102405 Promoter Prediction RNA Structure Function Prediction 2222021









- Slides: 46

10/24/05 Promoter Prediction RNA Structure & Function Prediction 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 1

Announcements Seminar (Mon Oct 24) (several additional seminars listed in email sent to class) 12: 10 PM IG Faculty Seminar in 101 Ind Ed II "Laser capture microdissection-facilitated transcriptional profiling of abscission zones in Arabidopsis" Coralie Lashbrook, EEOB http: //www. bb. iastate. edu/%7 Emarit/GEN 691. html Mark your calendars: 1: 10 PM Nov 14 Baker Seminar in Howe Hall Auditorium "Discovering transcription factor binding sites" Douglas Brutlag, Dept of Biochemistry & Medicine, Stanford University School of Medicine 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 2

Announcements 544 Semester Projects Thanks to all who sent already! Others: Information needed today! ddobbs@iastate. edu Briefly describe: • Your background & current grad research • Is there a problem related to your research you would like to learn more about & develop as project for this course? or • What would your ‘dream’ project be? 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 3

Announcements Exam 2 - this Friday Posted Online: Exam 2 Study Guide 544 Reading Assignment (2 papers) Office Hours: David Mon 1 -2 PM in 209 Atanasoff Drena Tues 10 -11 AM in 106 MBB Michael - none this week Thurs No Lab - Extra Office Hrs instead: David 1 -3 PM in 209 Atanasoff Drena 1 -3 PM in 106 MBB 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 4

Announcements • Updated PPTs & PDFs for Gene Prediction lectures (covered on Exam 2) will be posted today (changes are minor) • Is everyone on BCB 444/544 mailing list? Auditors? 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 5

Promoter Prediction & RNA Structure/Function Prediction Mon Wed Quite a few more words re: Gene prediction Promoter prediction RNA structure & function RNA structure prediction 2' & 3' structure prediction mi. RNA & target prediction Thurs No Lab Fri Exam 2 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 6

Reading Assignment - previous Mount Bioinformatics • Chp 9 Gene Prediction & Regulation • pp 361 -401 • Ck Errata: http: //www. bioinformaticsonline. org/help/errata 2. html * Brown Genomes 2 (NCBI textbooks online) • Sect 9 Overview: Assembly of Transcription Initiation Complex • http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=genomes. chapter. 7002 • Sect 9. 1 -9. 3 DNA binding proteins, Transcription initiation • http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=genomes. section. 7016 * NOTEs: Don’t worry about the details!! • See Study Guide for Exam 2 re: Sections covered 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 7

Optional - but very helpful reading: (that's a hint!) 1) Zhang MQ (2002) Computational prediction of eukaryotic proteincoding genes. Nat Rev Genet 3: 698 -709 http: //proxy. lib. iastate. edu: 2103/nrg/journal/v 3/n 9/full/nrg 890_fs. html 2) 1) Wasserman WW & Sandelin A (2004) Applied bioinformatics for identification of regulatory elements. Nat Rev Genet 5: 276 -287 http: //proxy. lib. iastate. edu: 2103/nrg/journal/v 5/n 4/full/nrg 1315_fs. html Check this out: http: //www. phylofoot. org/NRG_testcases/ 03489059922 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 8

Reading Assignment (for Wed) Mount Bioinformatics • Chp 8 Prediction of RNA Secondary Structure • pp. 327 -355 • Ck Errata: http: //www. bioinformaticsonline. org/help/errata 2. html Cates (Online) RNA Secondary Structure Prediction Module • http: //cnx. rice. edu/content/m 11065/latest/ 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 9

Review last lecture: Gene Prediction (formerly Gene Prediction - 3) • Overview of steps & strategies • Algorithms • Gene prediction software 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 10

Predicting Genes - Basic steps: • Obtain genomic DNA sequence • Translate in all 6 reading frames • Compare with protein sequence database • Also perform database similarity search with EST & c. DNA databases, if available • Use gene prediction programs to locate genes • Analyze gene regulatory sequences Note: 1. 2. 3. Several important details missing above: Mask to "remove" repetitive elements (ALUs, etc. )・ Perform database search on translated DNA (Blast. X, TFasta) Use several programs to predict genes (Gen. Scan, Gene. Mark. hmm) 4. Translate putative ORFs and search for functional motifs (Blocks, Motifs, etc. ) & regulatory sequences 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 11

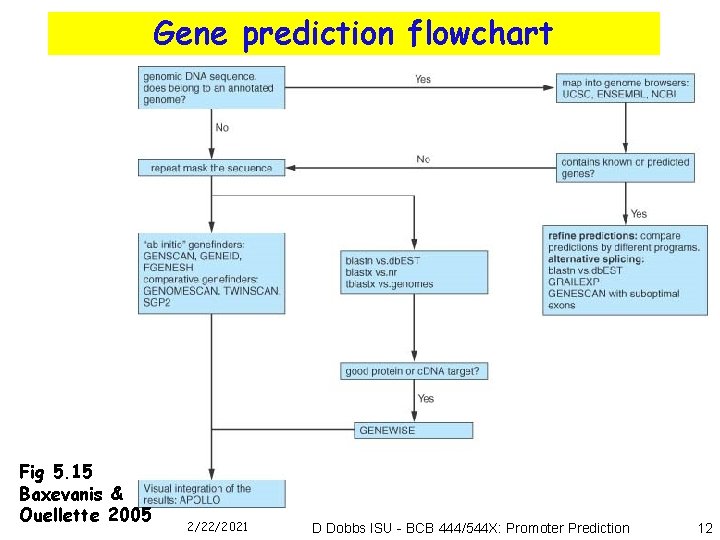
Gene prediction flowchart Fig 5. 15 Baxevanis & Ouellette 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 12

Overview of gene prediction strategies What sequence signals can be used? • Transcription: TF binding sites, promoter, initiation site, terminator • Processing signals: splice donor/acceptors, poly. A signal • Translation: start (AUG = Met) & stop (UGA, UUA, UAG) ORFs, codon usage What other types of information can be used? • c. DNAs & ESTs (pairwise alignment) • homology (sequence comparison, BLAST) 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 13

Examples of gene prediction software 1) Similarity-based or Comparative • • BLAST SGP 2 (extension of Gene. ID) • • • Gene. ID - (used in lab last week) GENSCAN - (used in lab last week) Gene. Mark. hmm - (should try this!) • Gene. Seqer (Brendel et al. , ISU) 2) Ab initio = “from the beginning” 3) Combined "evidence-based” BEST? GENSCAN, Gene. Mark. hmm, Gene. Seqer but depends on organism & specific task 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 14

Annotated lists of gene prediction software • URLs from Mount Chp 9, available online Table 9. 1 http: //www. bioinformaticsonline. org/links/ch_09_t_1. html • from Pevsner Chps 14 & 16 • Table in Zhang Nat Rev Genet article: • Another list: Kozar, Stanford http: //www. bioinfbook. org/chapt 14. htm - prokaryotic http: //www. bioinfbook. org/chapt 16. htm - eukaryotic hptt: //proxy. lib. iastate. edu: 2103/nrg/journal/v 3/n 9/full/nrg 890_fs. html http: //cmgm. stanford. edu/classes/genefind/ Ø Performance Evaluation? Guig�ó, Barcelona (& sites above) http: //www 1. imim. es/courses/Seq. Analysis/Gene. Identification/Evalua tion. html 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 15

Gene prediction: Eukaryotes vs prokaryotes Gene prediction is easier in microbial genomes Methods? Previously, mostly HMM-based see Mount Fig 9. 7 (E. coli gene) Now: similarity-based methods because so many genomes available Many microbial genomes have been fully sequenced & whole-genome "gene structure" and "gene function" annotations are available. e. g. , Gene. Mark. hmm TIGR Comprehensive Microbial Resource (CMR) NCBI Microbial Genomes 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 16

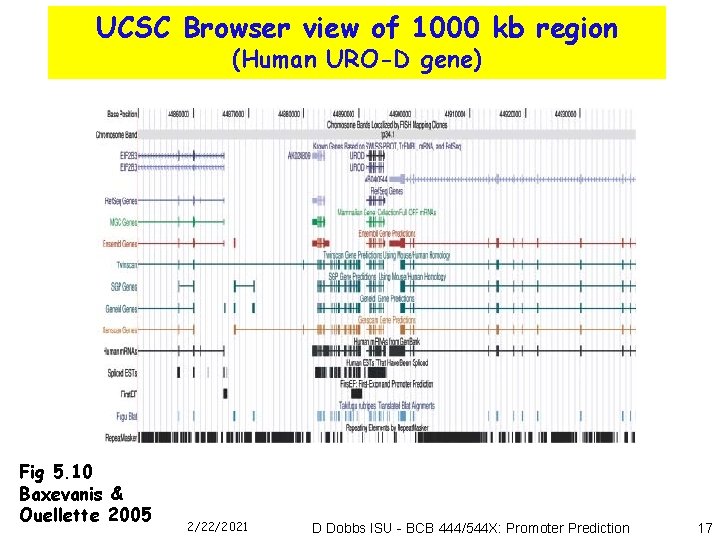
UCSC Browser view of 1000 kb region (Human URO-D gene) Fig 5. 10 Baxevanis & Ouellette 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 17

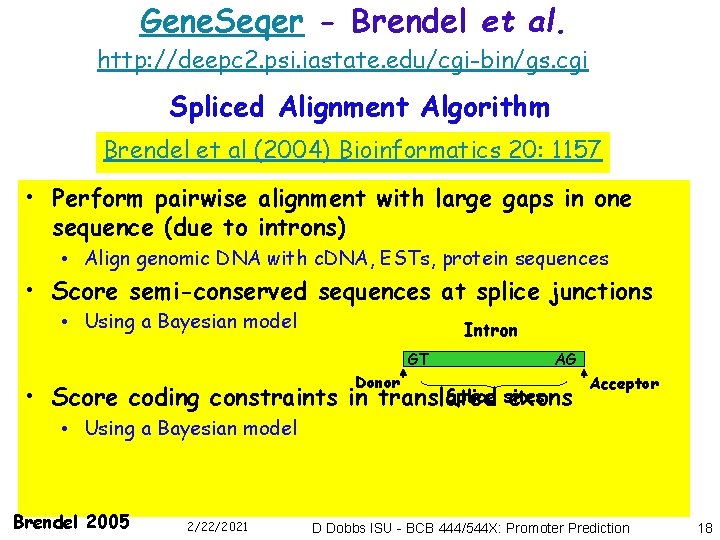
Gene. Seqer - Brendel et al. http: //deepc 2. psi. iastate. edu/cgi-bin/gs. cgi Spliced Alignment Algorithm Brendel et al (2004) Bioinformatics 20: 1157 • Perform pairwise alignment with large gaps in one sequence (due to introns) • Align genomic DNA with c. DNA, ESTs, protein sequences • Score semi-conserved sequences at splice junctions • Using a Bayesian model Intron GT Donor • Score coding constraints in AG Splice sites translated exons Acceptor • Using a Bayesian model Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 18

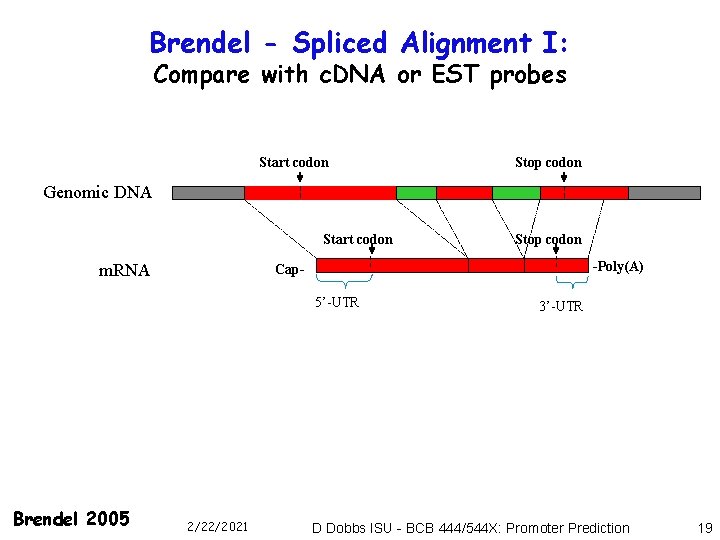
Brendel - Spliced Alignment I: Compare with c. DNA or EST probes Start codon Stop codon Genomic DNA Start codon m. RNA -Poly(A) Cap 5’-UTR Brendel 2005 Stop codon 2/22/2021 3’-UTR D Dobbs ISU - BCB 444/544 X: Promoter Prediction 19

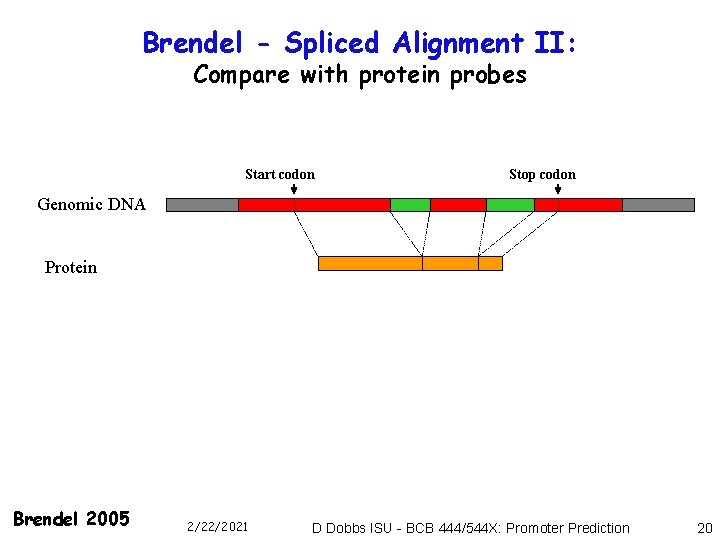
Brendel - Spliced Alignment II: Compare with protein probes Start codon Stop codon Genomic DNA Protein Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 20

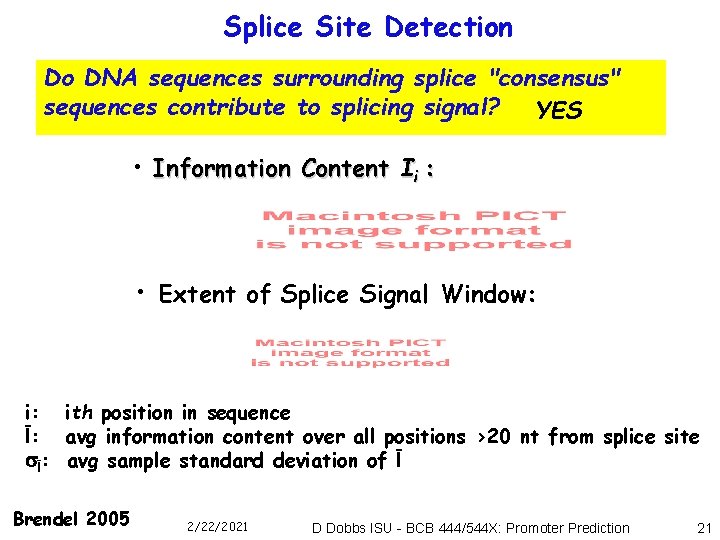
Splice Site Detection Do DNA sequences surrounding splice "consensus" sequences contribute to splicing signal? YES • Information Content Ii : • Extent of Splice Signal Window: i: ith position in sequence Ī: avg information content over all positions >20 nt from splice site Ī: avg sample standard deviation of Ī Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 21

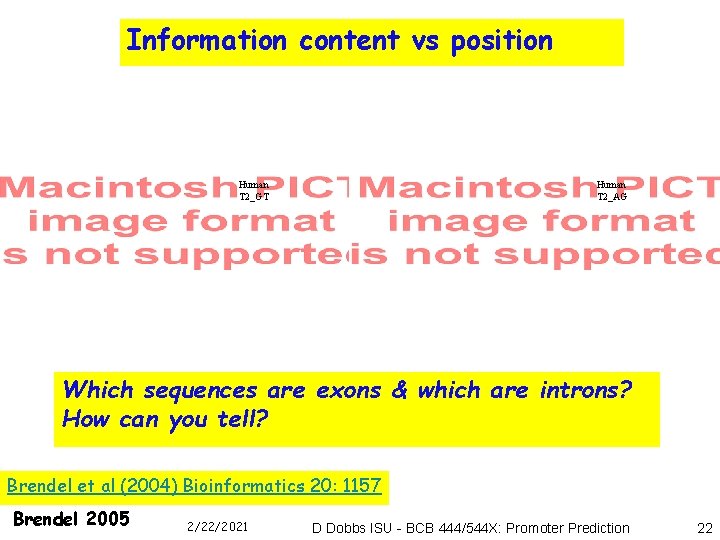
Information content vs position Human T 2_GT Human T 2_AG Which sequences are exons & which are introns? How can you tell? Brendel et al (2004) Bioinformatics 20: 1157 Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 22

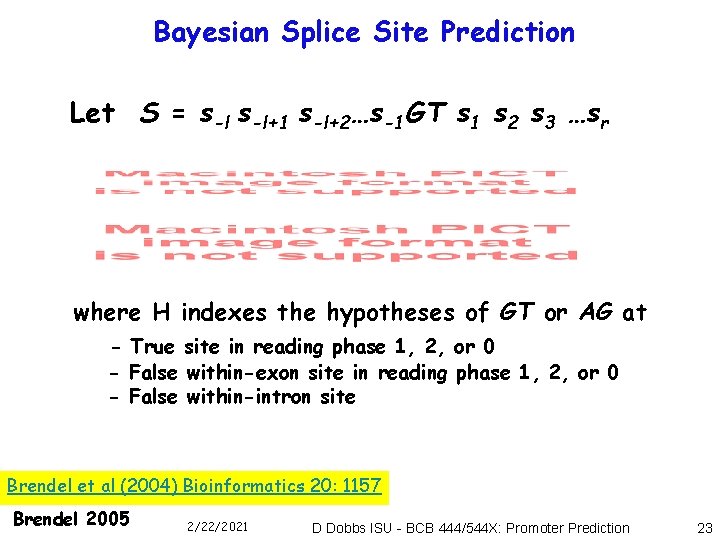
Bayesian Splice Site Prediction Let S = s-l+1 s-l+2…s-1 GT s 1 s 2 s 3 …sr where H indexes the hypotheses of GT or AG at - True site in reading phase 1, 2, or 0 - False within-exon site in reading phase 1, 2, or 0 - False within-intron site Brendel et al (2004) Bioinformatics 20: 1157 Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 23

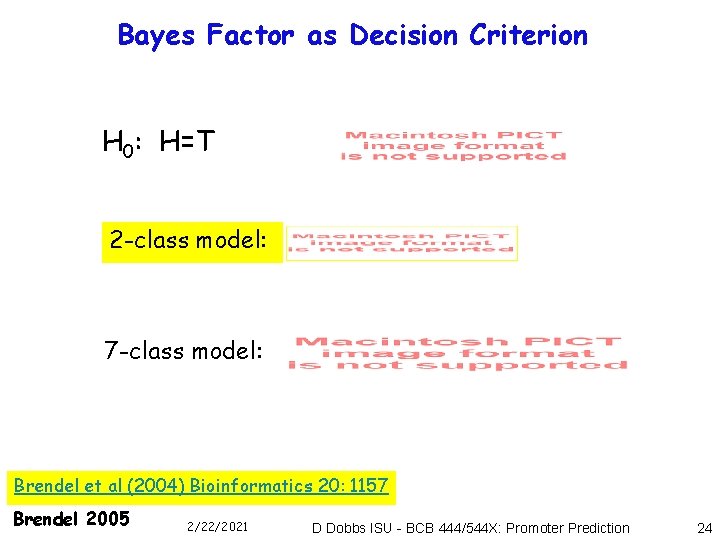
Bayes Factor as Decision Criterion H 0: H=T 2 -class model: 7 -class model: Brendel et al (2004) Bioinformatics 20: 1157 Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 24

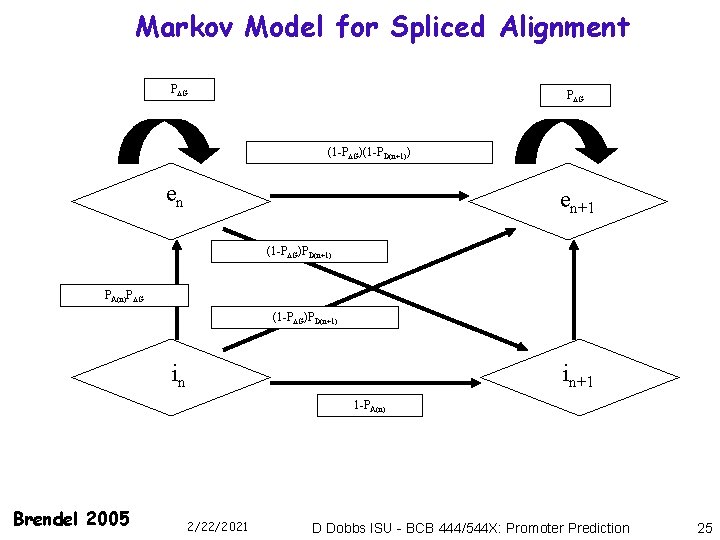
Markov Model for Spliced Alignment P G (1 -P G)(1 -PD(n+1)) en en+1 (1 -P G)PD(n+1) PA(n)P G (1 -P G)PD(n+1) in in+1 1 -PA(n) Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 25

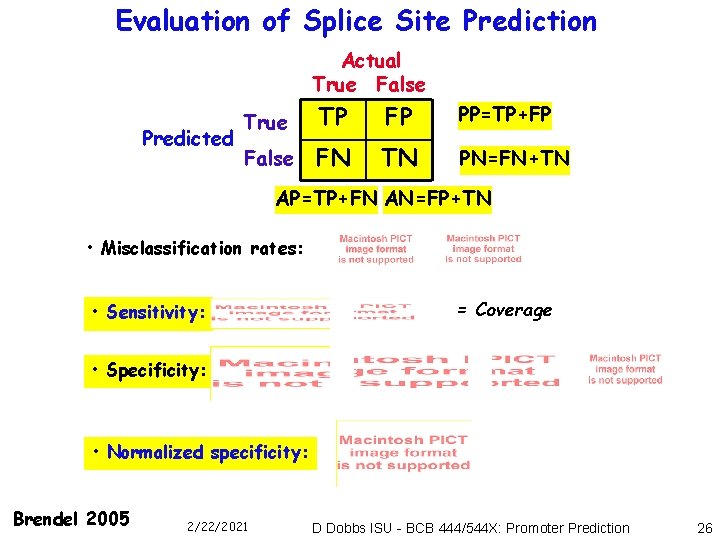
Evaluation of Splice Site Prediction Actual True False Predicted True TP FP PP=TP+FP False FN TN PN=FN+TN AP=TP+FN AN=FP+TN • Misclassification rates: • Sensitivity: = Coverage • Specificity: • Normalized specificity: Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 26

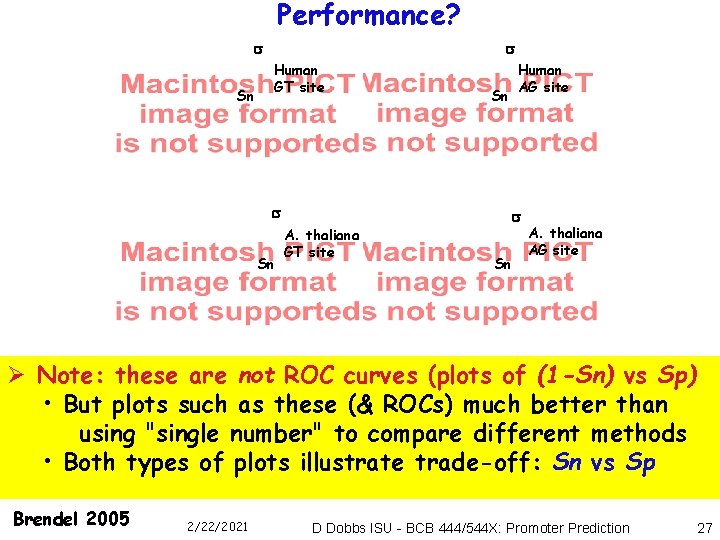
Performance? Sn Human GT site Sn Human AG site Sn A. thaliana GT site Sn A. thaliana AG site Ø Note: these are not ROC curves (plots of (1 -Sn) vs Sp) • But plots such as these (& ROCs) much better than using "single number" to compare different methods • Both types of plots illustrate trade-off: Sn vs Sp Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 27

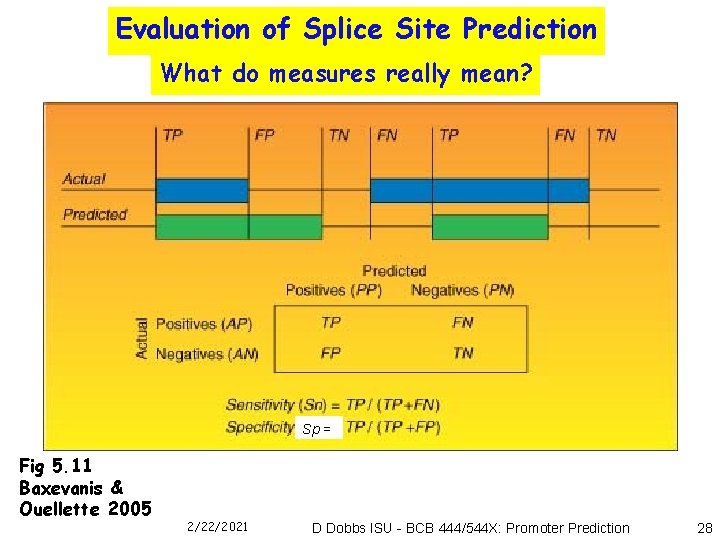
Evaluation of Splice Site Prediction What do measures really mean? Sp = Fig 5. 11 Baxevanis & Ouellette 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 28

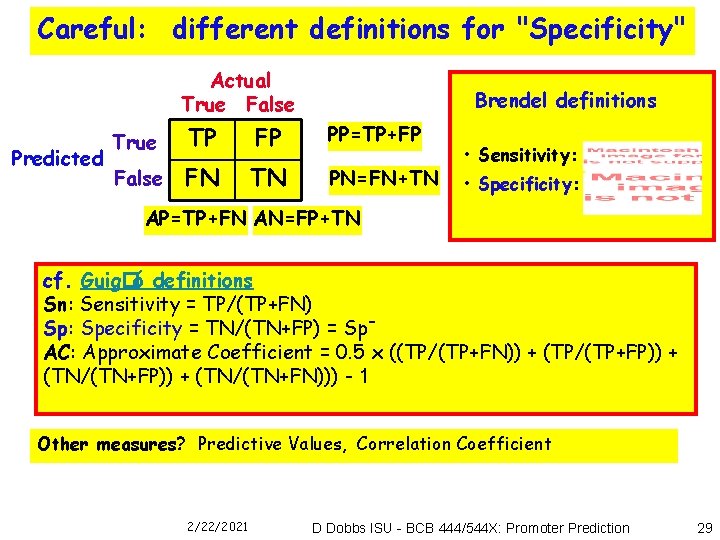
Careful: different definitions for "Specificity" Actual True False Predicted Brendel definitions True TP FP PP=TP+FP False FN TN PN=FN+TN • Sensitivity: • Specificity: AP=TP+FN AN=FP+TN cf. Guig�ó definitions Sn: Sensitivity = TP/(TP+FN) Sp: Specificity = TN/(TN+FP) = Sp. AC: Approximate Coefficient = 0. 5 x ((TP/(TP+FN)) + (TP/(TP+FP)) + (TN/(TN+FN))) - 1 Other measures? Predictive Values, Correlation Coefficient 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 29

Best measures for comparing different methods? • ROC curves (Receiver Operating Characteristic? !!) http: //www. anaesthetist. com/mnm/stats/roc/ "The Magnificent ROC" - has fun applets & quotes: "There is no statistical test, however intuitive and simple, which will not be abused by medical researchers" • Correlation Coefficient (Matthews correlation coefficient (MCC) Do not memorize this! MCC = 1 for a perfect prediction 0 for a completely random assignment -1 for a "perfectly incorrect" prediction 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 30

Performance of Gene. Seqer vs other methods? • Comparison with ab initio gene prediction (e. g. , GENESCAN) • Depends on: • Availability of ESTs • Availability of protein homologs Other Performance Evaluations? Guig�ó http: //www 1. imim. es/courses/Seq. Analysis/Gene. Identification /Evaluation. html Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 31

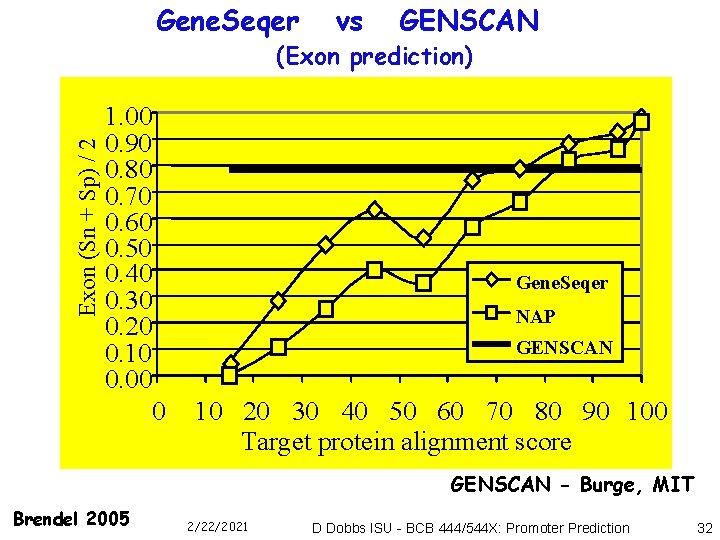
Gene. Seqer vs GENSCAN Exon (Sn + Sp) / 2 (Exon prediction) 1. 00 0. 90 0. 80 0. 70 0. 60 0. 50 0. 40 0. 30 0. 20 0. 10 0. 00 Gene. Seqer NAP GENSCAN 0 10 20 30 40 50 60 70 80 90 100 Target protein alignment score GENSCAN - Burge, MIT Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 32

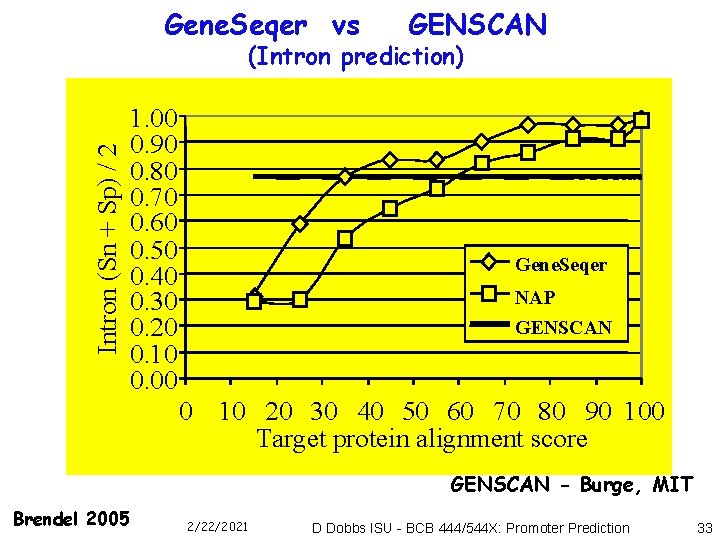
Gene. Seqer vs GENSCAN Intron (Sn + Sp) / 2 (Intron prediction) 1. 00 0. 90 0. 80 0. 70 0. 60 0. 50 0. 40 0. 30 0. 20 0. 10 0. 00 Gene. Seqer NAP GENSCAN 0 10 20 30 40 50 60 70 80 90 100 Target protein alignment score GENSCAN - Burge, MIT Brendel 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 33

Other Resources Current Protocols in Bioinformatics http: //www. 4 ulr. com/products/currentprotocols/bioinformatics. html Finding Genes 4. 1 An Overview of Gene Identification: Approaches, Strategies, and Considerations 4. 2 Using MZEF To Find Internal Coding Exons 4. 3 Using GENEID to Identify Genes 4. 4 Using Glimmer. M to Find Genes in Eukaryotic Genomes 4. 5 Prokaryotic Gene Prediction Using Gene. Mark and Gene. Mark. hmm 4. 6 Eukaryotic Gene Prediction Using Gene. Mark. hmm 4. 7 Application of First. EF to Find Promoters and First Exons in the Human Genome 4. 8 Using TWINSCAN to Predict Gene Structures in Genomic DNA Sequences 4. 9 Grail. EXP and Genome Analysis Pipeline for Genome Annotation 4. 10 Using Repeat. Masker to Identify Repetitive Elements in Genomic Sequences 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 34

New Today: Promoter Prediction • A few more words about Gene prediction • Predicting regulatory regions (focus on promoters) Brief review promoters & enhancers Predicting in eukaryotes vs prokaryotes Introduction to RNA Structure & function 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 35

Predicting Promoters What signals are there? Algorithms Promoter prediction software 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 36

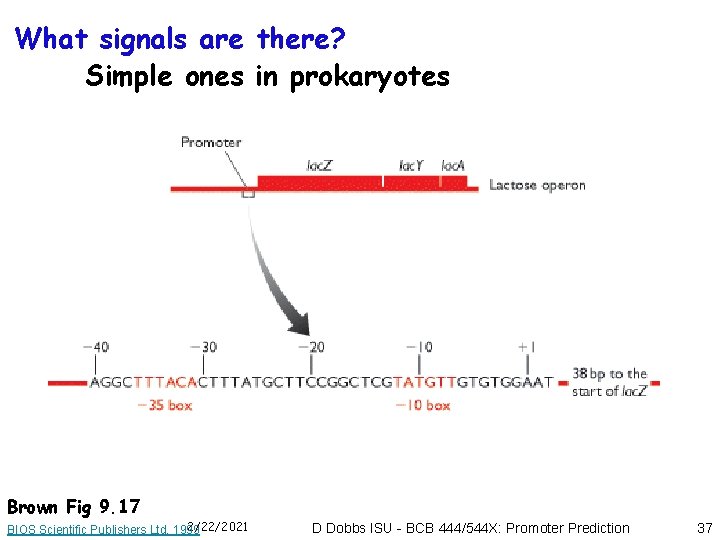
What signals are there? Simple ones in prokaryotes Brown Fig 9. 17 2/22/2021 BIOS Scientific Publishers Ltd, 1999 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 37

Prokaryotic promoters • RNA polymerase complex recognizes promoter sequences located very close to & on 5’ side (“upstream”) of initiation site • RNA polymerase complex binds directly to these. with no requirement for “transcription factors” • Prokaryotic promoter sequences are highly conserved • -10 region • -35 region 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 38

What signals are there? Complex ones in eukaryotes! Fig 9. 13 Mount 2004 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 39

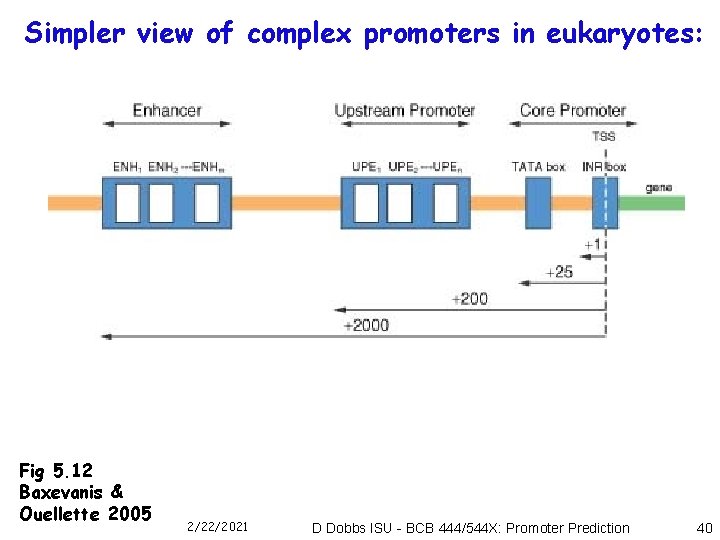
Simpler view of complex promoters in eukaryotes: Fig 5. 12 Baxevanis & Ouellette 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 40

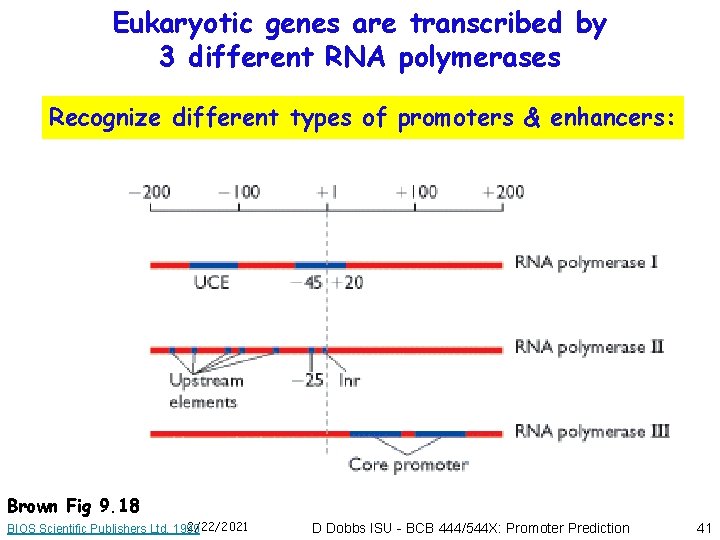
Eukaryotic genes are transcribed by 3 different RNA polymerases Recognize different types of promoters & enhancers: Brown Fig 9. 18 2/22/2021 BIOS Scientific Publishers Ltd, 1999 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 41

Eukaryotic promoters & enhancers • Promoters located “relatively” close to initiation site (but can be located within gene, rather than upstream!) • Enhancers also required for regulated transcription (these control expression in specific cell types, developmental stages, in response to environment) • RNA polymerase complexes do not specifically recognize promoter sequences directly • Transcription factors bind first and serve as “landmarks” for recognition by RNA polymerase complexes 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 42

Eukaryotic transcription factors • Transcription factors (TFs) are DNA binding proteins that also interact with RNA polymerase complex to activate or repress transcription • TFs contain characteristic “DNA binding motifs” http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=genomes. table. 7039 • TFs recognize specific short DNA sequence motifs “transcription factor binding sites” • Several databases for these, e. g. TRANSFAC http: //www. generegulation. com/cgibin/pub/databases/transfac 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 43

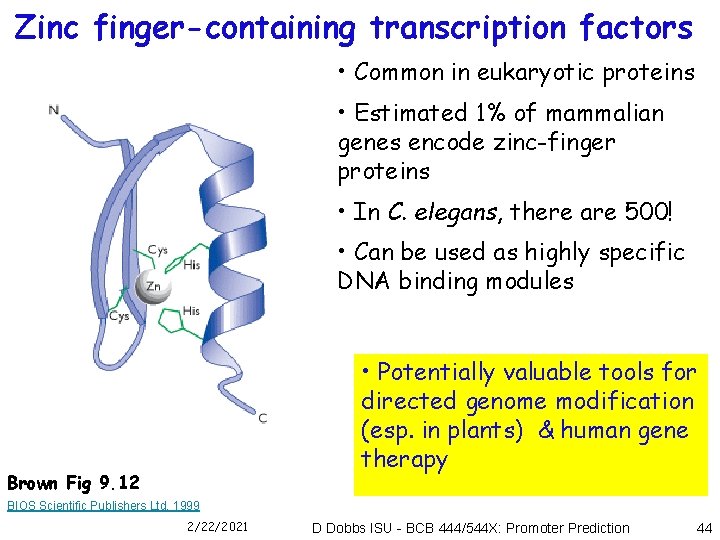
Zinc finger-containing transcription factors • Common in eukaryotic proteins • Estimated 1% of mammalian genes encode zinc-finger proteins • In C. elegans, there are 500! • Can be used as highly specific DNA binding modules • Potentially valuable tools for directed genome modification (esp. in plants) & human gene therapy Brown Fig 9. 12 BIOS Scientific Publishers Ltd, 1999 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 44

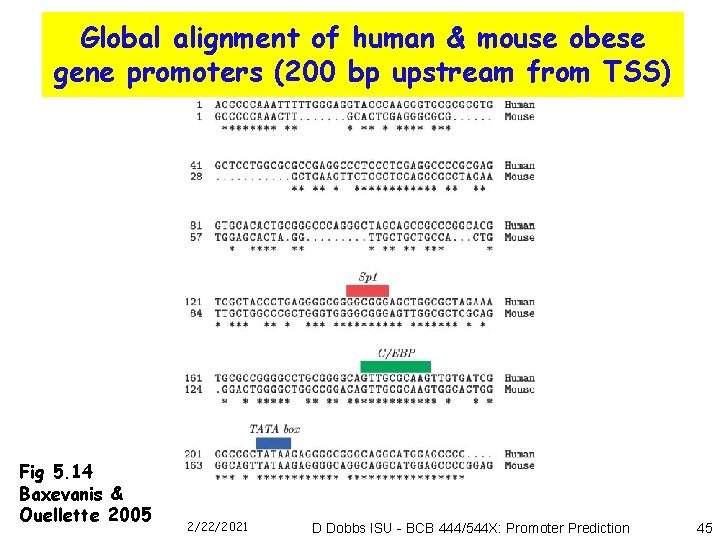
Global alignment of human & mouse obese gene promoters (200 bp upstream from TSS) Fig 5. 14 Baxevanis & Ouellette 2005 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 45

Reading Assignment (for Wed) Mount Bioinformatics • Chp 8 Prediction of RNA Secondary Structure • pp. 327 -355 • Ck Errata: http: //www. bioinformaticsonline. org/help/errata 2. html Cates (Online) RNA Secondary Structure Prediction Module • http: //cnx. rice. edu/content/m 11065/latest/ 2/22/2021 D Dobbs ISU - BCB 444/544 X: Promoter Prediction 46