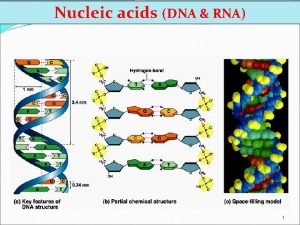
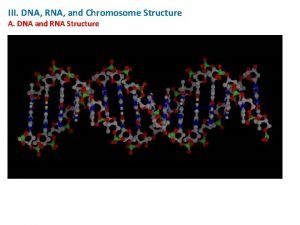
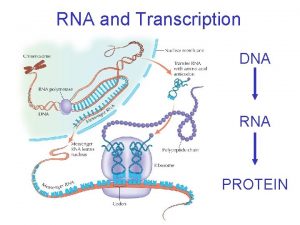
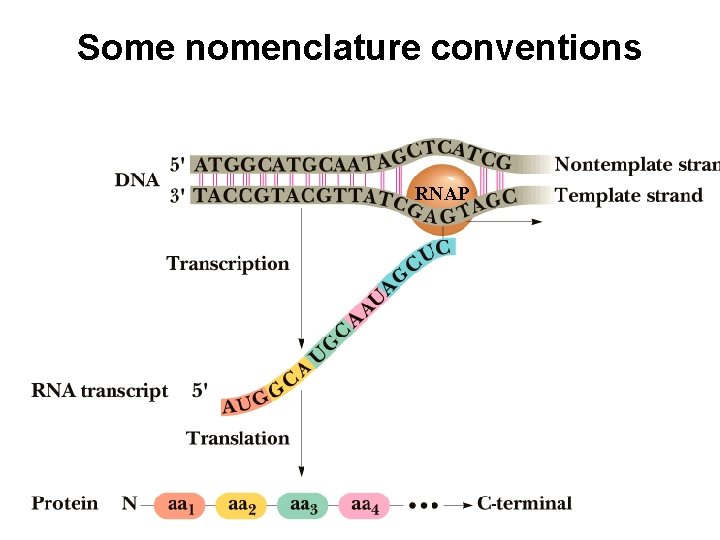
Some nomenclature conventions RNAP RNA DNA Similarities and






- Slides: 43



Some nomenclature conventions RNAP

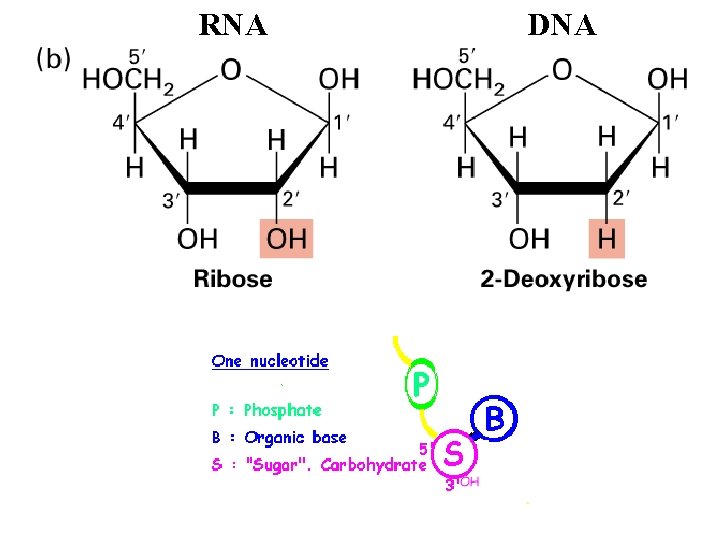
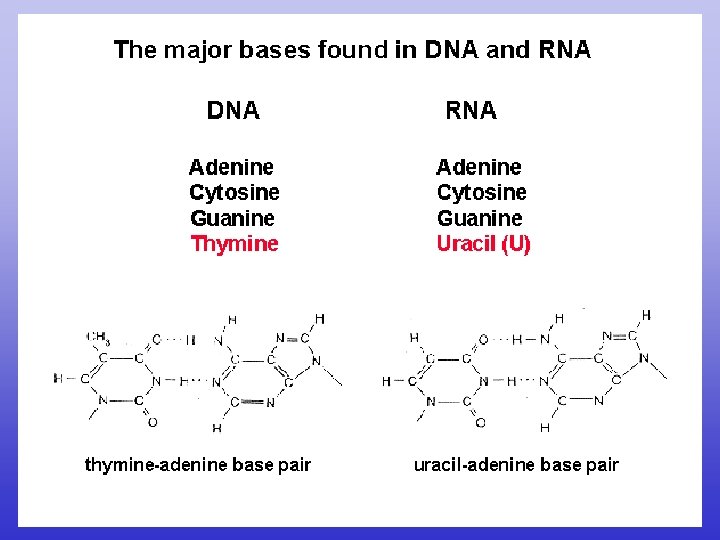
RNA DNA


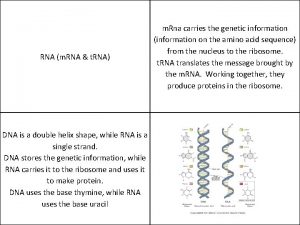
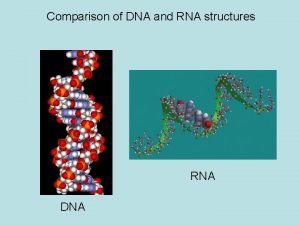
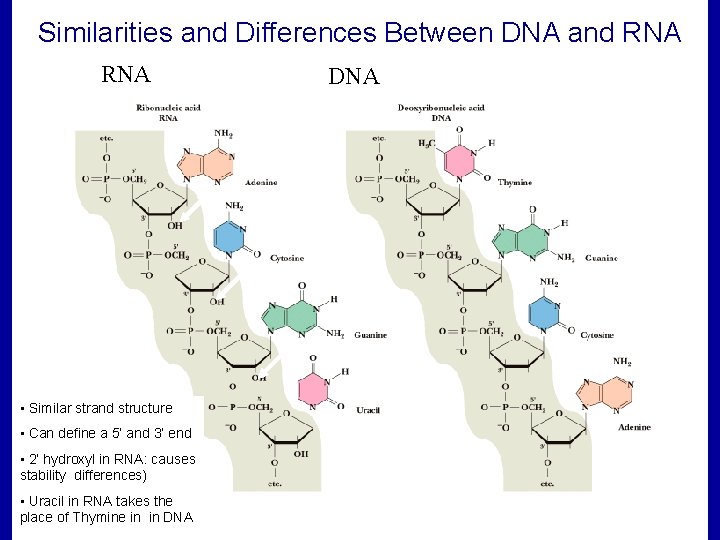
Similarities and Differences Between DNA and RNA • Similar strand structure • Can define a 5’ and 3’ end • 2’ hydroxyl in RNA: causes stability differences) • Uracil in RNA takes the place of Thymine in in DNA






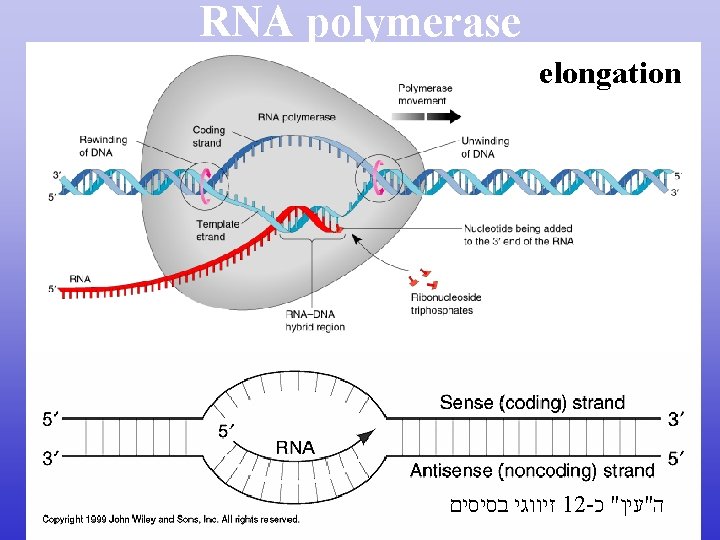
RNA polymerase

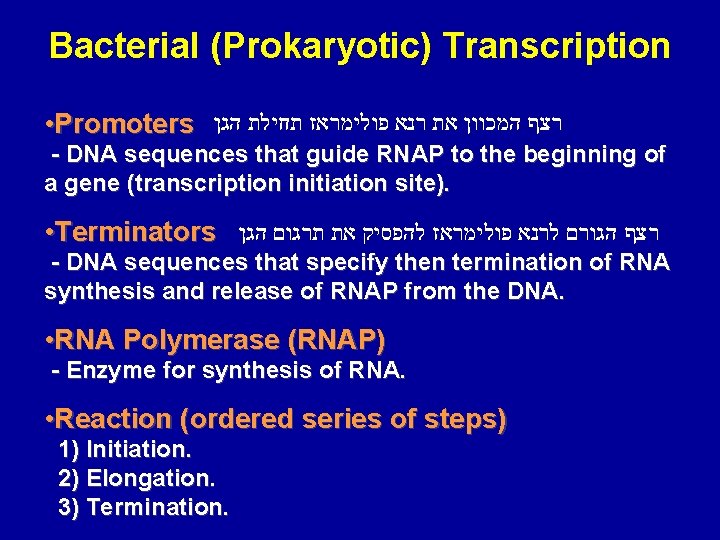
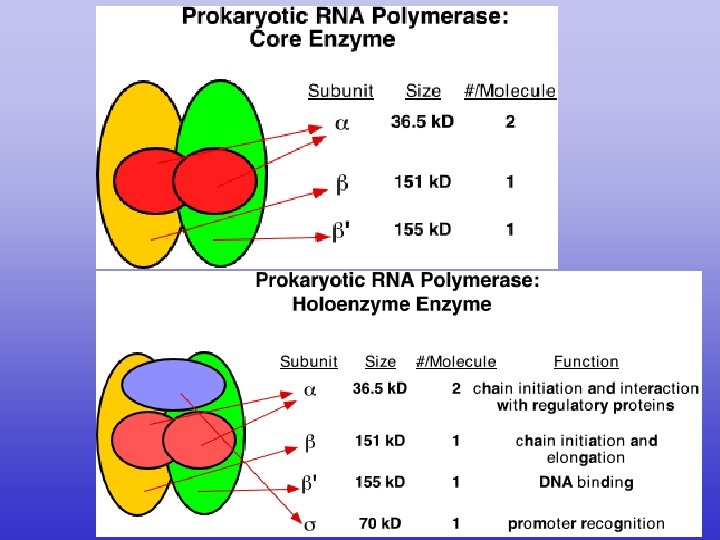
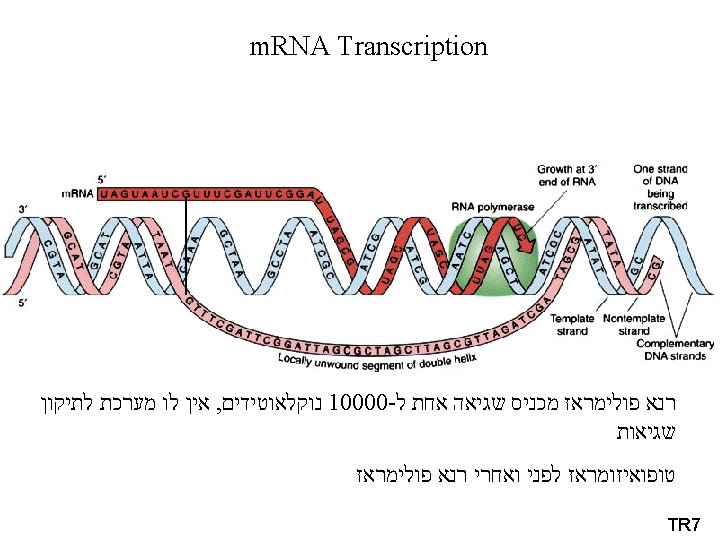
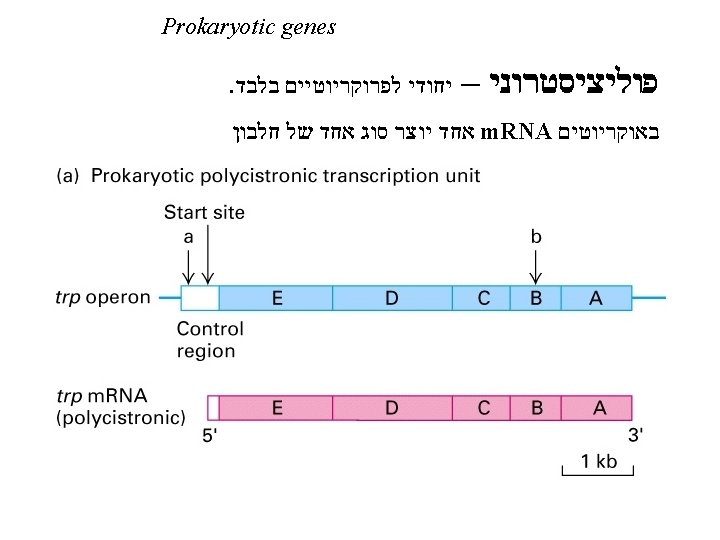
Bacterial (Prokaryotic) Transcription • Promoters רצף המכוון את רנא פולימראז תחילת הגן - DNA sequences that guide RNAP to the beginning of a gene (transcription initiation site). • Terminators רצף הגורם לרנא פולימראז להפסיק את תרגום הגן - DNA sequences that specify then termination of RNA synthesis and release of RNAP from the DNA. • RNA Polymerase (RNAP) - Enzyme for synthesis of RNA. • Reaction (ordered series of steps) 1) Initiation. 2) Elongation. 3) Termination.

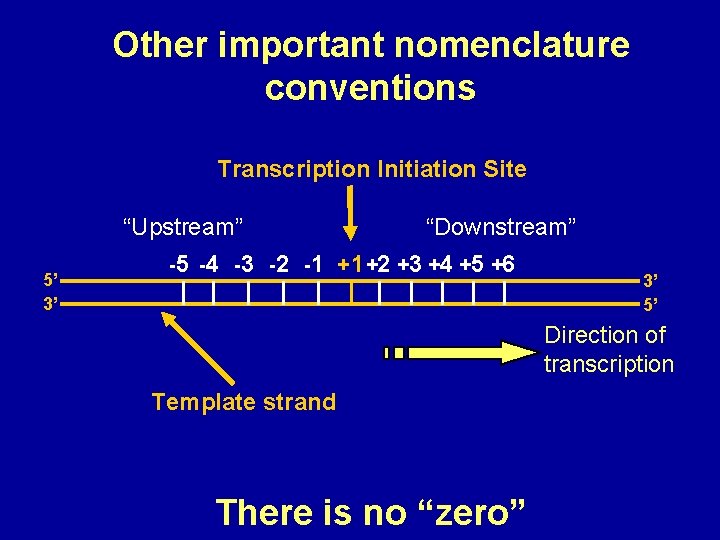
Other important nomenclature conventions Transcription Initiation Site “Upstream” 5’ 3’ -5 -4 -3 -2 -1 “Downstream” +1 +2 +3 +4 +5 +6 3’ 5’ Direction of transcription Template strand There is no “zero”

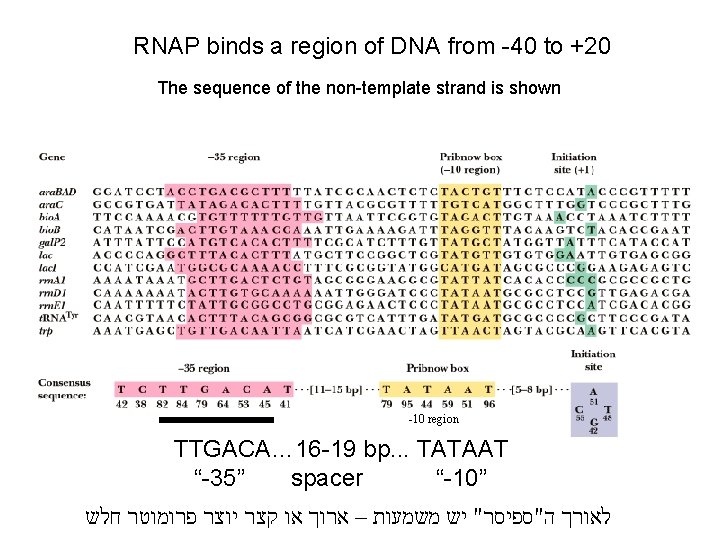
RNAP binds a region of DNA from -40 to +20 The sequence of the non-template strand is shown -10 region TTGACA… 16 -19 bp. . . TATAAT “-35” spacer “-10” לאורך ה"ספיסר" יש משמעות – ארוך או קצר יוצר פרומוטר חלש



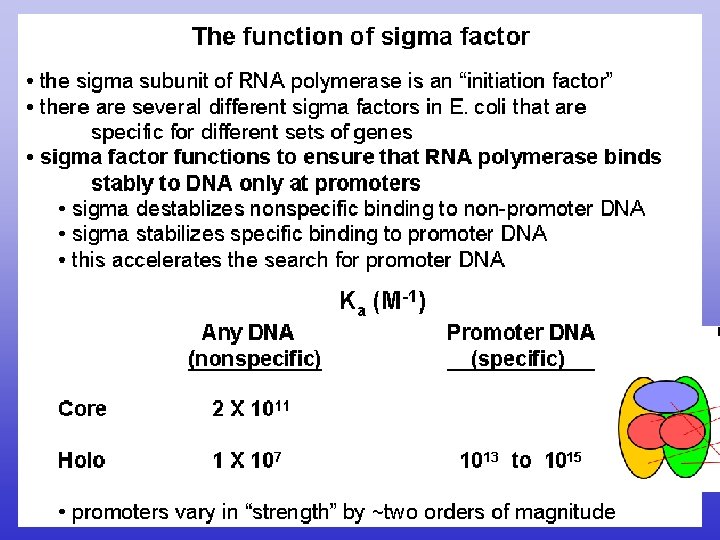
Sigma factor

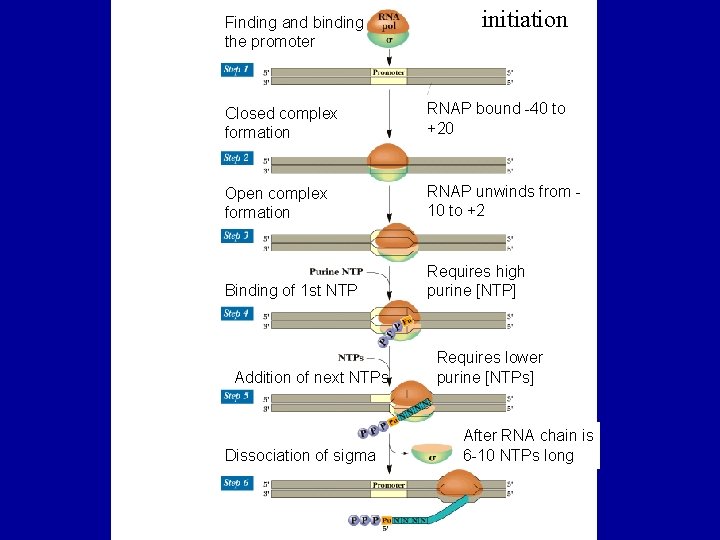
Finding and binding the promoter initiation Closed complex formation RNAP bound -40 to +20 Open complex formation RNAP unwinds from 10 to +2 Binding of 1 st NTP Requires high purine [NTP] Addition of next NTPs Dissociation of sigma Requires lower purine [NTPs] After RNA chain is 6 -10 NTPs long

RNA polymerase elongation זיווגי בסיסים 12 - ה"עין" כ


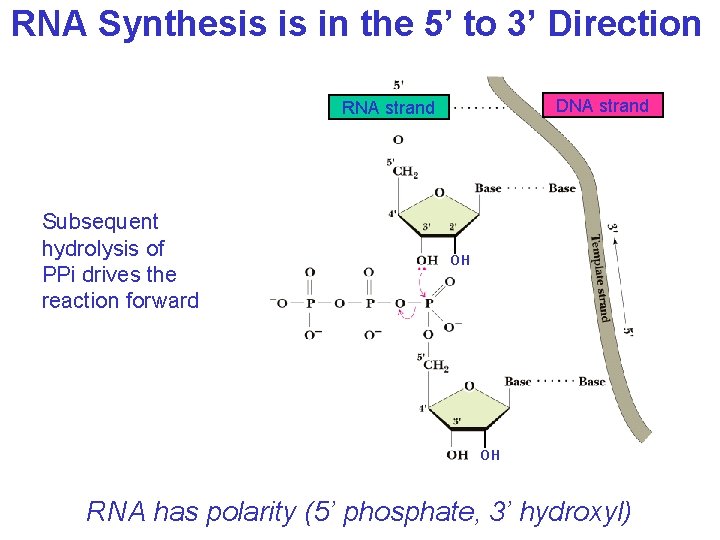
RNA Synthesis is in the 5’ to 3’ Direction DNA strand RNA strand Subsequent hydrolysis of PPi drives the reaction forward OH OH RNA has polarity (5’ phosphate, 3’ hydroxyl)

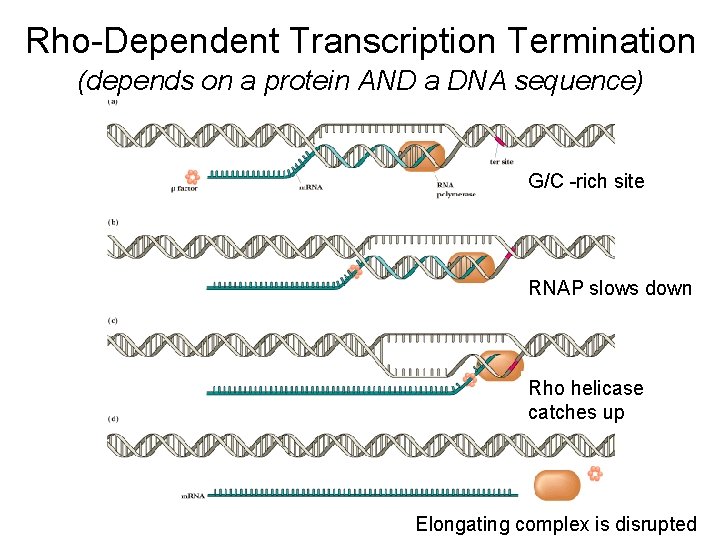
Rho-Dependent Transcription Termination (depends on a protein AND a DNA sequence) G/C -rich site RNAP slows down Rho helicase catches up Elongating complex is disrupted

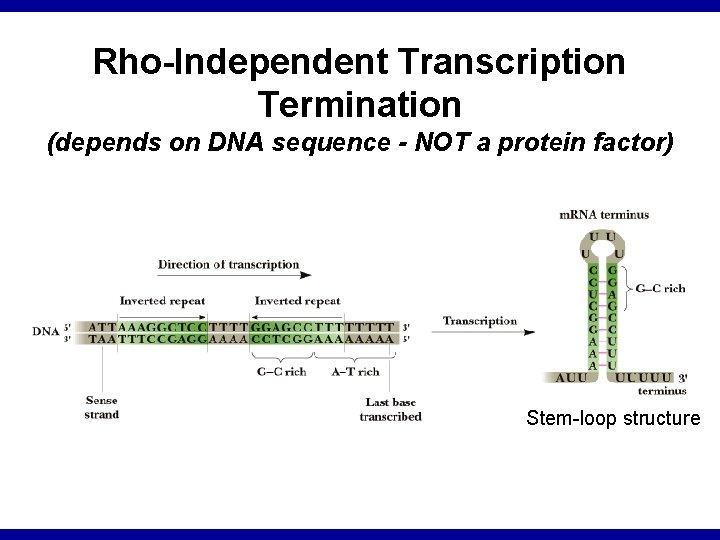
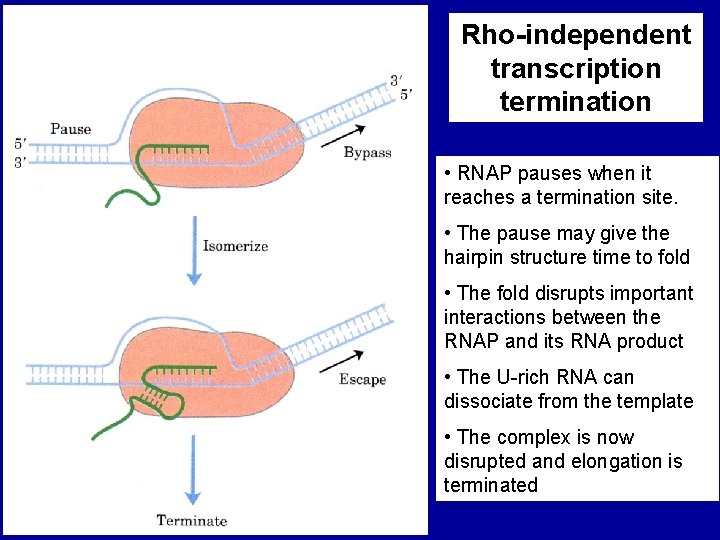
Rho-Independent Transcription Termination (depends on DNA sequence - NOT a protein factor) Stem-loop structure

Rho-independent transcription termination • RNAP pauses when it reaches a termination site. • The pause may give the hairpin structure time to fold • The fold disrupts important interactions between the RNAP and its RNA product • The U-rich RNA can dissociate from the template • The complex is now disrupted and elongation is terminated

RNA polymerase

Xmas tree – transcription




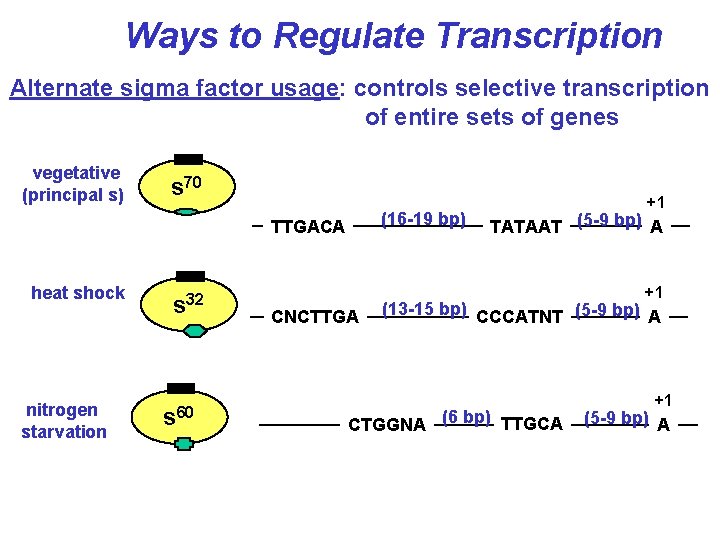
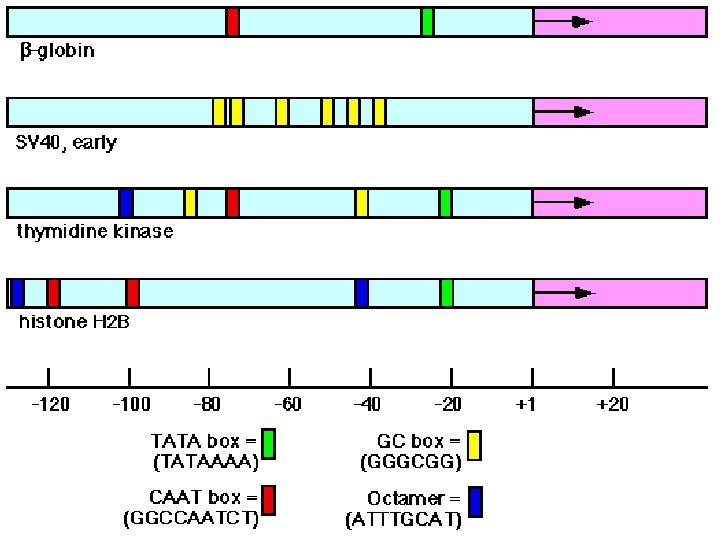
Ways to Regulate Transcription Alternate sigma factor usage: controls selective transcription of entire sets of genes vegetative (principal s) s 70 (16 -19 bp) TTGACA heat shock nitrogen starvation s 32 s 60 +1 TATAAT (5 -9 bp) A +1 CNCTTGA (13 -15 bp) CCCATNT (5 -9 bp) A CTGGNA (6 bp) TTGCA +1 (5 -9 bp) A




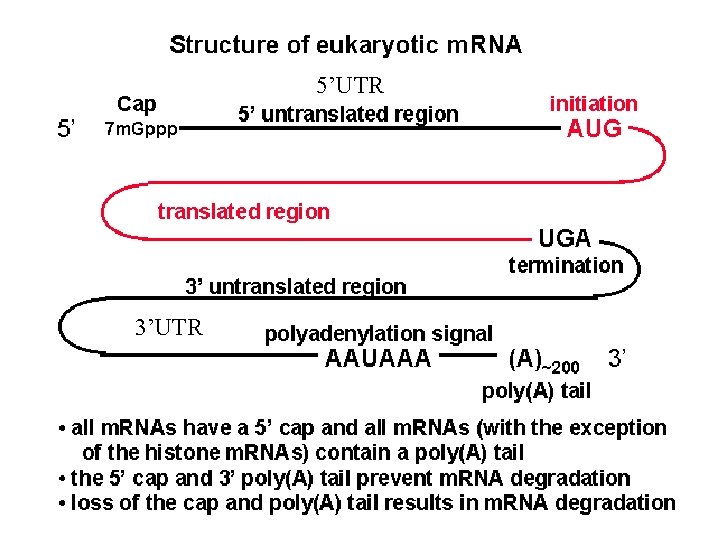
Pre-m. RNA


5’UTR 3’UTR






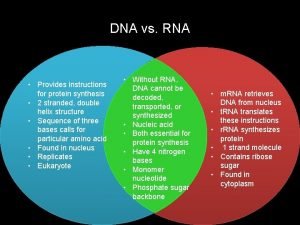
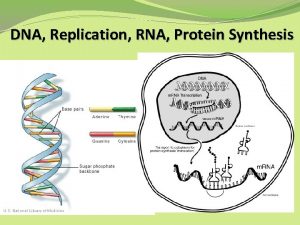
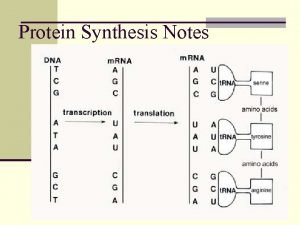
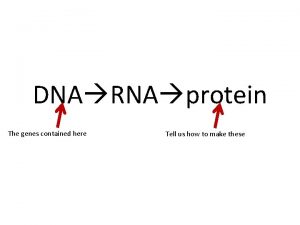
 What provides instructions for protein synthesis
What provides instructions for protein synthesis Dna and rna venn diagram
Dna and rna venn diagram Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Compare and contrast dna and rna.
Compare and contrast dna and rna. What is the goal of replication
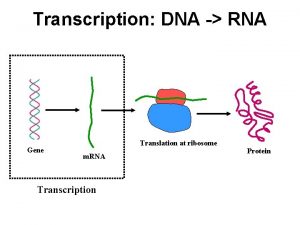
What is the goal of replication Dna to rna transcription
Dna to rna transcription Dna double helix coloring worksheet answer key
Dna double helix coloring worksheet answer key Transfer rna
Transfer rna Phosphodiester bond
Phosphodiester bond Dna and rna
Dna and rna Dna rna and proteins study guide answers
Dna rna and proteins study guide answers Fraction
Fraction Lenine
Lenine Chapter 12 dna and rna
Chapter 12 dna and rna Chapter 12 section 3 dna rna and protein
Chapter 12 section 3 dna rna and protein Rna and protein synthesis study guide
Rna and protein synthesis study guide Dna and rna
Dna and rna Nucleotide
Nucleotide Section 12-1 dna
Section 12-1 dna Aug amino acid
Aug amino acid Cell theory
Cell theory Dna dehydration synthesis
Dna dehydration synthesis 3 componets of dna
3 componets of dna Minor groove
Minor groove On and off
On and off Rna translation
Rna translation Cytoplasm structure
Cytoplasm structure Dna to rna rules
Dna to rna rules Dna vs rna nitrogen bases
Dna vs rna nitrogen bases Dna rna protein central dogma
Dna rna protein central dogma Ribosomem
Ribosomem Rna or dna
Rna or dna How does rna differ from dna? *
How does rna differ from dna? * Central dogma
Central dogma Virusmax
Virusmax Chargaff
Chargaff Protein dalam sel
Protein dalam sel What kingdom do viruses belong to
What kingdom do viruses belong to Microarray analysis
Microarray analysis Dna rna protein
Dna rna protein Dna rna protein diagram
Dna rna protein diagram Corona virus dna or rna
Corona virus dna or rna Dna rna
Dna rna Dna to rna transcription
Dna to rna transcription