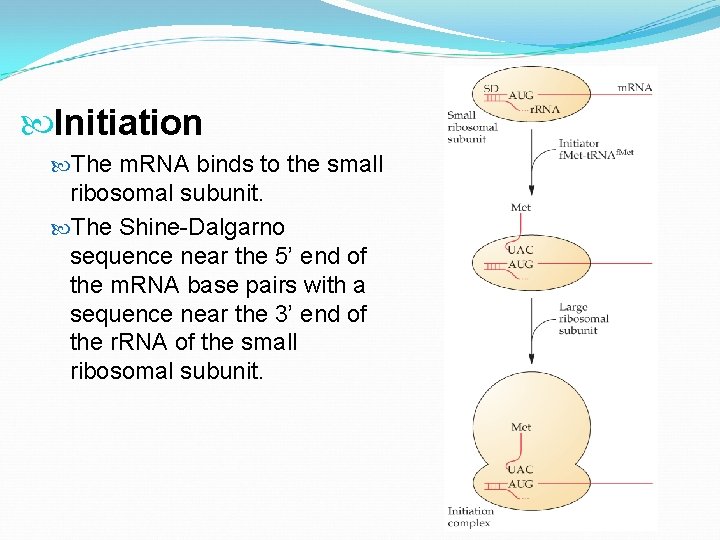
Initiation Start codon (AUG) designates first amino acid (methionine) in all proteins Large subunit then joins complex with t. RNA at P-site, all guided by initiation factors

Initiation The m. RNA binds to the small ribosomal subunit. The Shine-Dalgarno sequence near the 5’ end of the m. RNA base pairs with a sequence near the 3’ end of the r. RNA of the small ribosomal subunit.

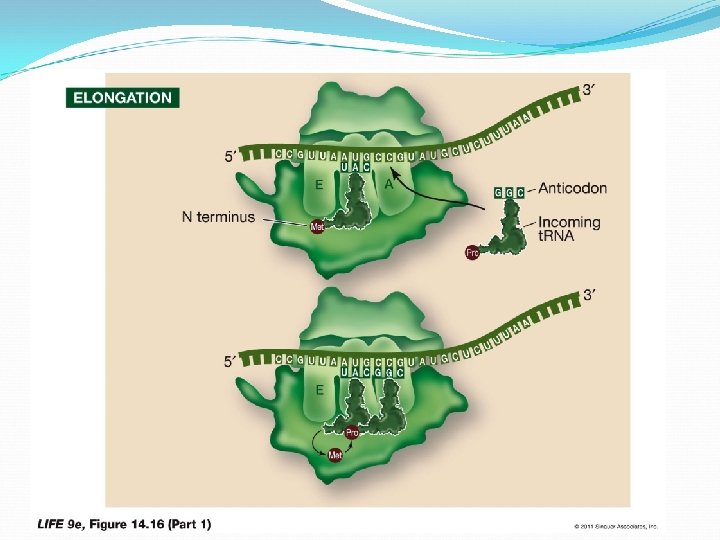
Elongation: The second charged t. RNA enters the A site. Large subunit catalyzes two reactions: It breaks bond between t. RNA in P site and its amino acid Peptide bond forms between that amino acid and the amino acid on t. RNA in the A site When the first t. RNA has released its methionine, it moves to the E site and dissociates from the ribosome—can then become charged again. Elongation occurs as the steps are repeated, assisted by proteins called elongation factors.



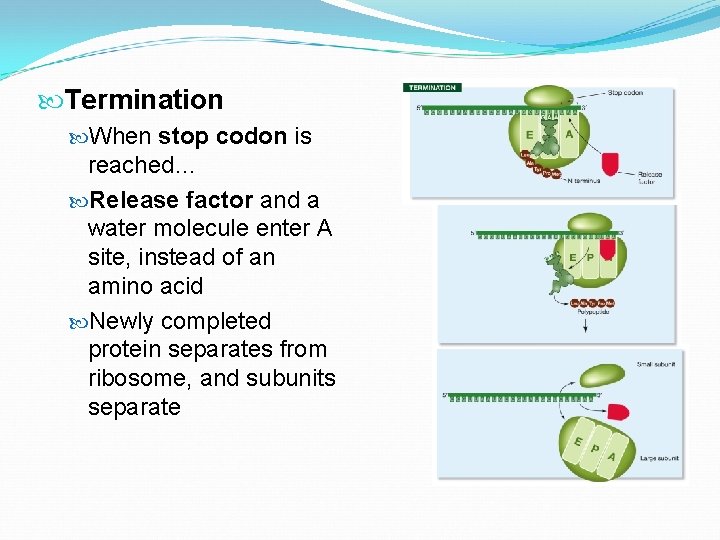
Termination When stop codon is reached… Release factor and a water molecule enter A site, instead of an amino acid Newly completed protein separates from ribosome, and subunits separate

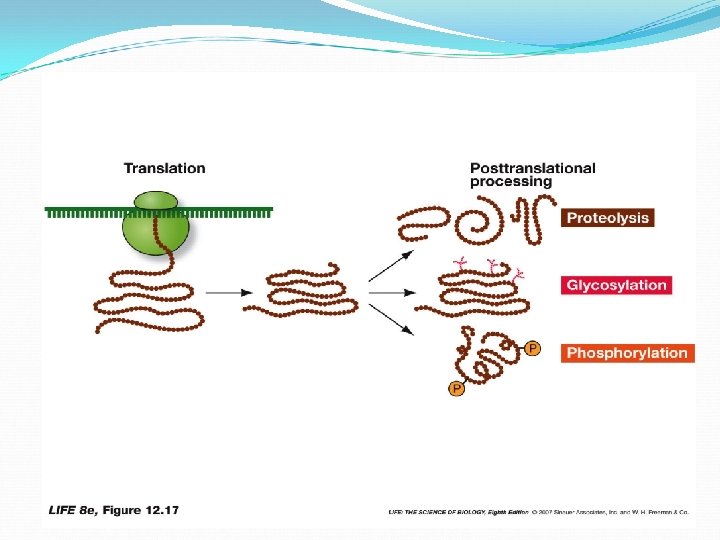
Protein modifications: Proteolysis: Cutting of a long polypeptide chain into final products, by proteases Glycosylation: Addition of sugars to form glycoproteins Phosphorylation: Addition of phosphate groups catalyzed by protein kinases— charged phosphate groups change the conformation


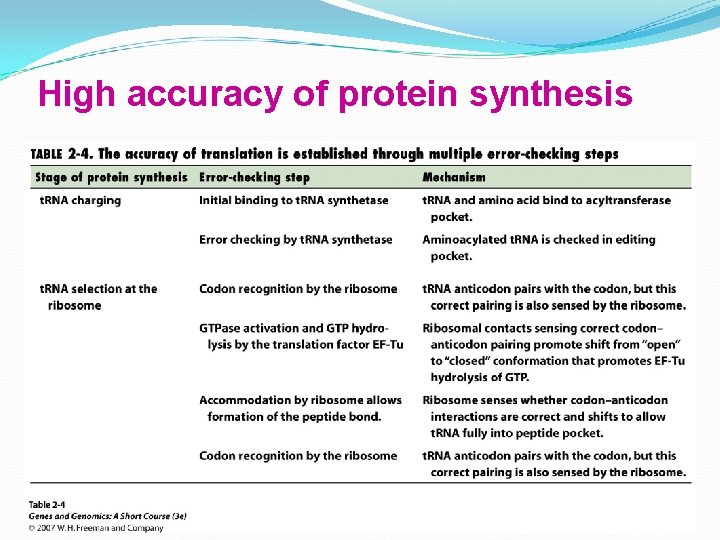
High accuracy of protein synthesis


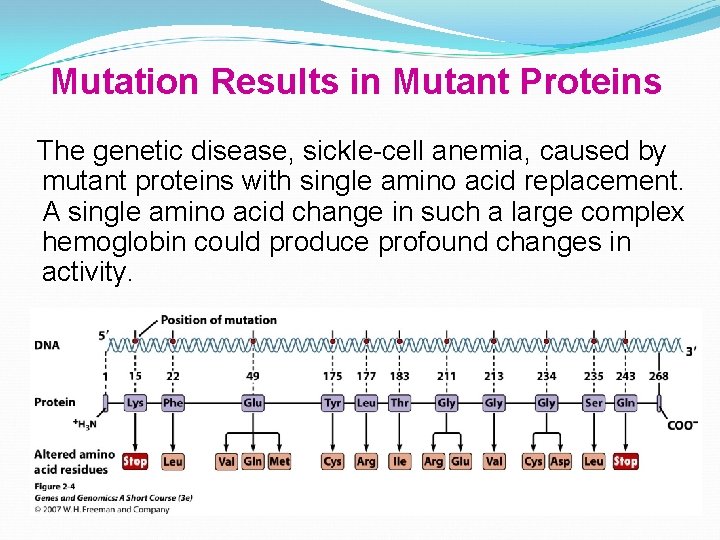
Mutation Results in Mutant Proteins The genetic disease, sickle-cell anemia, caused by mutant proteins with single amino acid replacement. A single amino acid change in such a large complex hemoglobin could produce profound changes in activity.


Control of Gene Expression Cells conserve energy and resources by making proteins only when needed. Intricate molecular mechanisms must exist in cells to control the numbers of their many proteins. There are three ways that might achieve this differential synthesis: transcriptional control, translational control, and post-transcriptional control.

Control of Gene Expression Cells regulate protein synthesis at any of four points Controlling the rate of m. RNA synthesis The stability of m. RNAs Changing the rate of protein synthesis The stability of proteins

Control of Gene Expression Cells regulate protein synthesis by several methods: Block transcription of the gene Hydrolyze the m. RNA after it is made Prevent translation of m. RNA at the ribosome Hydrolyze the protein after it is made Inhibit the function of the protein

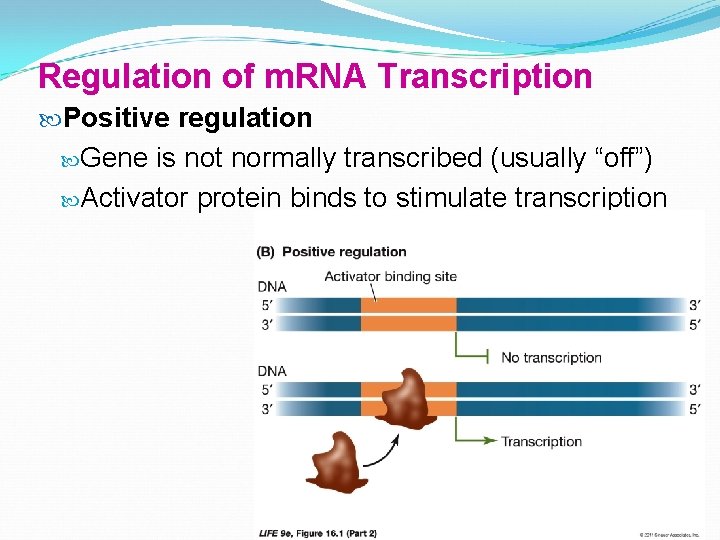
Regulation of m. RNA Transcription Positive regulation Gene is not normally transcribed (usually “off”) Activator protein binds to stimulate transcription

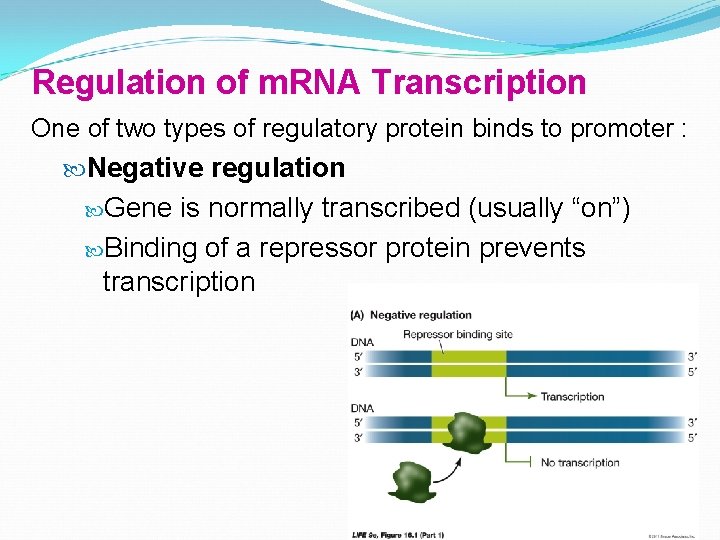
Regulation of m. RNA Transcription One of two types of regulatory protein binds to promoter : Negative regulation Gene is normally transcribed (usually “on”) Binding of a repressor protein prevents transcription

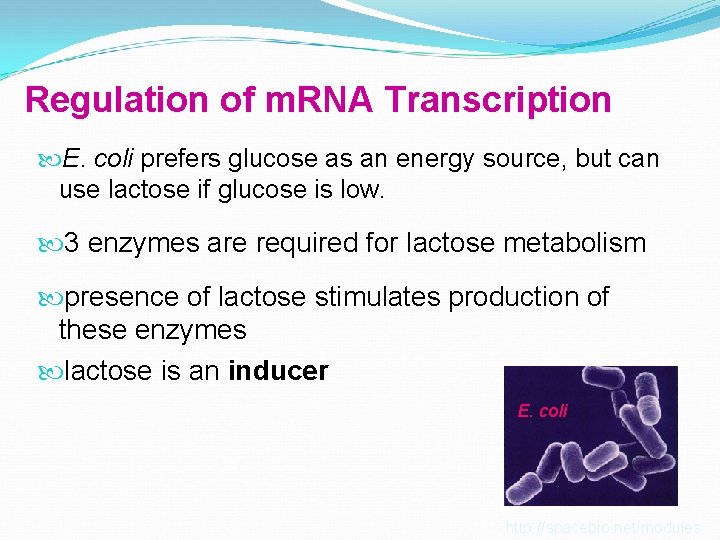
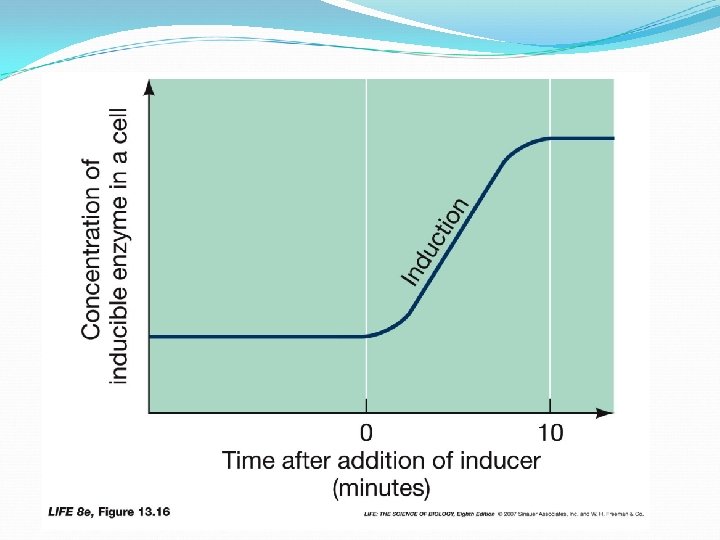
Regulation of m. RNA Transcription E. coli prefers glucose as an energy source, but can use lactose if glucose is low. 3 enzymes are required for lactose metabolism presence of lactose stimulates production of these enzymes lactose is an inducer E. coli http: //spacebio. net/modules

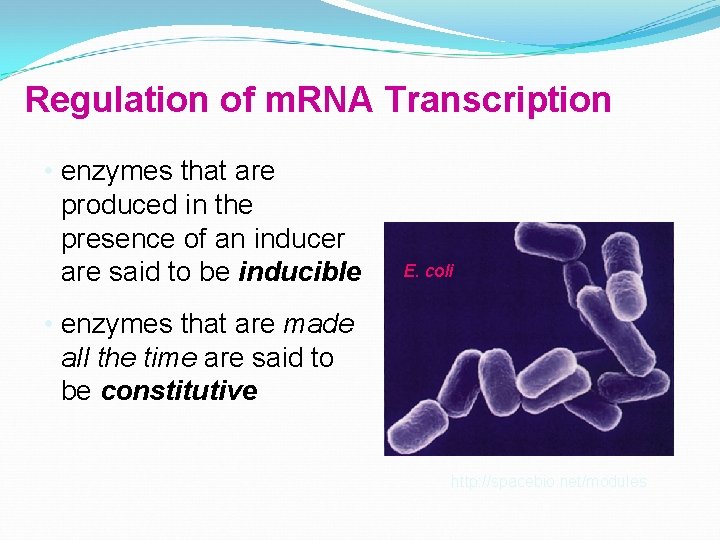
Regulation of m. RNA Transcription • enzymes that are produced in the presence of an inducer are said to be inducible E. coli • enzymes that are made all the time are said to be constitutive http: //spacebio. net/modules


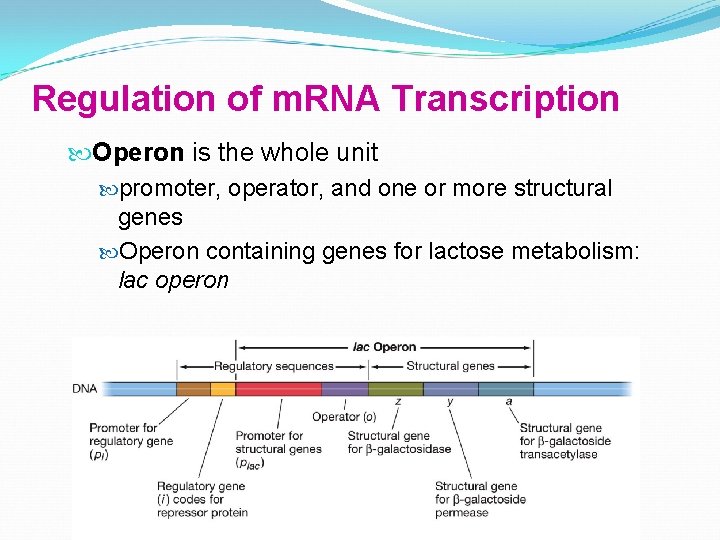
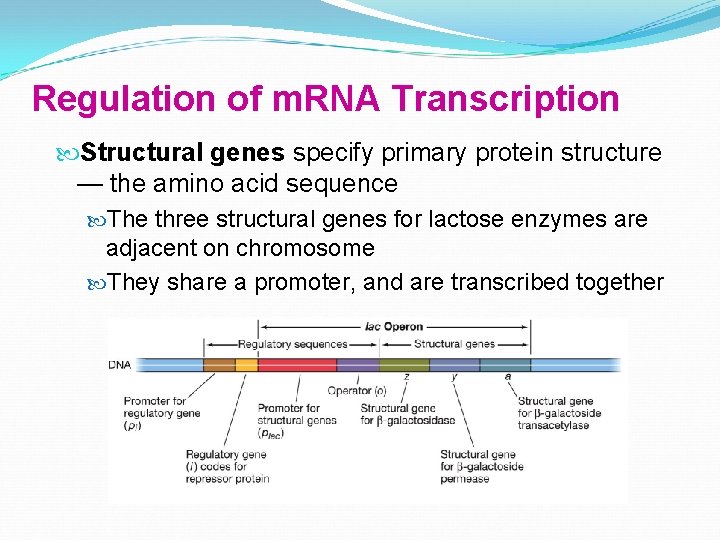
Regulation of m. RNA Transcription Operon is the whole unit promoter, operator, and one or more structural genes Operon containing genes for lactose metabolism: lac operon

Regulation of m. RNA Transcription Structural genes specify primary protein structure — the amino acid sequence The three structural genes for lactose enzymes are adjacent on chromosome They share a promoter, and are transcribed together

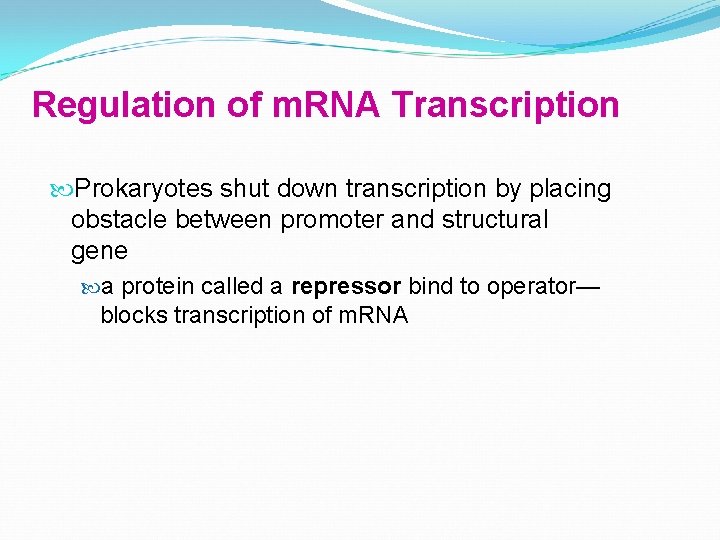
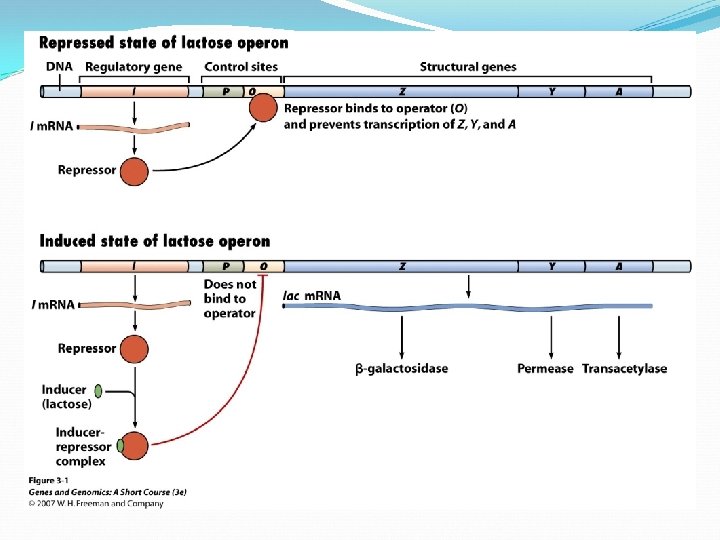
Regulation of m. RNA Transcription Prokaryotes shut down transcription by placing obstacle between promoter and structural gene a protein called a repressor bind to operator— blocks transcription of m. RNA

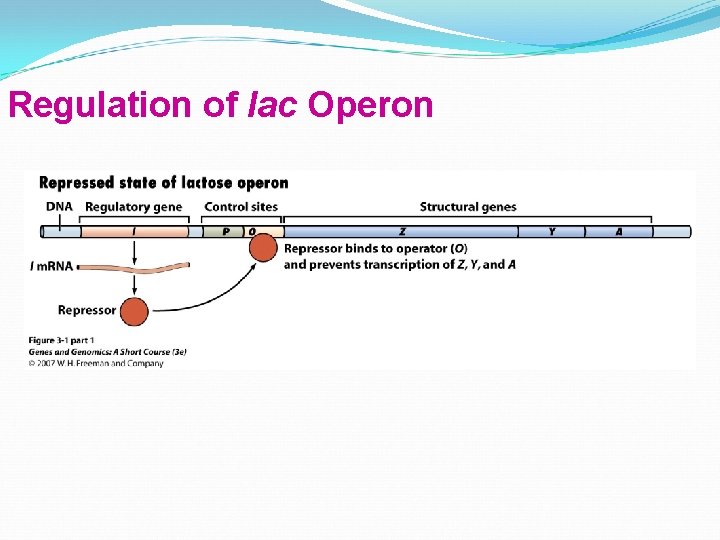
Regulation of lac Operon

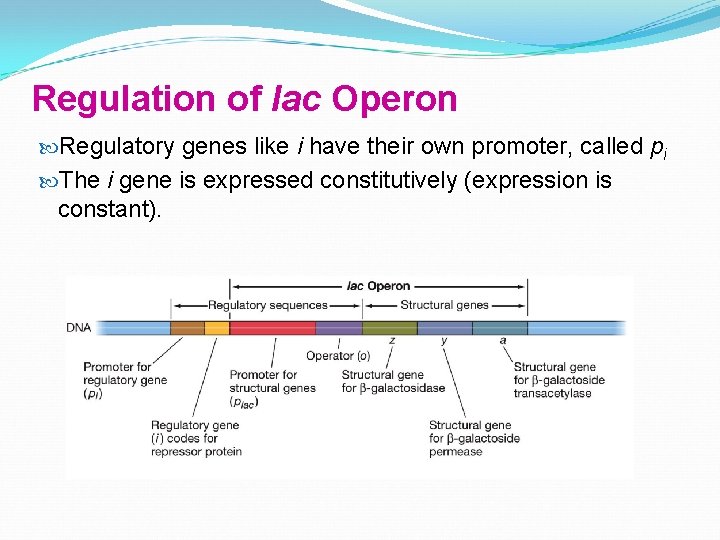
Regulation of lac Operon A repressor protein is coded by a regulatory gene. The regulatory gene that codes for the lac repressor is the i (inducibility) gene is near lac structural genes, but not all regulatory genes are near their operons

Regulation of lac Operon Regulatory genes like i have their own promoter, called pi The i gene is expressed constitutively (expression is constant).

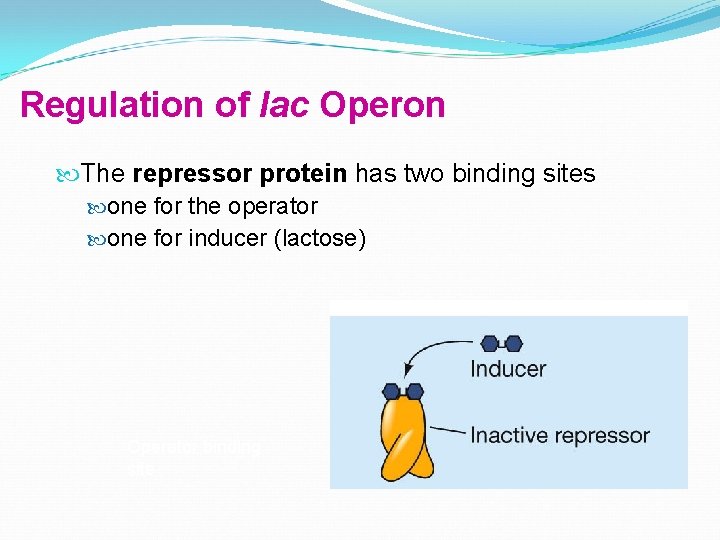
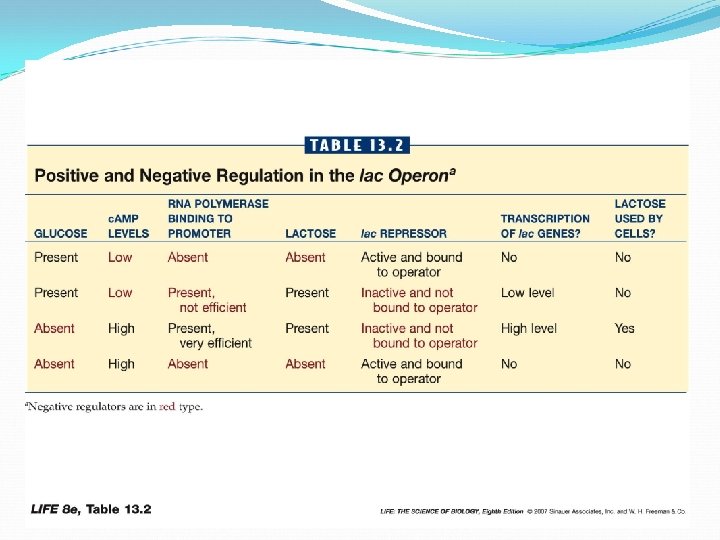
Regulation of lac Operon The repressor protein has two binding sites one for the operator one for inducer (lactose) Operator binding site

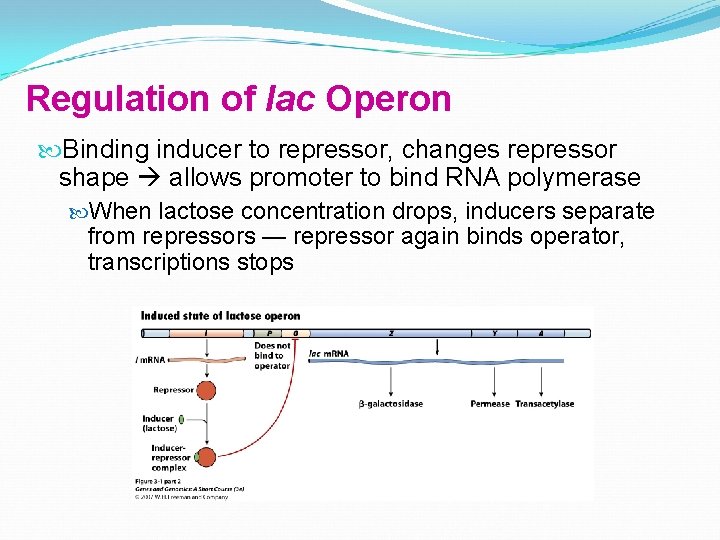
Regulation of lac Operon Binding inducer to repressor, changes repressor shape allows promoter to bind RNA polymerase When lactose concentration drops, inducers separate from repressors — repressor again binds operator, transcriptions stops


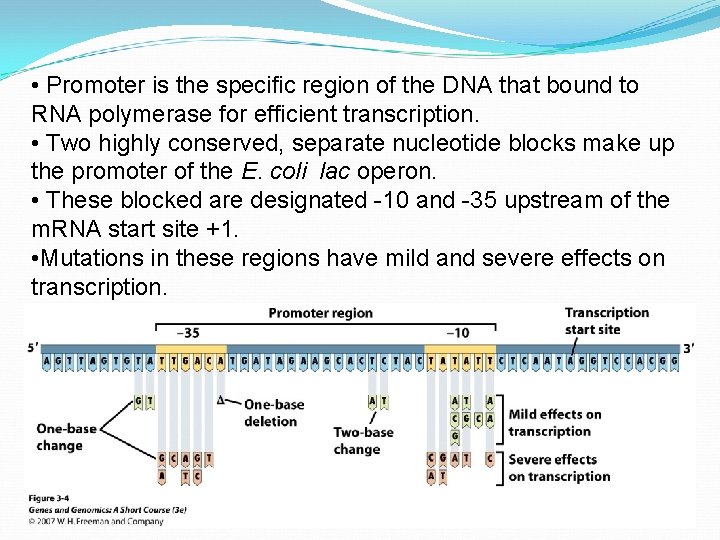
• Promoter is the specific region of the DNA that bound to RNA polymerase for efficient transcription. • Two highly conserved, separate nucleotide blocks make up the promoter of the E. coli lac operon. • These blocked are designated -10 and -35 upstream of the m. RNA start site +1. • Mutations in these regions have mild and severe effects on transcription.

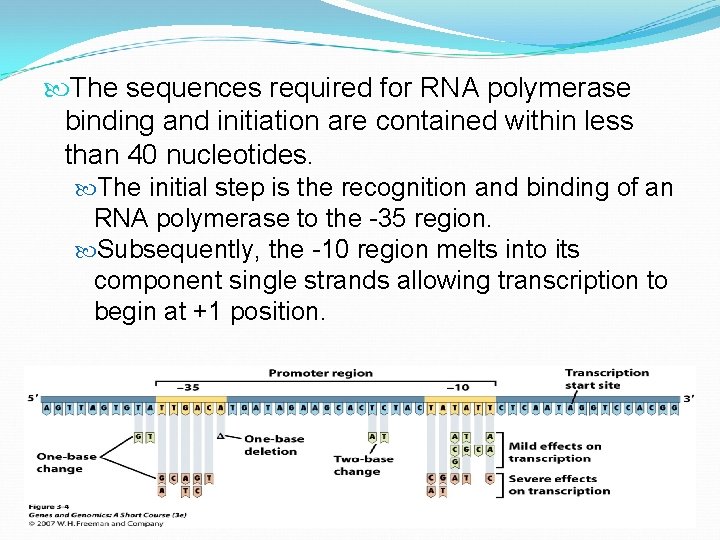
The sequences required for RNA polymerase binding and initiation are contained within less than 40 nucleotides. The initial step is the recognition and binding of an RNA polymerase to the -35 region. Subsequently, the -10 region melts into its component single strands allowing transcription to begin at +1 position.


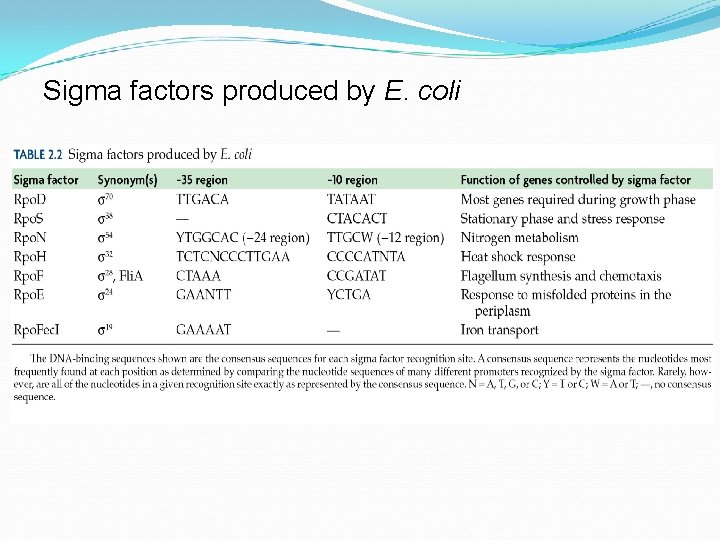
Bacterial RNA polymerase core complexes are bound by a single initiation factor σ (sigma). E coli can control gene expression by activation of σ factor in response to stimuli. σ is the most common initiation factor in E. coli genome that binds to many other promoters besides lac promoter. σ 32 is activated in response to “heat shock” and directs polymerase to transcribe specific genes to respond to this stress. σ 54 is activated when cells are starved for nitrogen, turning on genes that are responsible for uptake of nitrogen. 70

Sigma factors produced by E. coli

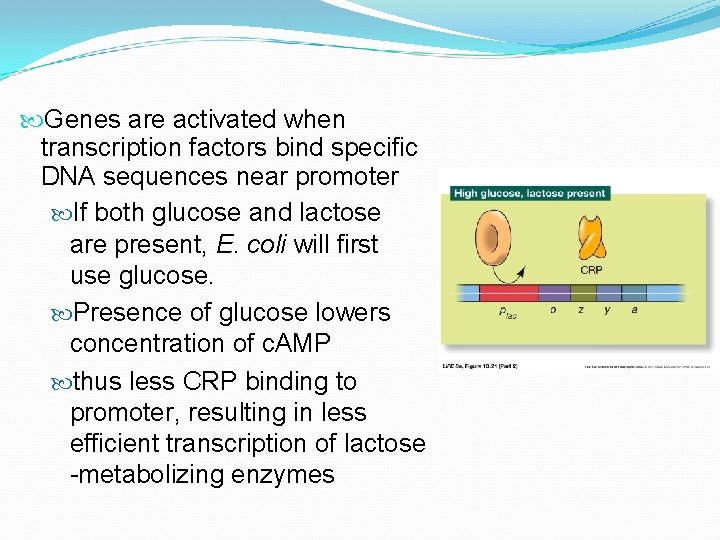
Genes are activated when transcription factors bind specific DNA sequences near promoter If both glucose and lactose are present, E. coli will first use glucose. Presence of glucose lowers concentration of c. AMP thus less CRP binding to promoter, resulting in less efficient transcription of lactose -metabolizing enzymes

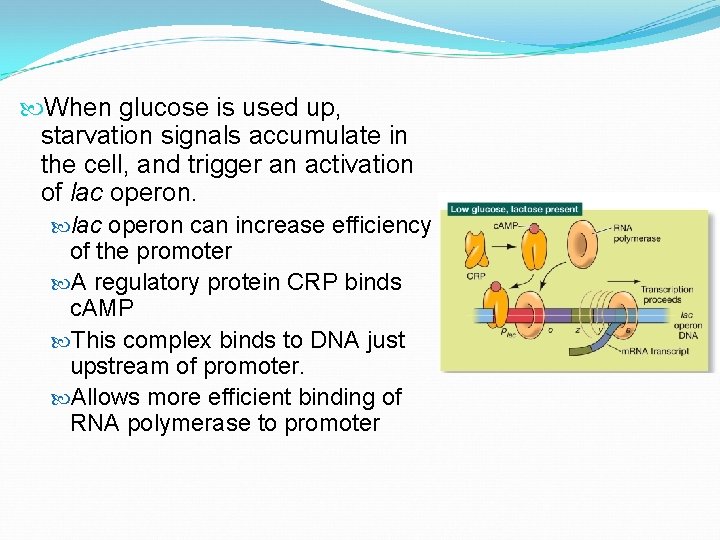
When glucose is used up, starvation signals accumulate in the cell, and trigger an activation of lac operon can increase efficiency of the promoter A regulatory protein CRP binds c. AMP This complex binds to DNA just upstream of promoter. Allows more efficient binding of RNA polymerase to promoter


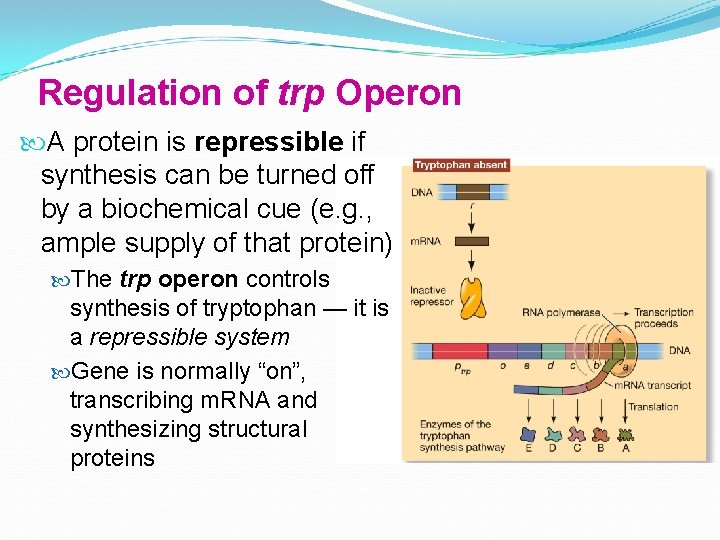
Regulation of trp Operon A protein is repressible if synthesis can be turned off by a biochemical cue (e. g. , ample supply of that protein) The trp operon controls synthesis of tryptophan — it is a repressible system Gene is normally “on”, transcribing m. RNA and synthesizing structural proteins


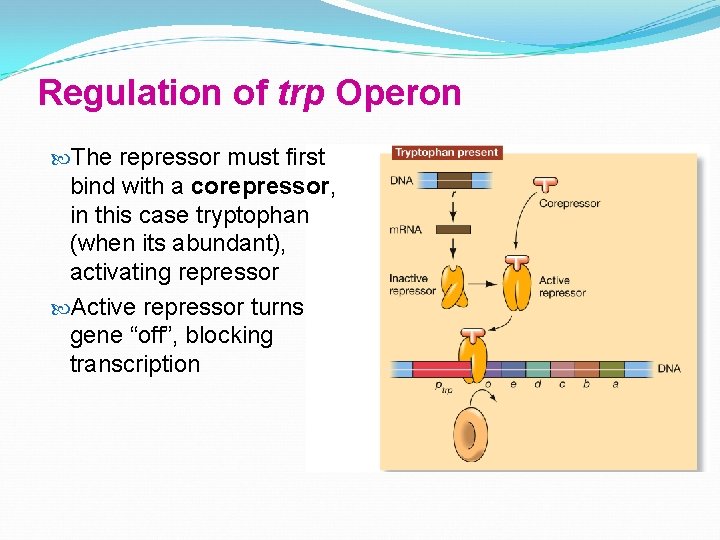
Regulation of trp Operon The repressor must first bind with a corepressor, in this case tryptophan (when its abundant), activating repressor Active repressor turns gene “off”, blocking transcription

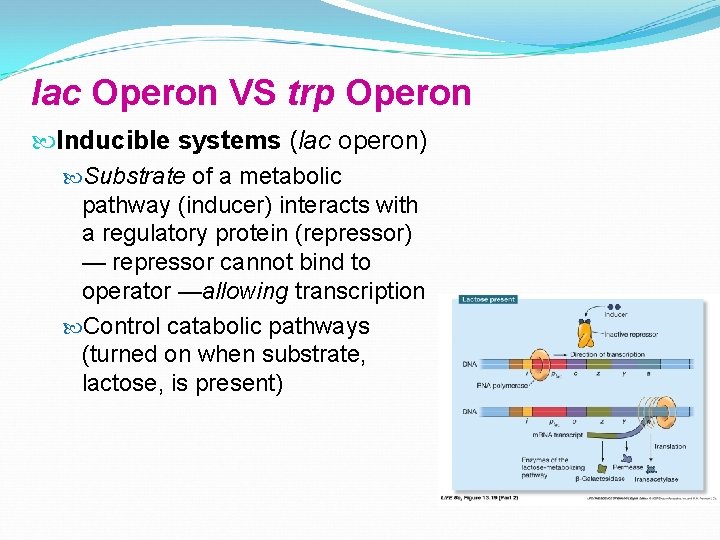
lac Operon VS trp Operon Inducible systems (lac operon) Substrate of a metabolic pathway (inducer) interacts with a regulatory protein (repressor) — repressor cannot bind to operator —allowing transcription Control catabolic pathways (turned on when substrate, lactose, is present)

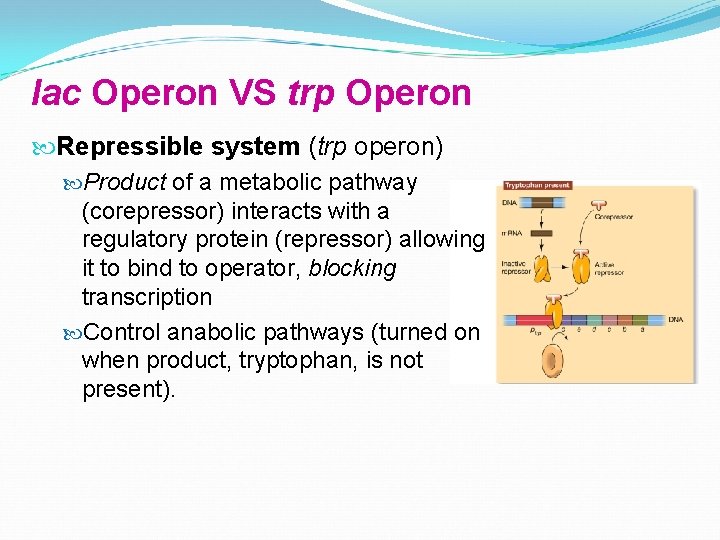
lac Operon VS trp Operon Repressible system (trp operon) Product of a metabolic pathway (corepressor) interacts with a regulatory protein (repressor) allowing it to bind to operator, blocking transcription Control anabolic pathways (turned on when product, tryptophan, is not present).

The Attenuation in trp operon • Attenuation is a method of regulating transcription by preventing complete m. RNA synthesis. – The 5’ end of trp operon m. RNA, the leader region (region 1) is rich in tryptophan codon. – When tryptophan is available, the translation of this region occurs. – As this happens, the trp m. RNA forms a stem-loop structure between region 3 and 4, transcription is attenuated. – When the tryptophan level drops, the attenuation is relieved. – The ribosome is stalled at the leader sequence, different stem-loop forms (2 and 3), allows the transcription to continue.

 Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Dna protein synthesis study guide answers
Dna protein synthesis study guide answers Section 12-3 rna and protein synthesis answer key
Section 12-3 rna and protein synthesis answer key Rna transfer
Rna transfer Chapter 12 section 3 dna rna and protein
Chapter 12 section 3 dna rna and protein Rna protein synthesis
Rna protein synthesis Protein synthesis
Protein synthesis Dna to protein steps
Dna to protein steps Mutations quiz
Mutations quiz Microarray analysis
Microarray analysis Dna rna protein
Dna rna protein Dna rna protein diagram
Dna rna protein diagram Protein
Protein Physical state of covalent compounds
Physical state of covalent compounds Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Zinc oxide + nitric acid → zinc nitrate + water
Zinc oxide + nitric acid → zinc nitrate + water Rna
Rna Rna transfer
Rna transfer Difference between dna and rna
Difference between dna and rna Dna and rna concept map
Dna and rna concept map Venn diagram of transcription and translation
Venn diagram of transcription and translation Dna and rna coloring worksheet
Dna and rna coloring worksheet Rna types
Rna types Phosphodiester bond
Phosphodiester bond Rnabases
Rnabases Dna rna and proteins study guide answers
Dna rna and proteins study guide answers Difference between dna and rna extraction
Difference between dna and rna extraction Dna and rna
Dna and rna Chapter 12 dna and rna
Chapter 12 dna and rna Dna and rna
Dna and rna Nucleotide
Nucleotide Section 12-1 dna
Section 12-1 dna Rna chart
Rna chart Dna and rna similarities
Dna and rna similarities Guac wheel
Guac wheel Dehydration synthesis dna
Dehydration synthesis dna Nitrogenous bases in dna and rna
Nitrogenous bases in dna and rna Chapter 12 dna the genetic material
Chapter 12 dna the genetic material Synthesis of rna
Synthesis of rna Protein synthesis and mutations
Protein synthesis and mutations Protein synthesis and mutations
Protein synthesis and mutations Protein synthesis and mutations
Protein synthesis and mutations Unlike dna rna contains
Unlike dna rna contains Cytoplasm structure
Cytoplasm structure Dna to rna rules
Dna to rna rules Rna base pairs vs dna
Rna base pairs vs dna Dna to rna transcription
Dna to rna transcription Rna or dna
Rna or dna Big q
Big q Overview of transcription and translation
Overview of transcription and translation Rna dna
Rna dna Rantai dna yang berperan sebagai beton untuk rna adalah
Rantai dna yang berperan sebagai beton untuk rna adalah What kingdom do viruses belong to
What kingdom do viruses belong to Corona virus dna or rna
Corona virus dna or rna Dna rna
Dna rna Dna to rna transcription
Dna to rna transcription Cytosio
Cytosio Dna rna
Dna rna Somatic vs germ cells
Somatic vs germ cells Rna vs dna
Rna vs dna Proteinas histonas
Proteinas histonas Dna rna
Dna rna Substansi genetik dna dan rna mempunyai kesamaan yaitu
Substansi genetik dna dan rna mempunyai kesamaan yaitu Dna replication
Dna replication Transcription and translation
Transcription and translation Protein synthesis gcse
Protein synthesis gcse Dna cookbook analogy
Dna cookbook analogy Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis Synthesis
Synthesis Protein synthesis animation mcgraw hill
Protein synthesis animation mcgraw hill Molecular genetics and biotechnology
Molecular genetics and biotechnology Translate
Translate Protein synthesis
Protein synthesis Ribosome
Ribosome Protein synthesis ppt
Protein synthesis ppt Picture transcription
Picture transcription Methoteraxate
Methoteraxate Which best summarizes the process of protein synthesis?
Which best summarizes the process of protein synthesis? Concept map of protein synthesis
Concept map of protein synthesis Protein synthesis
Protein synthesis Protein synthesis scramble
Protein synthesis scramble Fictional character μετάφραση
Fictional character μετάφραση Protein synthesis
Protein synthesis Sintese de proteinas na celula
Sintese de proteinas na celula Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis Double stranded dna
Double stranded dna Catalytic functions
Catalytic functions Protein pump vs protein channel
Protein pump vs protein channel Protein-protein docking
Protein-protein docking Coding dna and non coding dna
Coding dna and non coding dna Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing Dna-templated synthesis
Dna-templated synthesis Dna synthesis at replication fork
Dna synthesis at replication fork Telomerase
Telomerase