StructureFunction Analysis DNAProtein structurefunction analysis and prediction Proteinprotein
![Structure-Function Analysis Complexation Reaction • A + B AB Ka = [AB]/[A] • [B] Structure-Function Analysis Complexation Reaction • A + B AB Ka = [AB]/[A] • [B]](https://slidetodoc.com/presentation_image_h/2e0e7baa2dc7826b51367ab10b5d83af/image-4.jpg)


- Slides: 37

Structure-Function Analysis DNA/Protein structure-function analysis and prediction • Protein-protein Interaction (PPI): • Protein-protein Interaction – Interfaces – Solvation – Energetics – Conformational change – Allostery • Prediction: – Gene Cluster – Phylogenetic Profile: – Rosetta Stone – Sequence co-evolution – Random Decision forest • Docking [ Examples ] 17 Jan 2006 1

Structure-Function Analysis PPI Characteristics • Universal – Cell functionality based on protein-protein interactions • Cyto-skeleton • Ribosome • RNA polymerase • Numerous – Yeast: • ~6. 000 proteins • at least 3 interactions each ~18. 000 interactions – Human: • estimated ~100. 000 interactions • Network – simplest: homodimer (two) – common: hetero-oligomer (more) – holistic: protein network (all) 17 Jan 2006 2

Structure-Function Analysis Interface Area • Contact area – usually >1100 Å2 – each partner >550 Å2 • each partner loses ~800 Å2 of solvent accessible surface area – ~20 amino acids lose ~40 Å2 – ~100 -200 J per Å2 • Average buried accessible surface area: – 12% for dimers – 17% for trimers – 21% for tetramers • 83 -84% of all interfaces are flat • Secondary structure: – 50% -helix – 20% -sheet – 20% coil – 10% mixed • Less hydrophobic than core, more hydrophobic than exterior 17 Jan 2006 3
![StructureFunction Analysis Complexation Reaction A B AB Ka ABA B Structure-Function Analysis Complexation Reaction • A + B AB Ka = [AB]/[A] • [B]](https://slidetodoc.com/presentation_image_h/2e0e7baa2dc7826b51367ab10b5d83af/image-4.jpg)
Structure-Function Analysis Complexation Reaction • A + B AB Ka = [AB]/[A] • [B] association Kd = [A] • [B]/[AB] dissociation • Free energy: DGd = -RT • ln Kd Kd = exp(-DGd / RT) (R = 8. 3144 J mol-1 K-1 ) 17 Jan 2006 4

Structure-Function Analysis Experimental Methods • 2 D (poly-acrylamide) gel electrophoresis mass spectrometry • Liquid chromatography – e. g. gel permeation chromatography • Binding study with one immobilized partner – e. g. surface plasmon resonance • In vivo by two-hybrid systems or FRET • Binding constants by ultra-centrifugation, micro-calorimetry or competition • experiments with labelled ligand – e. g. fluorescence, radioactivity • Role of individual amino acids by site directed mutagenesis • Structural studies – e. g. NMR or X-ray 17 Jan 2006 5

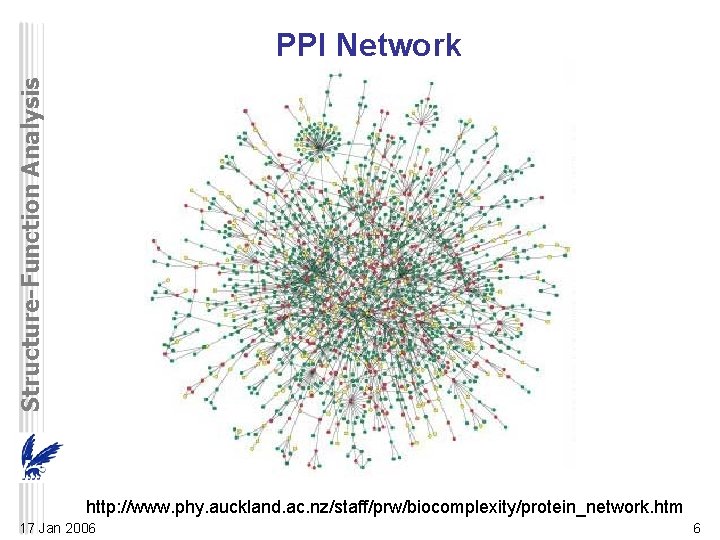
Structure-Function Analysis PPI Network http: //www. phy. auckland. ac. nz/staff/prw/biocomplexity/protein_network. htm 17 Jan 2006 6

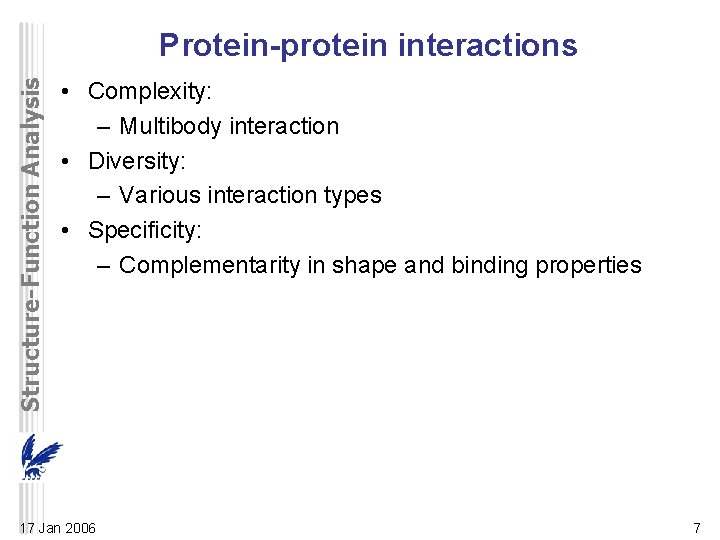
Structure-Function Analysis Protein-protein interactions • Complexity: – Multibody interaction • Diversity: – Various interaction types • Specificity: – Complementarity in shape and binding properties 17 Jan 2006 7

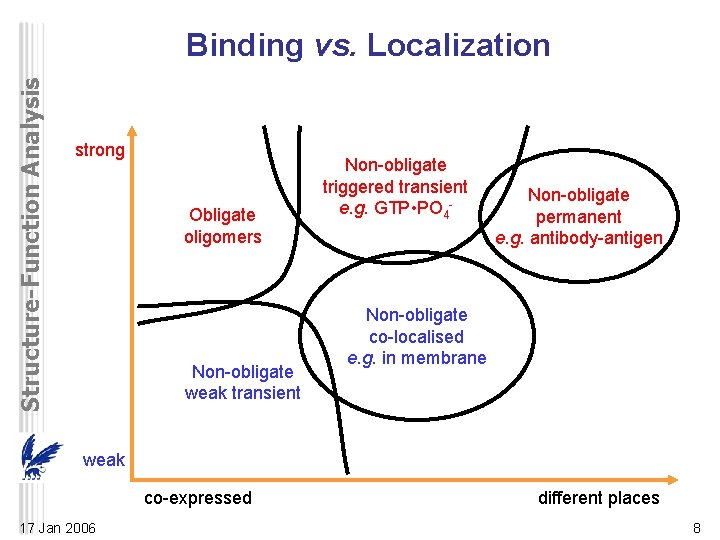
Structure-Function Analysis Binding vs. Localization strong Obligate oligomers Non-obligate weak transient Non-obligate triggered transient e. g. GTP • PO 4 - Non-obligate permanent e. g. antibody-antigen Non-obligate co-localised e. g. in membrane weak co-expressed 17 Jan 2006 different places 8

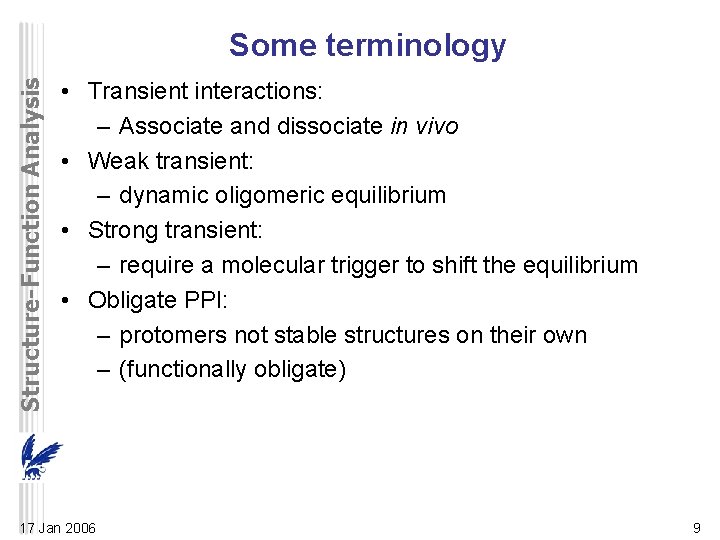
Structure-Function Analysis Some terminology • Transient interactions: – Associate and dissociate in vivo • Weak transient: – dynamic oligomeric equilibrium • Strong transient: – require a molecular trigger to shift the equilibrium • Obligate PPI: – protomers not stable structures on their own – (functionally obligate) 17 Jan 2006 9

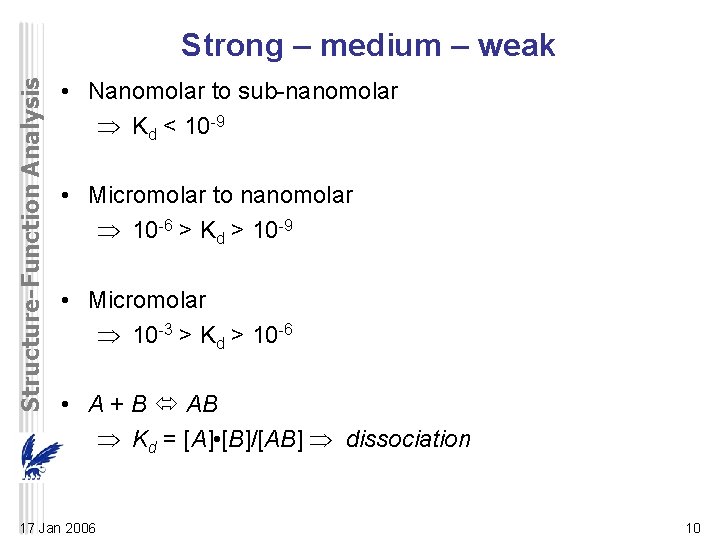
Structure-Function Analysis Strong – medium – weak • Nanomolar to sub-nanomolar Kd < 10 -9 • Micromolar to nanomolar 10 -6 > Kd > 10 -9 • Micromolar 10 -3 > Kd > 10 -6 • A + B AB Kd = [A] • [B]/[AB] dissociation 17 Jan 2006 10

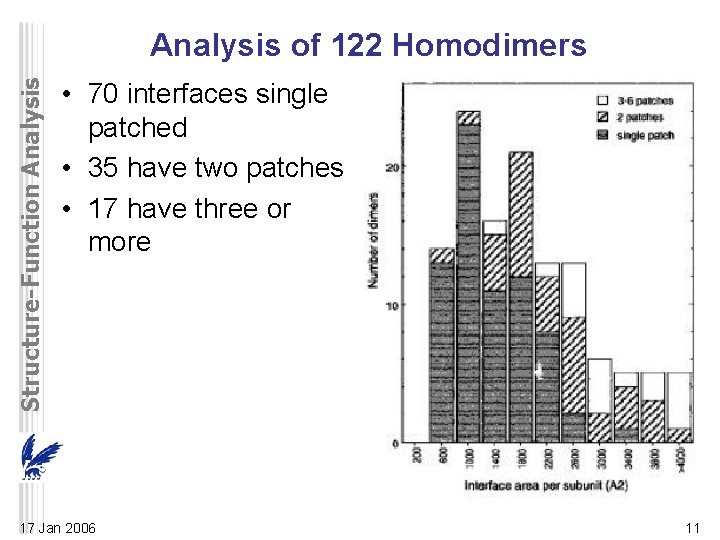
Structure-Function Analysis of 122 Homodimers • 70 interfaces single patched • 35 have two patches • 17 have three or more 17 Jan 2006 11

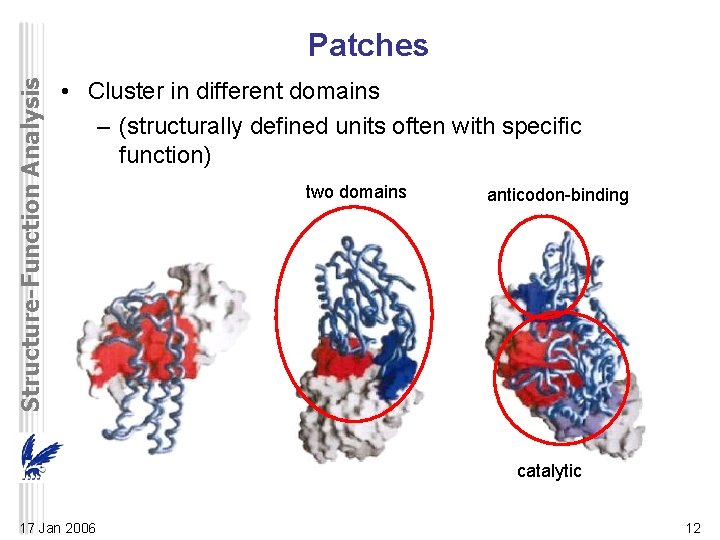
Structure-Function Analysis Patches • Cluster in different domains – (structurally defined units often with specific function) two domains anticodon-binding catalytic 17 Jan 2006 12

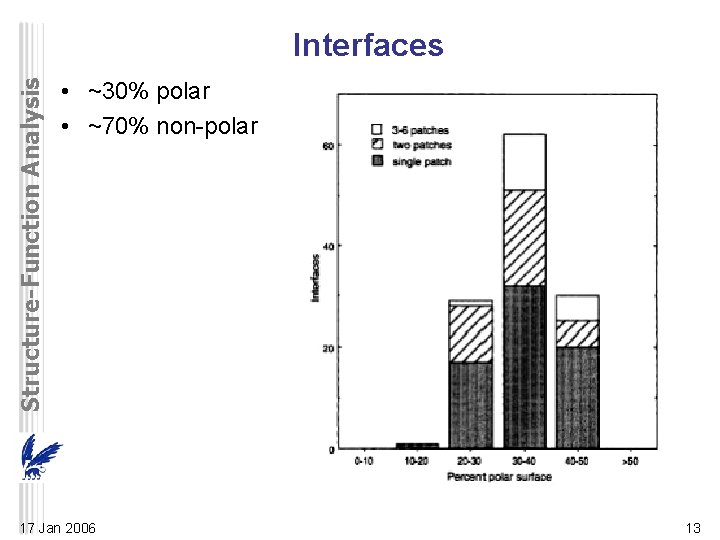
Structure-Function Analysis Interfaces • ~30% polar • ~70% non-polar 17 Jan 2006 13

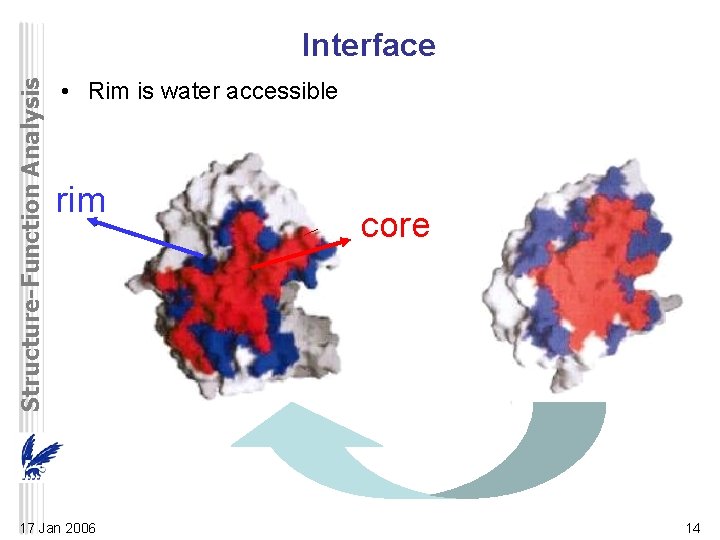
Structure-Function Analysis Interface • Rim is water accessible rim 17 Jan 2006 core 14

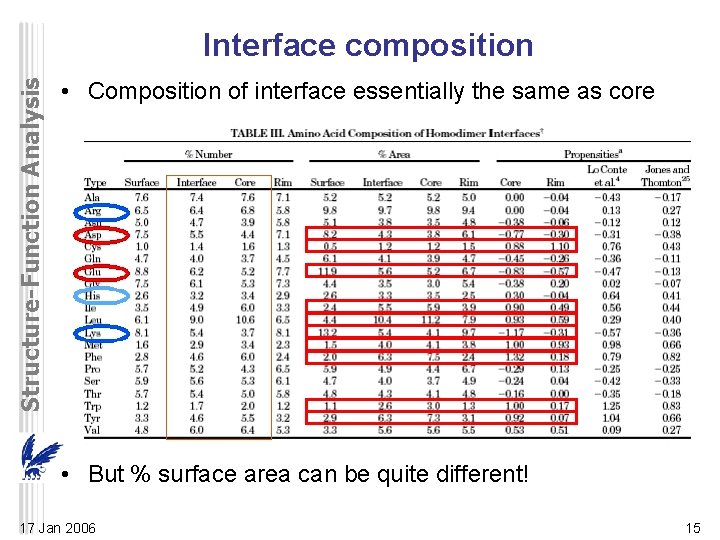
Structure-Function Analysis Interface composition • Composition of interface essentially the same as core • But % surface area can be quite different! 17 Jan 2006 15

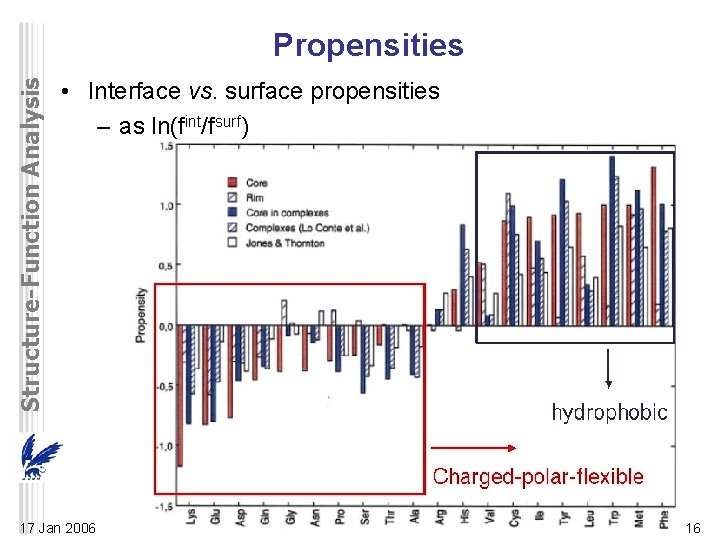
Structure-Function Analysis Propensities • Interface vs. surface propensities – as ln(fint/fsurf) 17 Jan 2006 16

Structure-Function Analysis Conformational Change • Chaperones – extreme conformational changes upon complexation ligand unfolds within the chaperone Gro. EL/Gro. ES • Allosteric proteins – conformational change at 'active' site – ligand binds to 'regulating' site • Peptides – often adopt 'bound' conformation – different from the 'free' conformation 17 Jan 2006 17

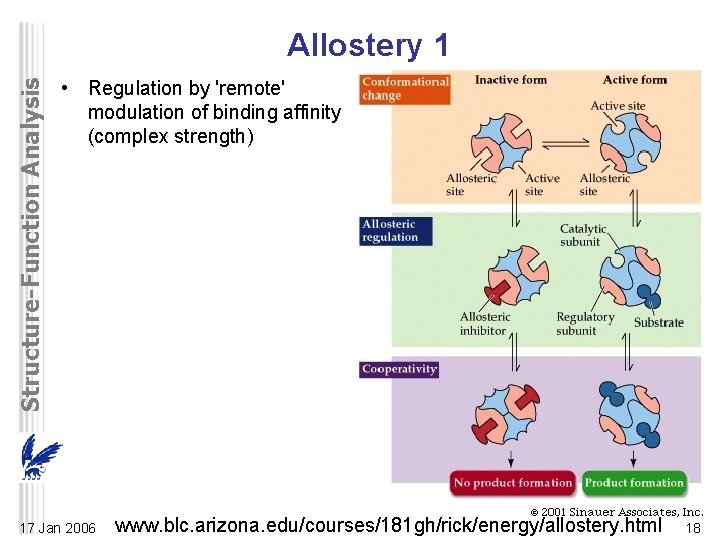
Structure-Function Analysis Allostery 1 • Regulation by 'remote' modulation of binding affinity (complex strength) 17 Jan 2006 www. blc. arizona. edu/courses/181 gh/rick/energy/allostery. html 18

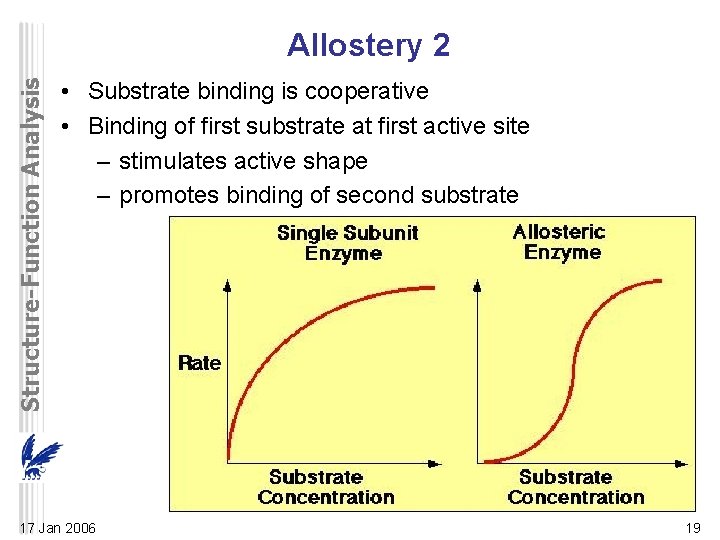
Structure-Function Analysis Allostery 2 • Substrate binding is cooperative • Binding of first substrate at first active site – stimulates active shape – promotes binding of second substrate 17 Jan 2006 19

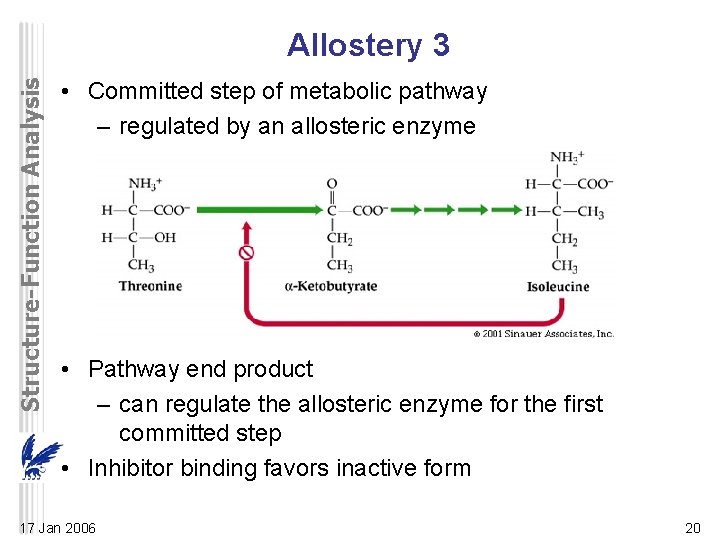
Structure-Function Analysis Allostery 3 • Committed step of metabolic pathway – regulated by an allosteric enzyme • Pathway end product – can regulate the allosteric enzyme for the first committed step • Inhibitor binding favors inactive form 17 Jan 2006 20

Structure-Function Analysis DNA/Protein structure-function analysis and prediction • Protein-protein Interaction (PPI): • Protein-protein Interaction – Interfaces – Solvation – Energetics – Conformational change – Allostery • Prediction: – Gene Cluster – Phylogenetic Profile: – Rosetta Stone – Sequence co-evolution – Random Decision forest • Docking [ Examples ] 17 Jan 2006 21

Structure-Function Analysis Predicting Protein-Protein Interactions: • Gene Cluster: – Gene neighborhood • Phylogenetic Profile: – Co-occurrence across species/genomes • Rosetta Stone: – Occurrence of protein with domains linked • Sequence co-evolution: – Tree correlation indicated functional relation • Random Decision forest: – Using data on domain interactions [ Shoemaker & Panchenko, PLOS-CB 2007 3 e 43 ] 17 Jan 2006 22

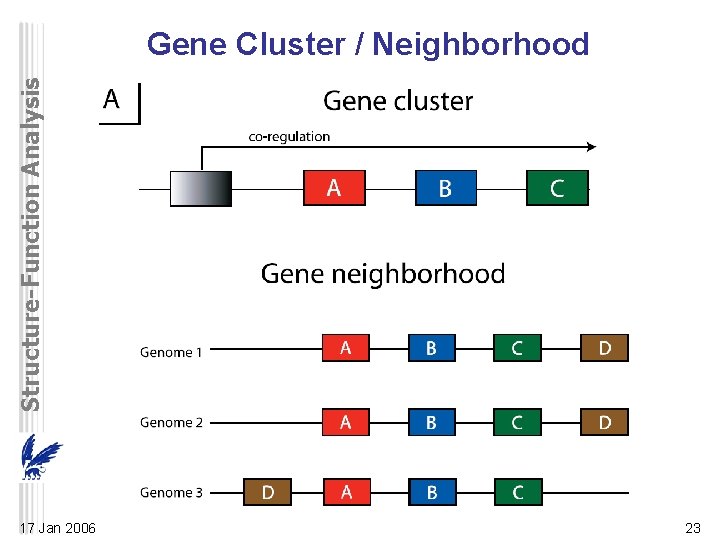
Structure-Function Analysis Gene Cluster / Neighborhood 17 Jan 2006 23

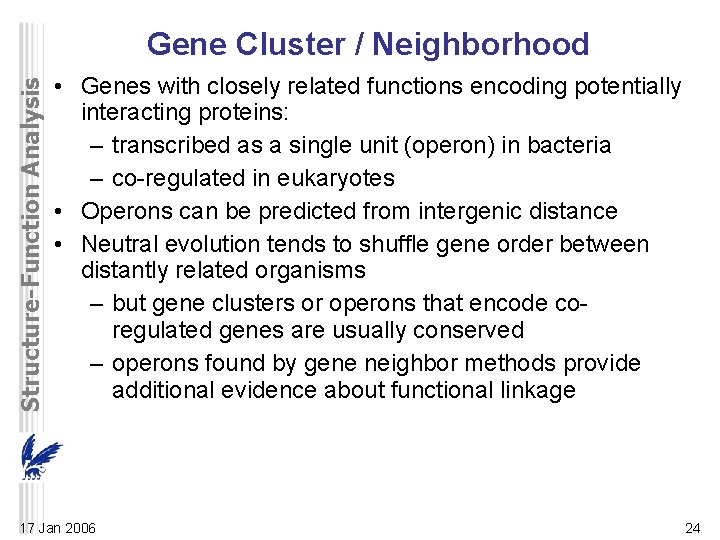
Structure-Function Analysis Gene Cluster / Neighborhood • Genes with closely related functions encoding potentially interacting proteins: – transcribed as a single unit (operon) in bacteria – co-regulated in eukaryotes • Operons can be predicted from intergenic distance • Neutral evolution tends to shuffle gene order between distantly related organisms – but gene clusters or operons that encode coregulated genes are usually conserved – operons found by gene neighbor methods provide additional evidence about functional linkage 17 Jan 2006 24

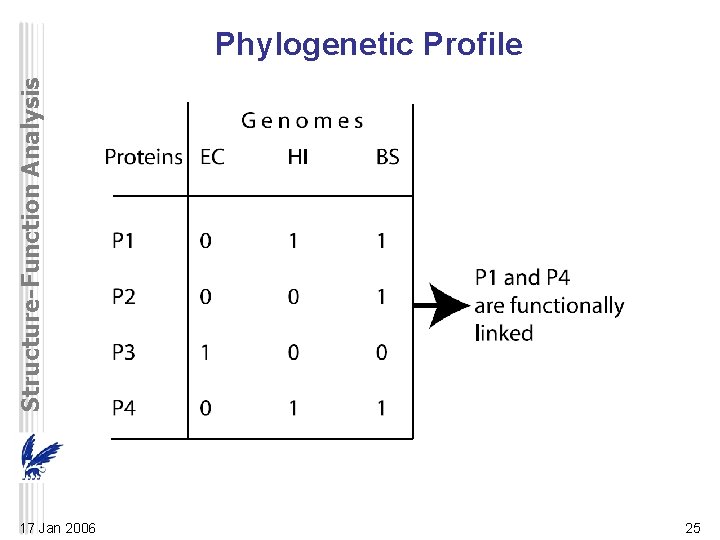
Structure-Function Analysis Phylogenetic Profile 17 Jan 2006 25

Structure-Function Analysis Phylogenetic Profile • hypothesis that functionally linked and potentially interacting nonhomologous proteins co-evolve and have orthologs in the same subset of organisms – components of complexes and pathways should be present simultaneously in order to perform their functions. • phylogenetic profile is a vector of N elements (number of genomes) – presence/absence of protein in genome is ‘‘ 1’’ or ‘‘ 0’’ at each position of a profile. • clustered using bit-distance measure – proteins in a cluster are considered functionally related. • also for protein domains instead of entire proteins 17 Jan 2006 26

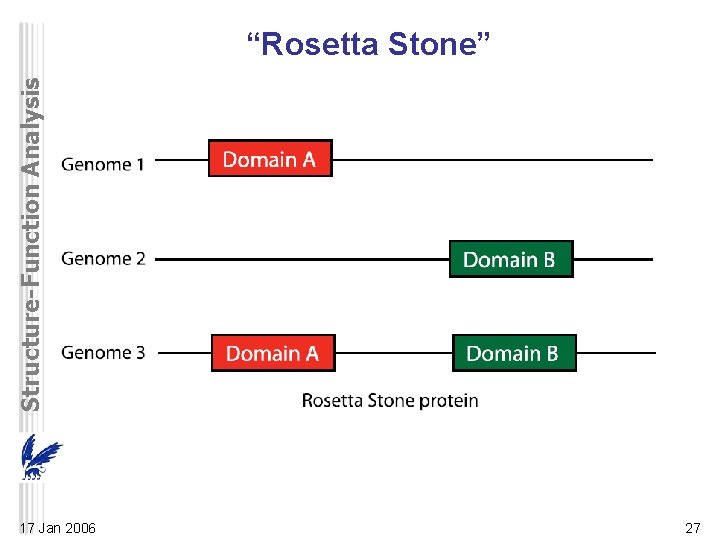
Structure-Function Analysis “Rosetta Stone” 17 Jan 2006 27

Structure-Function Analysis “Rosetta Stone” • infer protein interactions from sequences in different genomes – some interacting proteins/domains have homologs that are fused into one protein a so-called “Rosetta Stone” protein • Apparently, gene fusion can occur to optimize coexpression of genes encoding for interacting proteins. 17 Jan 2006 28

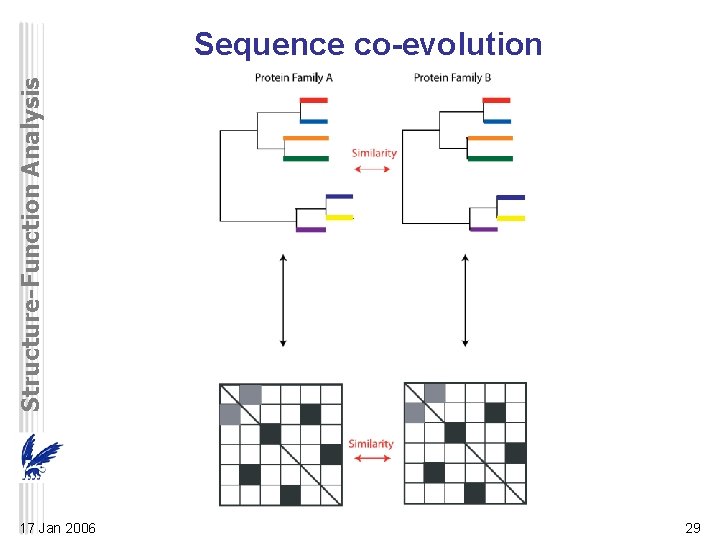
Structure-Function Analysis Sequence co-evolution 17 Jan 2006 29

Structure-Function Analysis Sequence co-evolution • interacting proteins often co-evolve so changes in one protein leading to the loss of function or interaction can be compensated by changes in the other – orthologs of coevolving proteins also tend to interact • infer unknown interactions in other genomes • similarity between phylogenetic trees of two nonhomologous interacting protein families – correlation coefficient between the distance matrices – requires correspondence between the matrix elements / tree branches (i. e. ortholog relations) • align distance matrices to minimize difference • predicted interactions correspond to aligned col’s • max. ~30 proteins in a family 17 Jan 2006 30

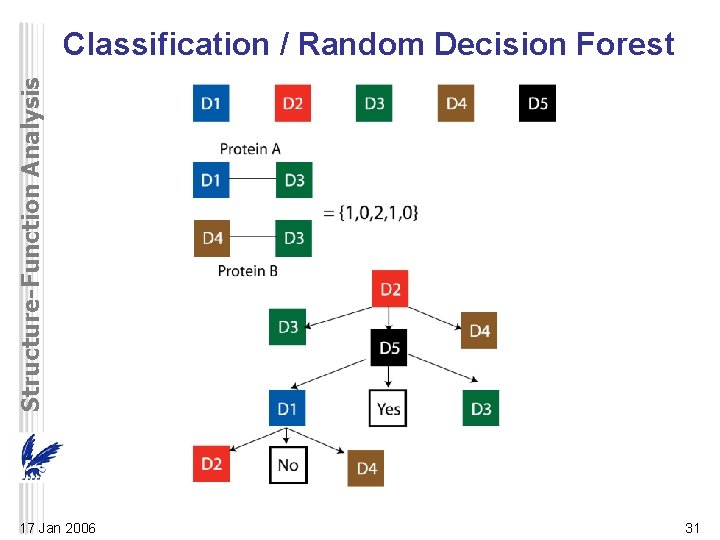
Structure-Function Analysis Classification / Random Decision Forest 17 Jan 2006 31

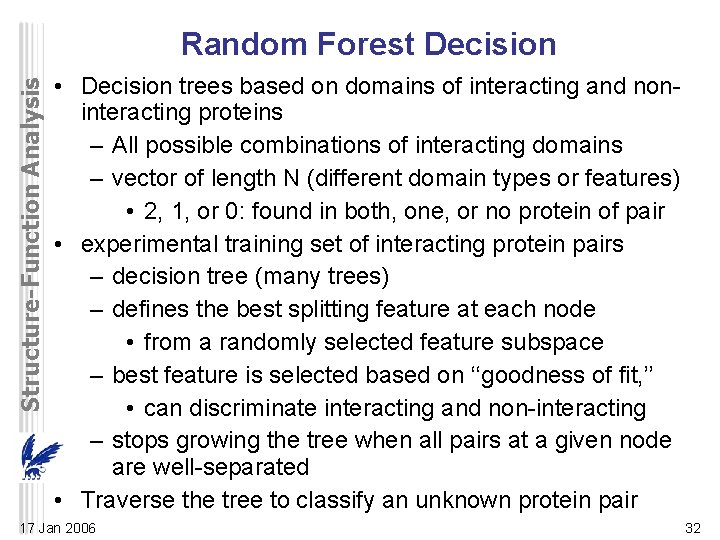
Structure-Function Analysis Random Forest Decision • Decision trees based on domains of interacting and noninteracting proteins – All possible combinations of interacting domains – vector of length N (different domain types or features) • 2, 1, or 0: found in both, one, or no protein of pair • experimental training set of interacting protein pairs – decision tree (many trees) – defines the best splitting feature at each node • from a randomly selected feature subspace – best feature is selected based on ‘‘goodness of fit, ’’ • can discriminate interacting and non-interacting – stops growing the tree when all pairs at a given node are well-separated • Traverse the tree to classify an unknown protein pair 17 Jan 2006 32

Structure-Function Analysis DNA/Protein structure-function analysis and prediction • Protein-protein Interaction (PPI) and Docking: • Protein-protein Interaction – Interfaces – Solvation – Energetics – Conformational change – Allostery 17 Jan 2006 • Prediction • Docking – Search space – Docking methods [ Examples ] 33

Structure-Function Analysis Docking - ZDOCK • Protein-protein docking – 3 -dimensional (3 D) structure of protein complex – starting from 3 D structures of receptor and ligand • Rigid-body docking algorithm (ZDOCK) – pairwise shape complementarity function – all possible binding modes – using Fast Fourier Transform algorithm • Refinement algorithm (RDOCK) – top 2000 predicted structures – three-stage energy minimization – electrostatic and desolvation energies • molecular mechanical software (CHARMM) • statistical energy method (Atomic Contact Energy) • 49 non-redundant unbound test cases: – near-native structure (<2. 5Å) for 37% test cases • for 49% within top 4 17 Jan 2006 34

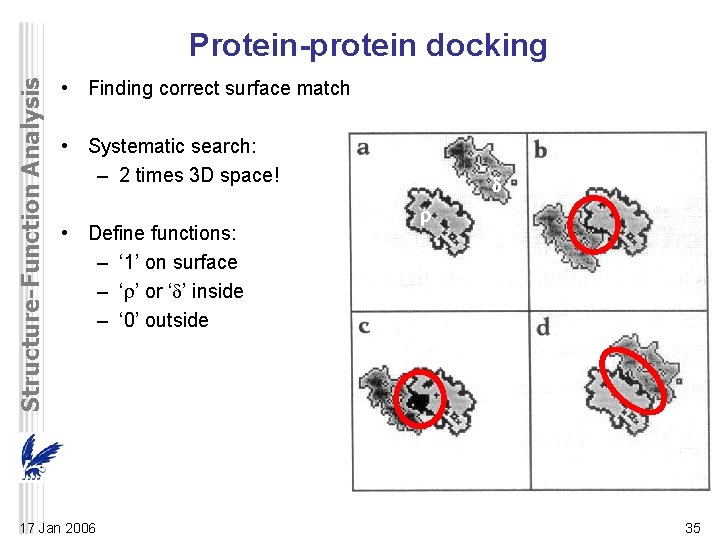
Structure-Function Analysis Protein-protein docking • Finding correct surface match • Systematic search: – 2 times 3 D space! • Define functions: – ‘ 1’ on surface – ‘ ’ or ‘ ’ inside – ‘ 0’ outside 17 Jan 2006 35

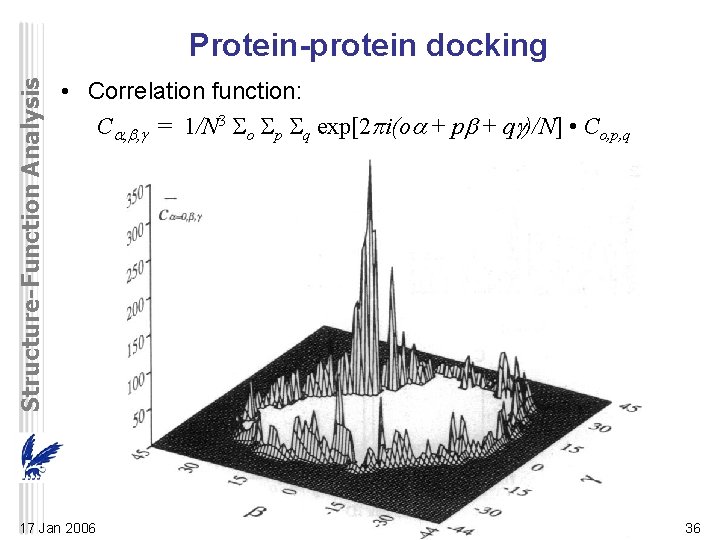
Structure-Function Analysis Protein-protein docking • Correlation function: C = 1/N 3 o p q exp[2 i(o + p + q )/N] • Co, p, q 17 Jan 2006 36

Structure-Function Analysis Docking Programs • • • • ZDOCK, RDOCK Auto. Dock Bielefeld Protein Docking DOCK DOT FTDock, RPScore and Multi. Dock GRAMM Hex 3. 0 ICM Protein-Protein docking KORDO Mol. Fit MPI Protein Docking Nussinov-Wolfson Structural Bioinformatics Group … 17 Jan 2006 37