CSE 182 L 7 Protein Sequence Analysis using




















![A geometric approach • Plot the following vectors – – E= [10, 20] I A geometric approach • Plot the following vectors – – E= [10, 20] I](https://slidetodoc.com/presentation_image_h/19d9105a4627c78c5f37b869cad8472f/image-65.jpg)

![Combining Signals • We can compute the following: – – – E-score[i, j] i Combining Signals • We can compute the following: – – – E-score[i, j] i](https://slidetodoc.com/presentation_image_h/19d9105a4627c78c5f37b869cad8472f/image-70.jpg)









- Slides: 97

CSE 182 -L 7 Protein Sequence Analysis using HMMs, Gene Finding

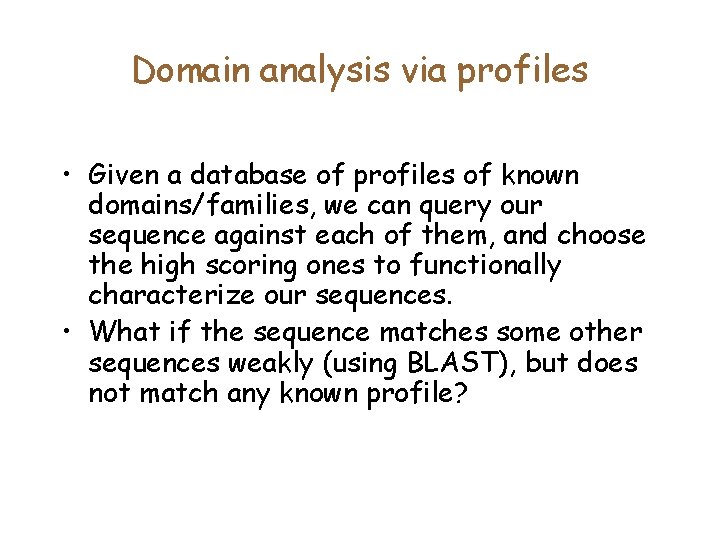
Domain analysis via profiles • Given a database of profiles of known domains/families, we can query our sequence against each of them, and choose the high scoring ones to functionally characterize our sequences. • What if the sequence matches some other sequences weakly (using BLAST), but does not match any known profile?

Psi-BLAST idea Seq Db --In the next iteration, the red sequence will be thrown out. --It matches the query in non-essential residues • Iterate: – – Find homologs using Blast on query Discard very similar homologs Align, make a profile, search with profile. Why is this more sensitive?

Psi-BLAST speed • Two time consuming steps. 1. Multiple alignment of homologs 2. Searching with Profiles. 1. Does the keyword search idea work? • • Multiple alignment: – Use ungapped multiple alignments only Pigeonhole principle again: – – If profile of length m must score >= T Then, a sub-profile of length l must score >= l. T|/m Generate all l-mers that score at least l. T|/M Search using an automaton

Representation 3: HMMs • Building good profiles relies upon good alignments. – Difficult if there are gaps in the alignment. – Psi-BLAST/BLOCKS etc. work with gapless alignments. • An HMM representation of Profiles helps put the alignment construction/membership query in a uniform framework. • Also allows for position specific gap scoring. V

QUIZ! • Question: • your ‘friend’ likes to gamble. • He tosses a coin: HEADS, he gives you a dollar. TAILS, you give him a dollar. • Usually, he uses a fair coin, but ‘once in a while’, he uses a loaded coin. • Can you say what fraction of the times he loads the coin?

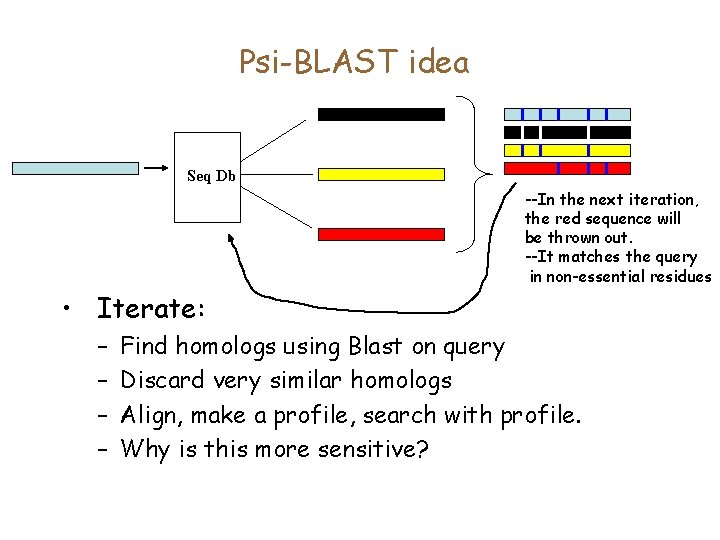
The generative model • Think of each column in the alignment as generating a distribution. • For each column, build a node that outputs a residue with the appropriate distribution 0. 71 Pr[F]=0. 71 Pr[Y]=0. 14

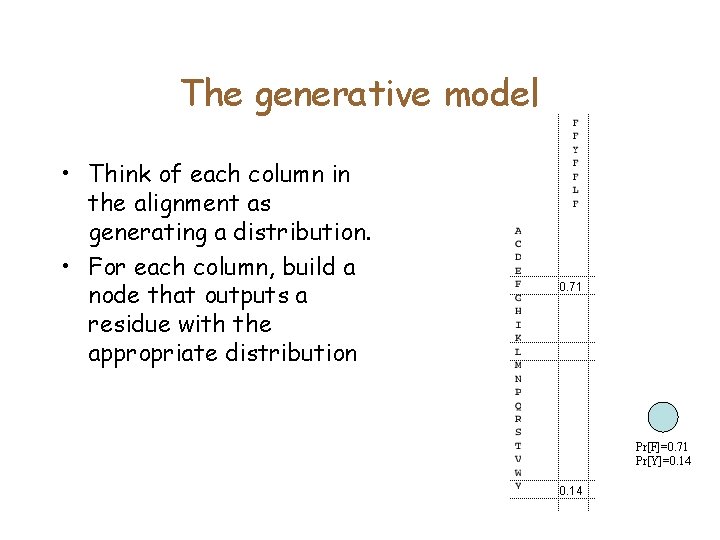
A simple Profile HMM • Connect nodes for each column into a chain. Thie chain generates random sequences. • What is the probability of generating FKVVGQVILD? • In this representation – Prob [New sequence S belongs to a family]= Prob[HMM generates sequence S] • What is the difference with Profiles?

Profile HMMs can handle gaps • The match states are the same as on the previous page. • Insertion and deletion states help introduce gaps. • A sequence may be generated using different paths.

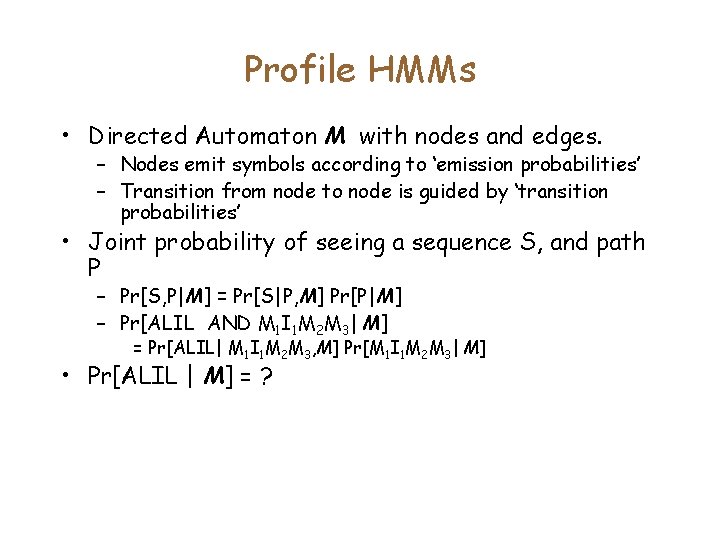
Example A L - L A I V L A I - L • Probability [ALIL] is part of the family? • Note that multiple paths can generate this sequence. – M 1 I 1 M 2 M 3 – M 1 M 2 I 2 M 3 • In order to compute the probabilities, we must assign probabilities of transition between states

Profile HMMs • Directed Automaton M with nodes and edges. – Nodes emit symbols according to ‘emission probabilities’ – Transition from node to node is guided by ‘transition probabilities’ • Joint probability of seeing a sequence S, and path P – Pr[S, P|M] = Pr[S|P, M] Pr[P|M] – Pr[ALIL AND M 1 I 1 M 2 M 3| M] = Pr[ALIL| M 1 I 1 M 2 M 3, M] Pr[M 1 I 1 M 2 M 3| M] • Pr[ALIL | M] = ?

Formally • • The emitted sequence is S=S 1 S 2…Sm The path traversed id P 1 P 2 P 3. . ej(s) = emission probability of symbol s in state Pj Transition probability T[j, k] : Probability of transitioning from state j to state k. • Pr(P, S|M) = e. P 1(S 1) T[P 1, P 2] e. P 2(S 2) …… • What is Pr(S|M)?

Two solutions • An unknown (hidden) path is traversed to produce (emit) the sequence S. • The probability that M emits S can be either – The sum over the joint probabilities over all paths. • Pr(S|M) = ∑P Pr(S, P|M) – OR, it is the probability of the most likely path • Pr(S|M) = max. P Pr(S, P|M) • Both are appropriate ways to model, and have similar algorithms to solve them.

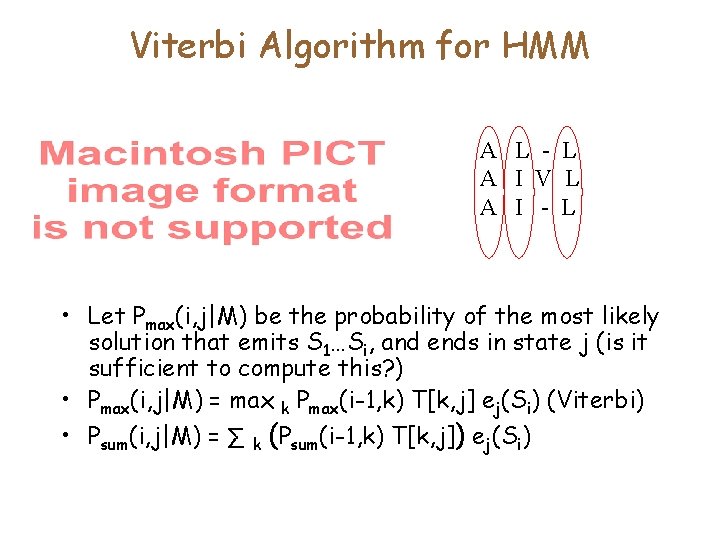
Viterbi Algorithm for HMM A L - L A I V L A I - L • Let Pmax(i, j|M) be the probability of the most likely solution that emits S 1…Si, and ends in state j (is it sufficient to compute this? ) • Pmax(i, j|M) = max k Pmax(i-1, k) T[k, j] ej(Si) (Viterbi) • Psum(i, j|M) = ∑ k (Psum(i-1, k) T[k, j]) ej(Si)

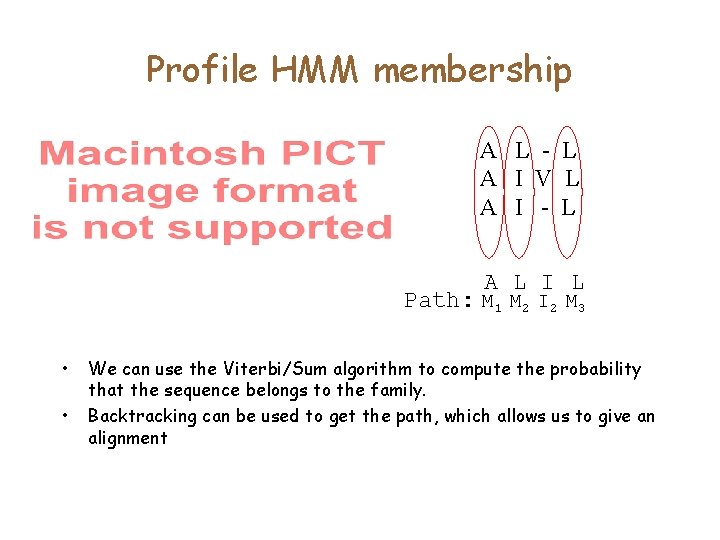
Profile HMM membership A L - L A I V L A I - L A L I L Path: M 1 M 2 I 2 M 3 • • We can use the Viterbi/Sum algorithm to compute the probability that the sequence belongs to the family. Backtracking can be used to get the path, which allows us to give an alignment

Summary • HMMs allow us to model position specific gap penalties, and allow for automated training to get a good alignment. • Patterns/Profiles/HMMs allow us to represent families and foucs on key residues • Each has its advantages and disadvantages, and needs special algorithms to query efficiently.

Protein Domain databases • • • HMM A number of databases capture proteins (domains) using various representations Each domain is also associated with structure/function information, parsed from the literature. Each database has specific query mecahnisms that allow us to compare our seqeunces against them, and assign function 3 D

Gene Finding What is a Gene?

Gene • We define a gene as a location on the genome that codes for proteins. • The genic information is used to manufacture proteins through transcription, and translation. • There is a unique mapping from triplets to amino-acids

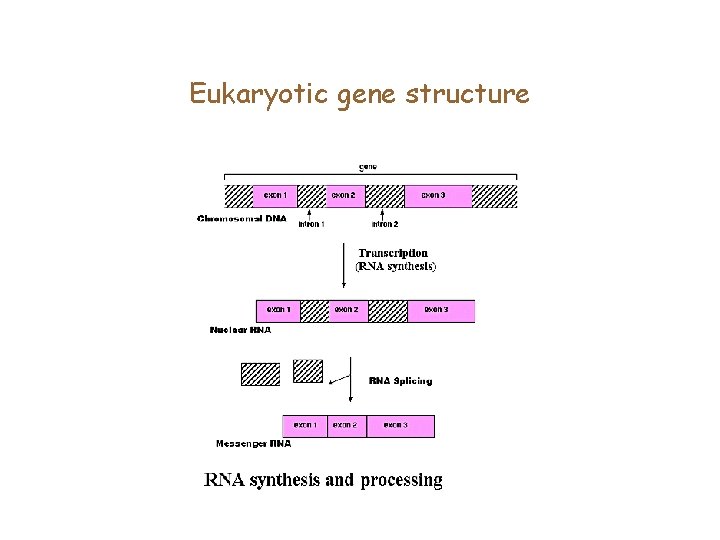
Eukaryotic gene structure

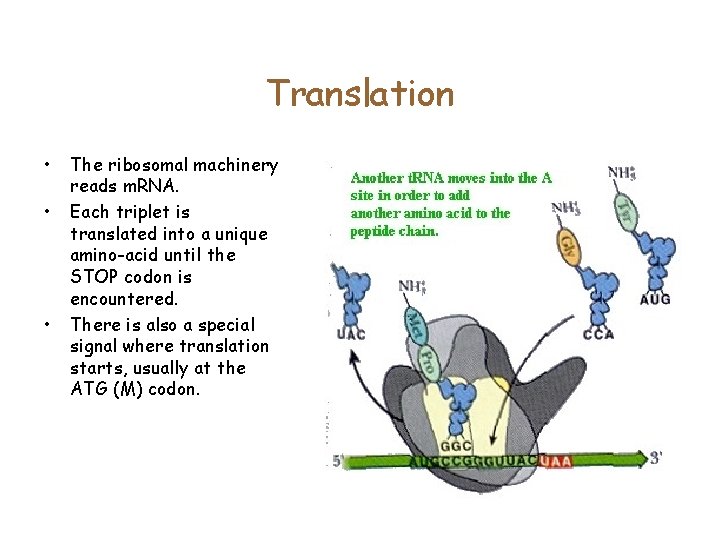
Translation • • • The ribosomal machinery reads m. RNA. Each triplet is translated into a unique amino-acid until the STOP codon is encountered. There is also a special signal where translation starts, usually at the ATG (M) codon.

Translation • • The ribosomal machinery reads m. RNA. Each triplet is translated into a unique amino-acid until the STOP codon is encountered. There is also a special signal where translation starts, usually at the ATG (M) codon. Given a DNA sequence, how many ways can you translate it?

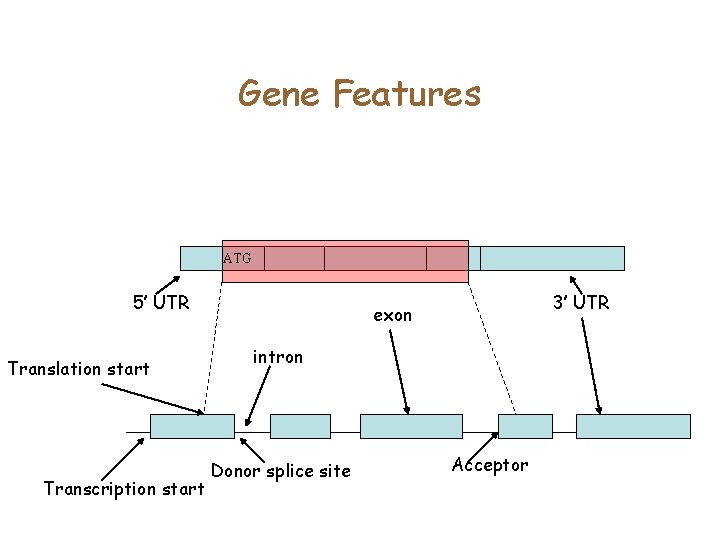
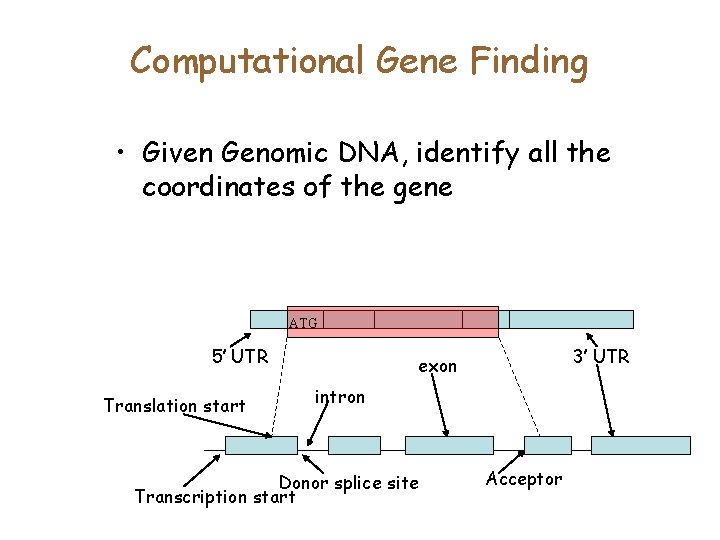
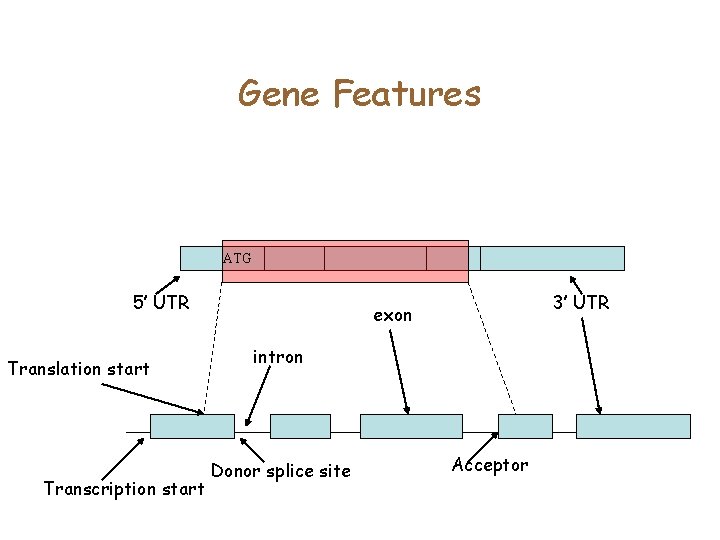
Gene Features ATG 5’ UTR Translation start Transcription start 3’ UTR exon intron Donor splice site Acceptor

Gene identification • Eukaryotic gene definitions: – Location that codes for a protein – The transcript sequence(s) that encodes the protein – The protein sequence(s) • Suppose you want to know all of the genes in an organism. • This was a major problem in the 70 s. Ph. Ds, and careers were spent isolating a single gene sequence. • All of that changed with better reagents and the development of high throughput methods like EST sequencing

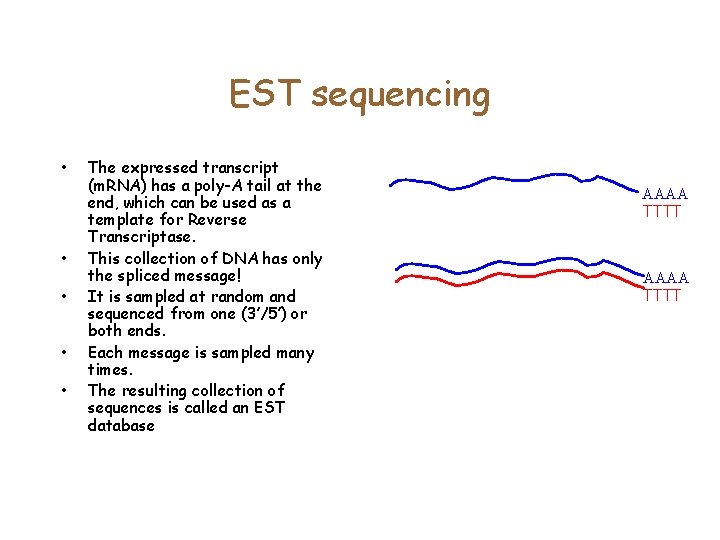
Expressed Sequence Tags • • It is possible to extract all of the m. RNA from a cell. However, m. RNA is unstable An enzyme called reverse transcriptase is used to make a DNA copy of the RNA. Use DNA polymerase to get a complementary DNA strand. Sequence the (stable) c. DNA from both ends. This leads to a collection of transcripts/expressed sequences (ESTs). Many might be from the same gene AAAA TTTT

EST sequencing • • • The expressed transcript (m. RNA) has a poly-A tail at the end, which can be used as a template for Reverse Transcriptase. This collection of DNA has only the spliced message! It is sampled at random and sequenced from one (3’/5’) or both ends. Each message is sampled many times. The resulting collection of sequences is called an EST database AAAA TTTT

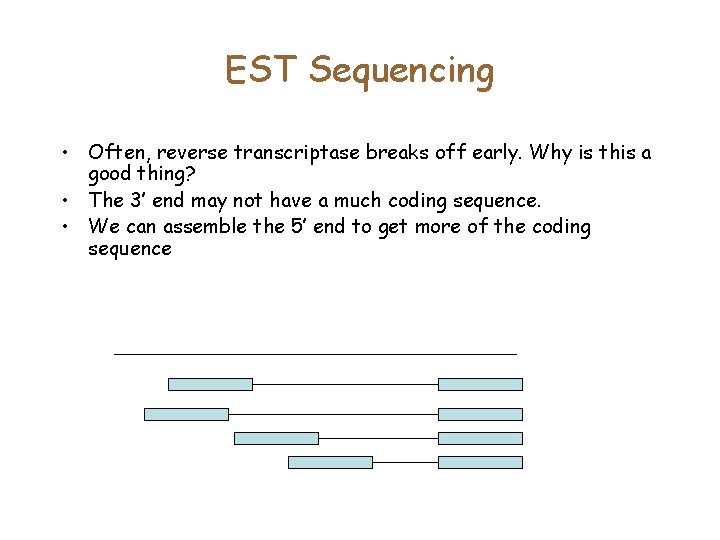
EST Sequencing • Often, reverse transcriptase breaks off early. Why is this a good thing? • The 3’ end may not have a much coding sequence. • We can assemble the 5’ end to get more of the coding sequence

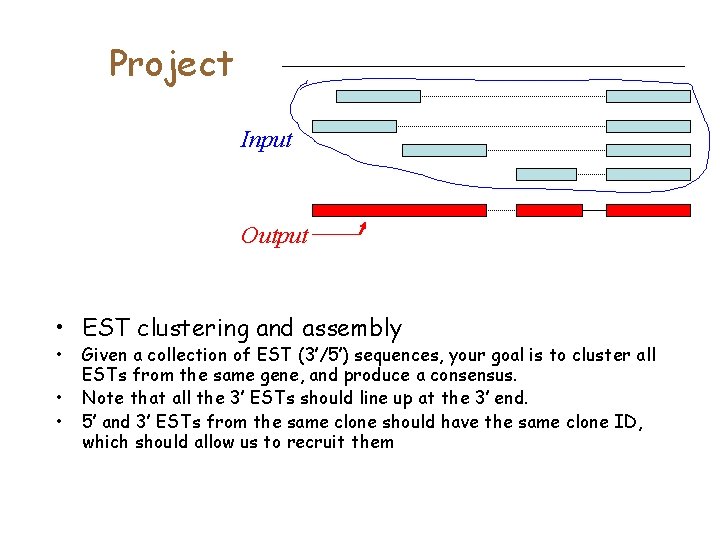
Project Input Output • EST clustering and assembly • • • Given a collection of EST (3’/5’) sequences, your goal is to cluster all ESTs from the same gene, and produce a consensus. Note that all the 3’ ESTs should line up at the 3’ end. 5’ and 3’ ESTs from the same clone should have the same clone ID, which should allow us to recruit them

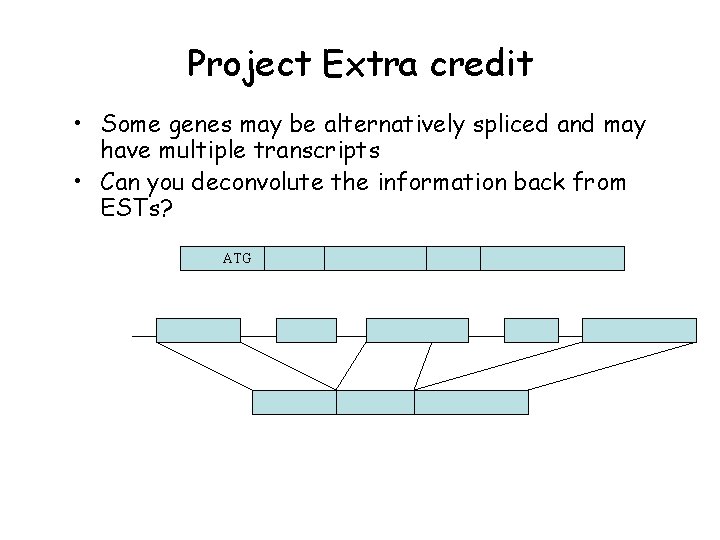
Project Extra credit • Some genes may be alternatively spliced and may have multiple transcripts • Can you deconvolute the information back from ESTs? ATG

Computational Gene Finding • Given Genomic DNA, identify all the coordinates of the gene ATG 5’ UTR Translation start 3’ UTR exon intron Donor splice site Transcription start Acceptor

Gene Finding: The 1 st generation • Given genomic DNA, does it contain a gene (or not)? • Key idea: The distributions of nucleotides is different in coding (translated exons) and noncoding regions. • Therefore, a statistical test can be used to discriminate between coding and non-coding regions.

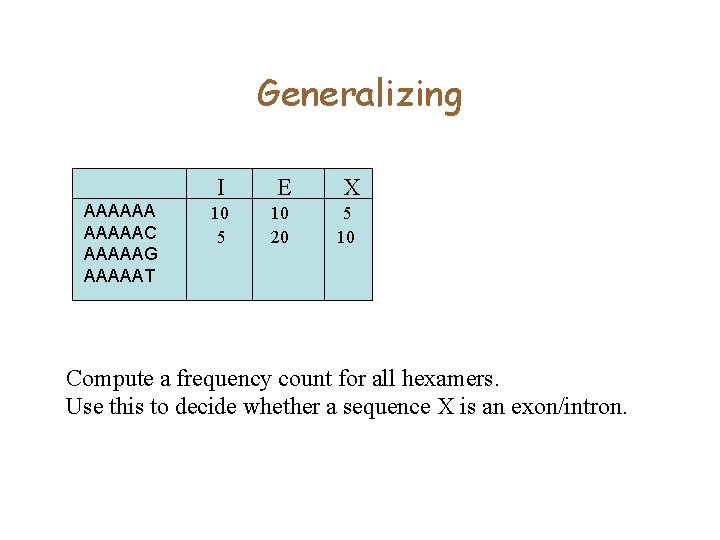
Coding versus Non-coding • You are given a collection of exons, and a collection of intergenic sequence. • Count the number of occurrences of ATGATG in Introns and Exons. – Suppose 1% of the hexamers in Exons are ATGATG – Only 0. 01% of the hexamers in Intons are ATGATG • How can you use this idea to find genes?

Generalizing I E AAAAAAC AAAAAG AAAAAT Compute a frequency count for all hexamers. Use this to decide whether a sequence is an exon/intron

Coding versus non-coding • Fickett and Tung (1992) compared various measures • Measures that preserve the triplet frame are the most successful. • Genscan: 5 th order Markov Model • Conservation across species

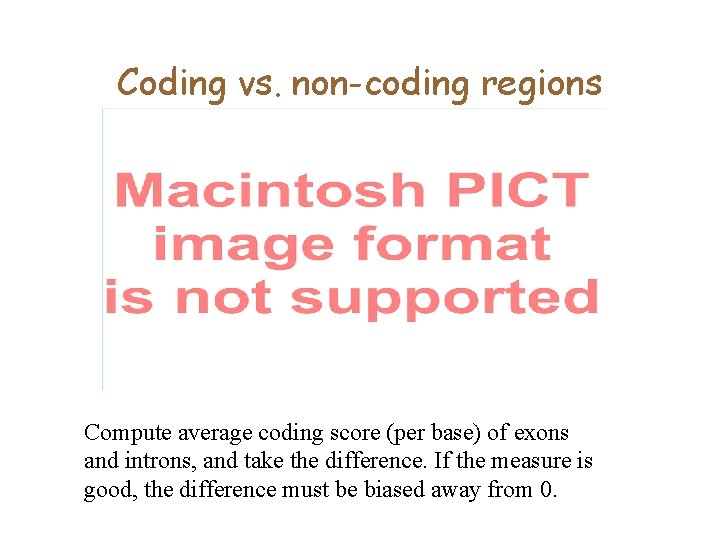
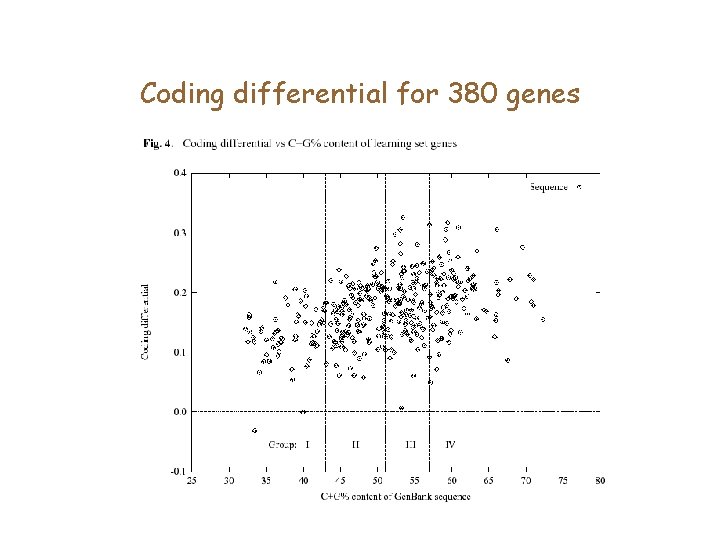
Coding vs. non-coding regions Compute average coding score (per base) of exons and introns, and take the difference. If the measure is good, the difference must be biased away from 0.

Coding differential for 380 genes

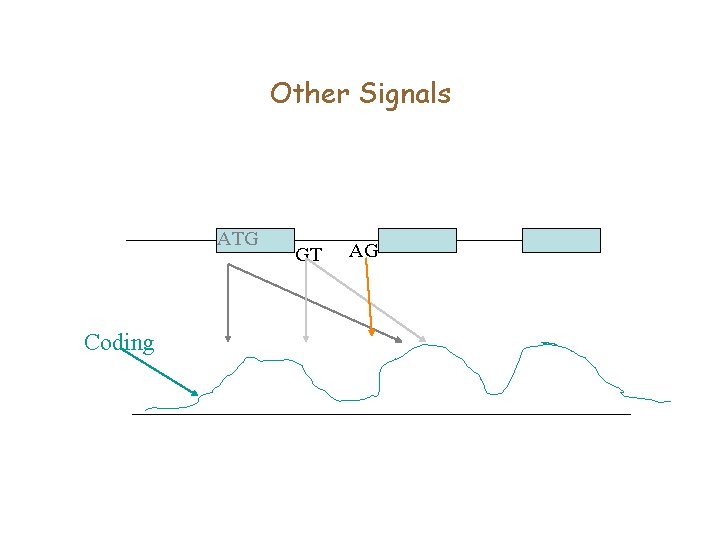
Other Signals ATG Coding GT AG

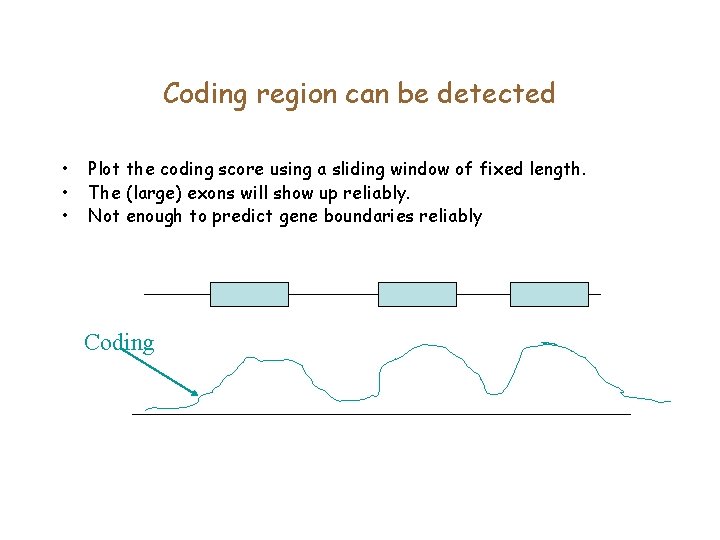
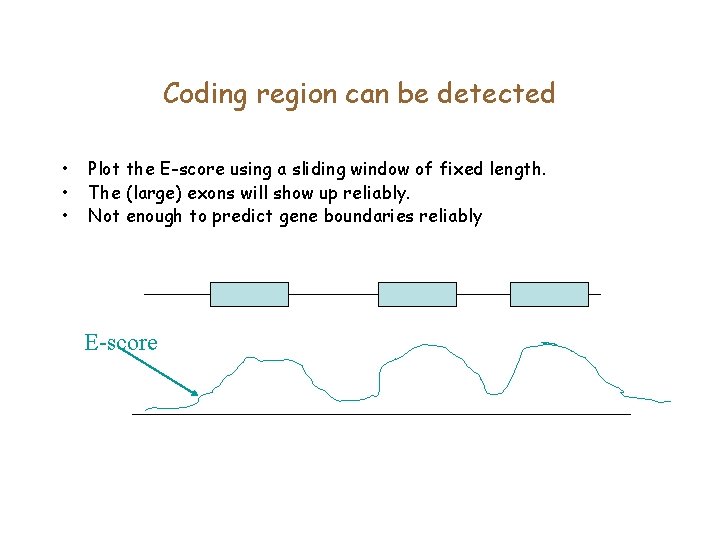
Coding region can be detected • • • Plot the coding score using a sliding window of fixed length. The (large) exons will show up reliably. Not enough to predict gene boundaries reliably Coding

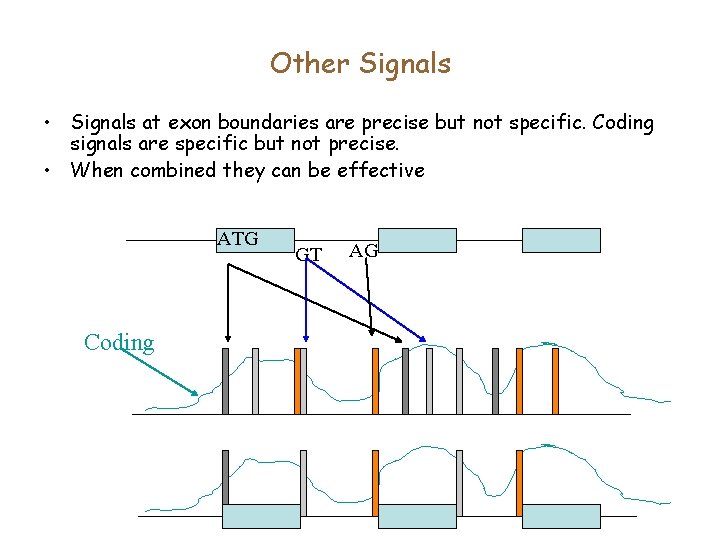
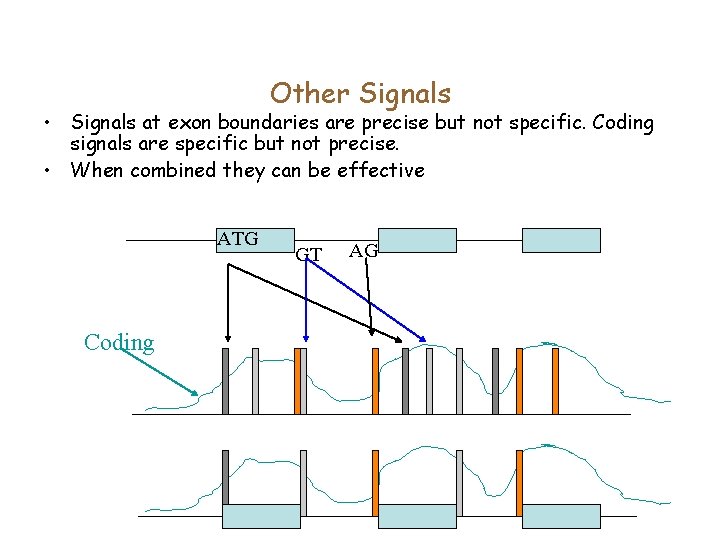
Other Signals • Signals at exon boundaries are precise but not specific. Coding signals are specific but not precise. • When combined they can be effective ATG Coding GT AG

The second generation of Gene finding • Ex: Grail II. Used statistical techniques to combine various signals into a coherent gene structure. • It was not easy to train on many parameters. Guigo & Bursett test revealed that accuracy was still very low. • Problem with multiple genes in a genomic region


HMMs and gene finding • HMMs allow for a systematic approach to merging many signals. • They can model multiple genes, partial genes in a genomic region, as also genes on both strands.

The Viterbi Algorithm

HMMs and gene finding • The Viterbi algorithm (and backtracking) allows us to parse a string through the states of an HMM • Can we describe Eukaryotic gene structure by the states of an HMM? • This could be a solution to the GF problem.

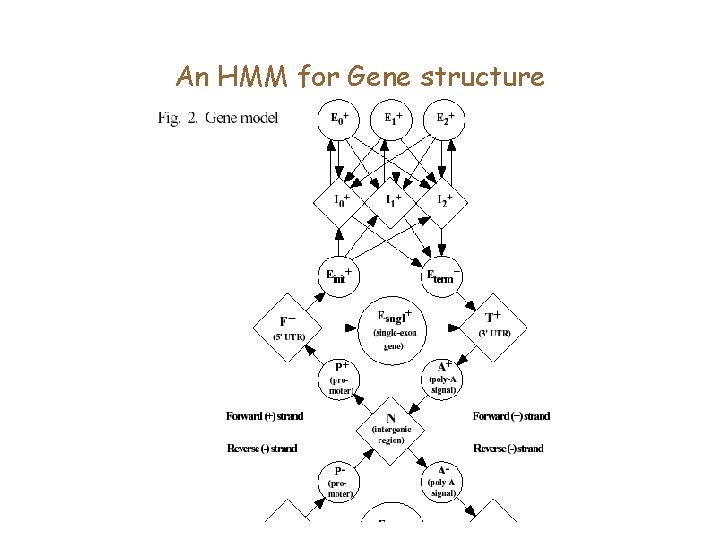
An HMM for Gene structure

Generalized HMMs, and other refinements • A probabilistic model for each of the states (ex: Exon, Splice site) needs to be described • In standard HMMs, there is an exponential distribution on the duration of time spent in a state. • This is violated by many states of the gene structure HMM. Solution is to model these using generalized HMMs.

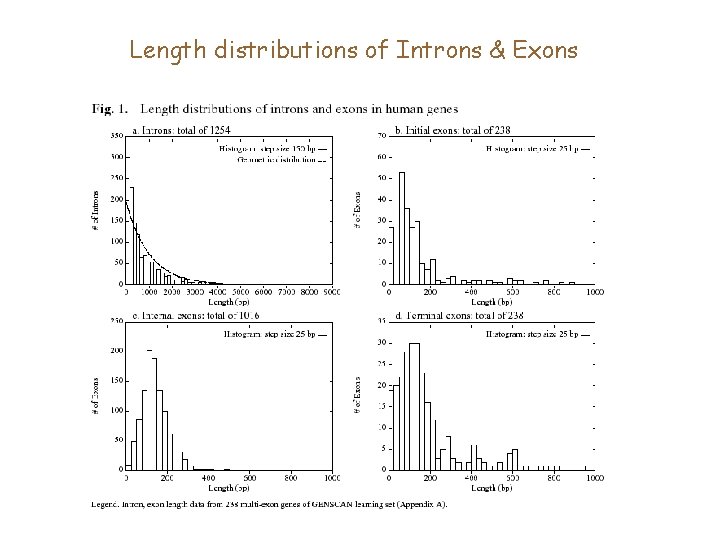
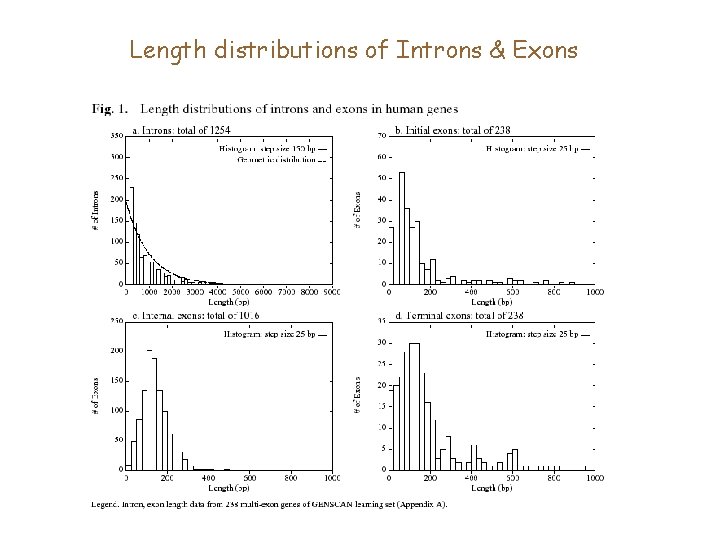
Length distributions of Introns & Exons

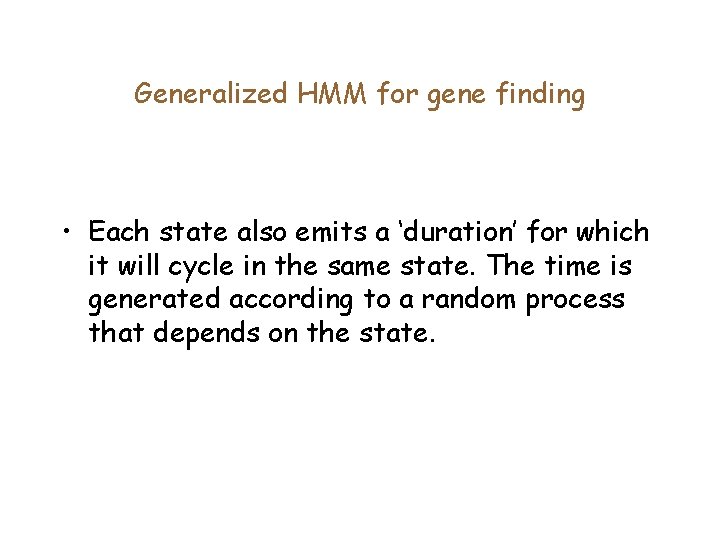
Generalized HMM for gene finding • Each state also emits a ‘duration’ for which it will cycle in the same state. The time is generated according to a random process that depends on the state.

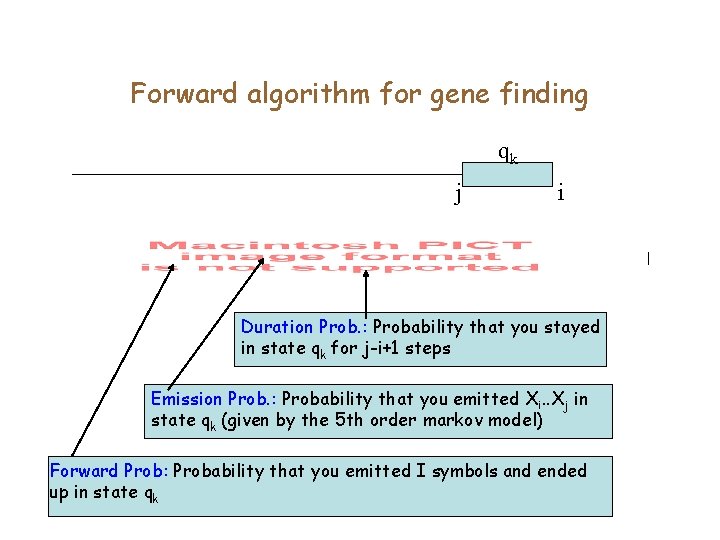
Forward algorithm for gene finding qk j i Duration Prob. : Probability that you stayed in state qk for j-i+1 steps Emission Prob. : Probability that you emitted Xi. . Xj in state qk (given by the 5 th order markov model) Forward Prob: Probability that you emitted I symbols and ended up in state qk

HMMs and Gene finding • Generalized HMMs are an attractive model for computational gene finding – Allow incorporation of various signals – Quality of gene finding depends upon quality of signals.

DNA Signals • Coding versus non-coding • Splice Signals • Translation start

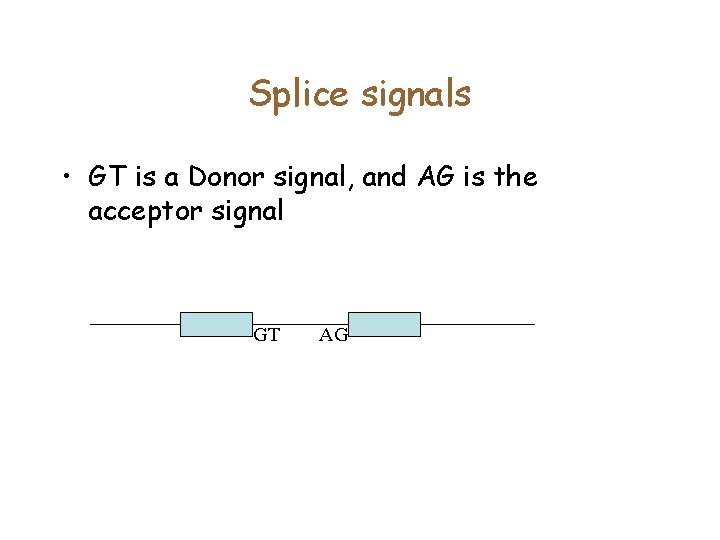
Splice signals • GT is a Donor signal, and AG is the acceptor signal GT AG

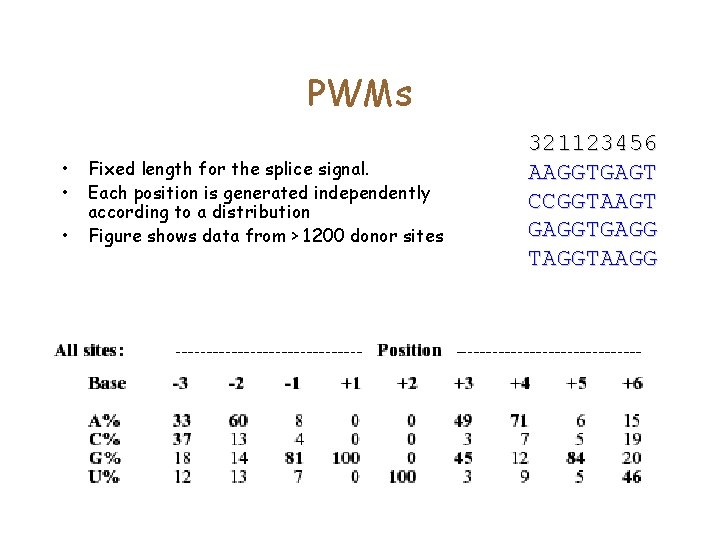
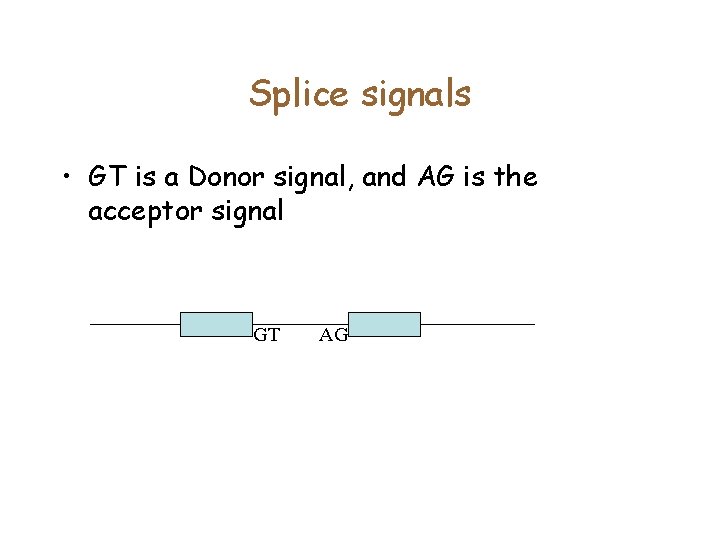
PWMs • • • Fixed length for the splice signal. Each position is generated independently according to a distribution Figure shows data from > 1200 donor sites 321123456 AAGGTGAGT CCGGTAAGT GAGGTGAGG TAGGTAAGG

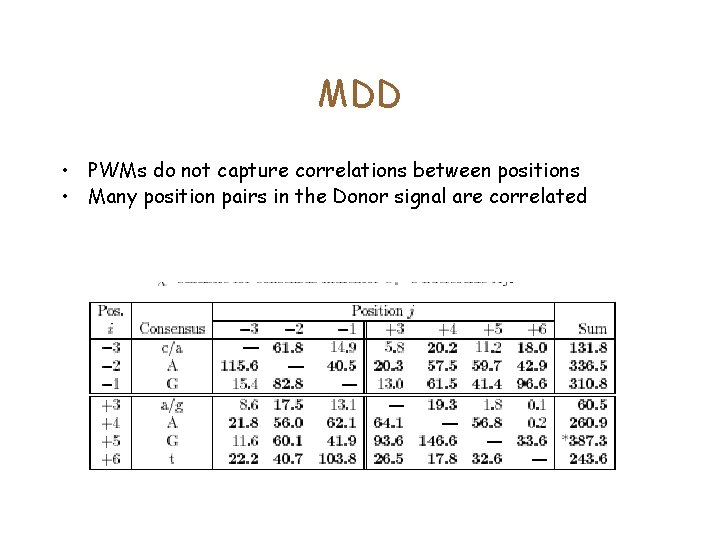
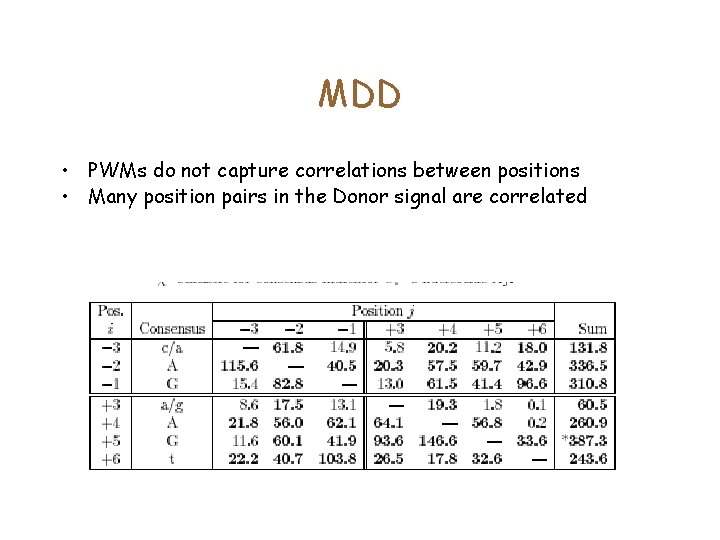
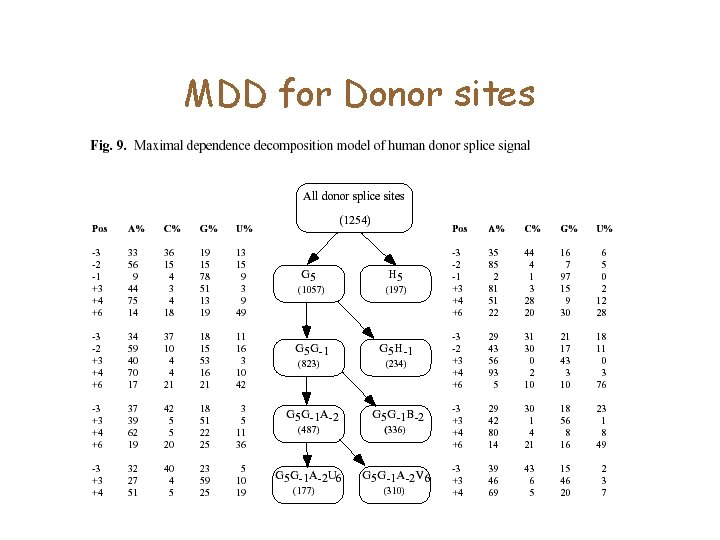
MDD • PWMs do not capture correlations between positions • Many position pairs in the Donor signal are correlated

• Choose the position which has the highest correlation score. • Split sequences into two: those which have the consensus at position I, and the remaining. • Recurse until <Terminating conditions>

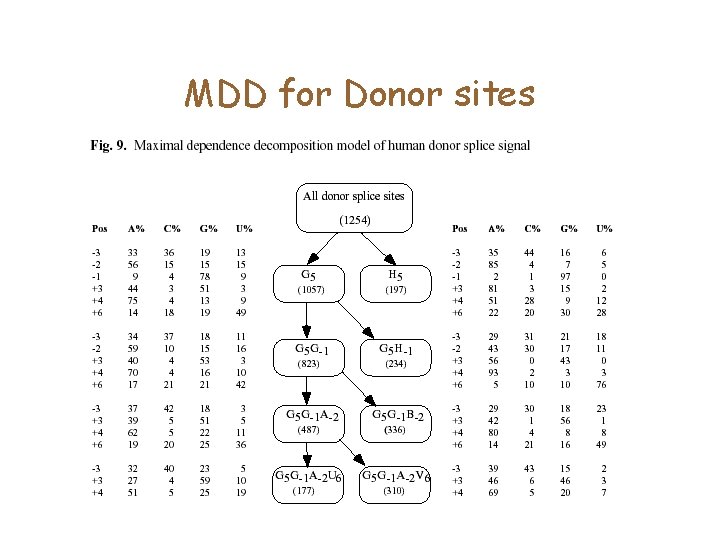
MDD for Donor sites

De novo Gene prediction: Sumary • Various signals distinguish coding regions from non-coding • HMMs are a reasonable model for Gene structures, and provide a uniform method for combining various signals. • Further improvement may come from improved signal detection

How many genes do we have? Nature Science

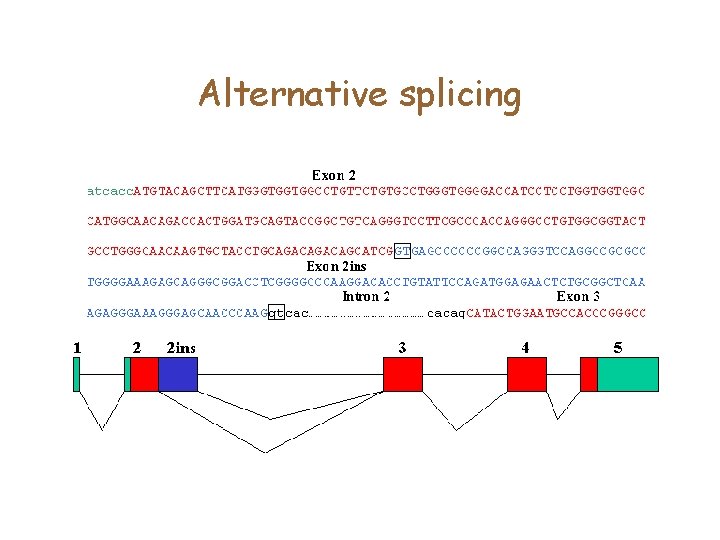
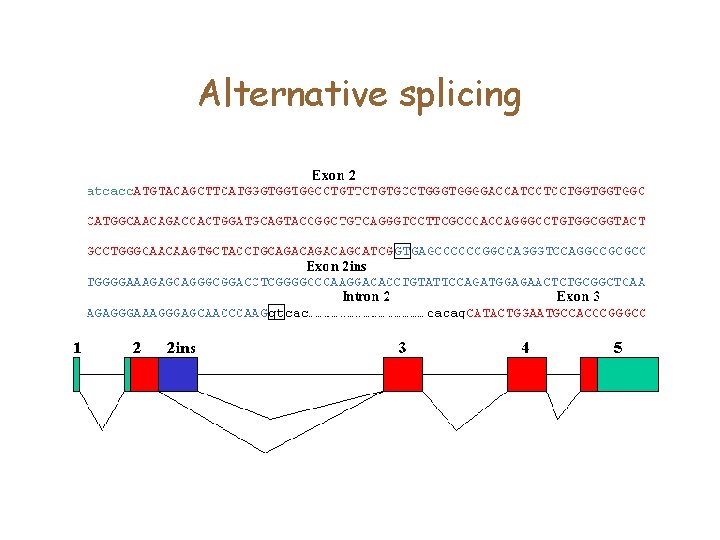
Alternative splicing

Comparative methods • Gene prediction is harder with alternative splicing. • One approach might be to use comparative methods to detect genes • Given a similar m. RNA/protein (from another species, perhaps? ), can you find the best parse of a genomic sequence that matches that target sequence • Yes, with a variant on alignment algorithms that penalize separately for introns, versus other gaps.

Gene Features ATG 5’ UTR Translation start Transcription start 3’ UTR exon intron Donor splice site Acceptor

Gene Finding • Given genomic DNA

Coding versus Non-coding • You are given a collection of exons, and a collection of intergenic sequence. • Count the number of occurrences of ATGATG in Introns and Exons. – Suppose 1% of the hexamers in Exons are ATGATG – Only 0. 01% of the hexamers in Intons are ATGATG • How can you use this idea to find genes?

Generalizing AAAAAAC AAAAAG AAAAAT I E X 10 5 10 20 5 10 Compute a frequency count for all hexamers. Use this to decide whether a sequence X is an exon/intron.
![A geometric approach Plot the following vectors E 10 20 I A geometric approach • Plot the following vectors – – E= [10, 20] I](https://slidetodoc.com/presentation_image_h/19d9105a4627c78c5f37b869cad8472f/image-65.jpg)
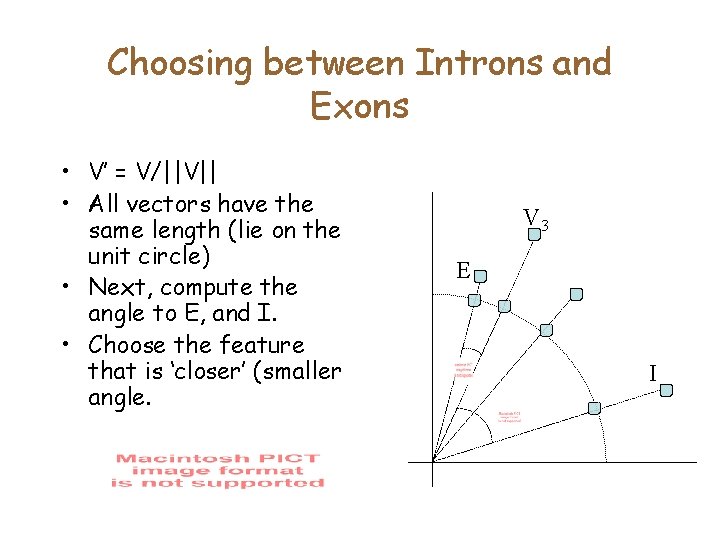
A geometric approach • Plot the following vectors – – E= [10, 20] I = [10, 5] V 3 = [5, 10] V 4 = [9, 15] • Is V 3 more like E or more like I? 20 15 10 5 5 10 15

Choosing between Introns and Exons • V’ = V/||V|| • All vectors have the same length (lie on the unit circle) • Next, compute the angle to E, and I. • Choose the feature that is ‘closer’ (smaller angle. V 3 E I

Coding versus non-coding • Fickett and Tung (1992) compared various measures • Measures that preserve the triplet frame are the most successful. • Genscan: 5 th order Markov Model • Conservation across species

Coding region can be detected • • • Plot the E-score using a sliding window of fixed length. The (large) exons will show up reliably. Not enough to predict gene boundaries reliably E-score

Other Signals • Signals at exon boundaries are precise but not specific. Coding signals are specific but not precise. • When combined they can be effective ATG Coding GT AG
![Combining Signals We can compute the following Escorei j i Combining Signals • We can compute the following: – – – E-score[i, j] i](https://slidetodoc.com/presentation_image_h/19d9105a4627c78c5f37b869cad8472f/image-70.jpg)
Combining Signals • We can compute the following: – – – E-score[i, j] i j I-score[i, j] D-score[i] I-score[i] Goal is to find coordinates that maximize the total score

The second generation of Gene finding • Ex: Grail II. Used statistical techniques to combine various signals into a coherent gene structure. • It was not easy to train on many parameters. Guigo & Bursett test revealed that accuracy was still very low. • Problem with multiple genes in a genomic region

Combining signals using D. P. • An HMM is the best way to model and optimize the combination of signals • Here, we will use a simpler approach which is essentially the same as the Viterbi algorithm for HMMs, but without the formalism.

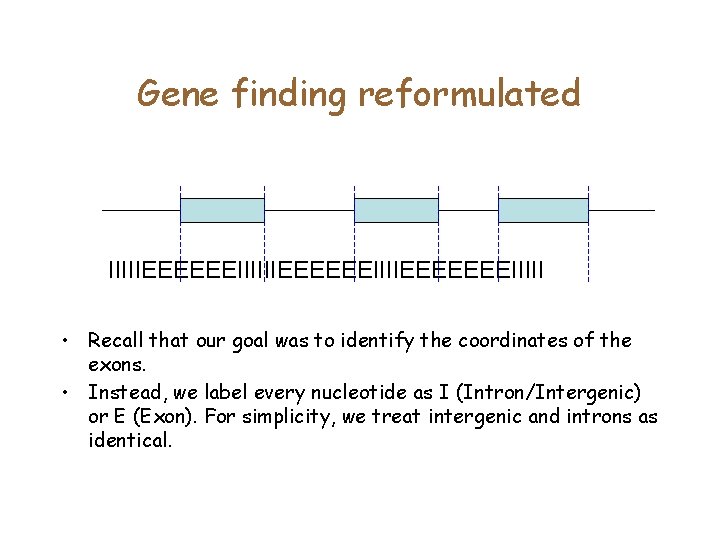
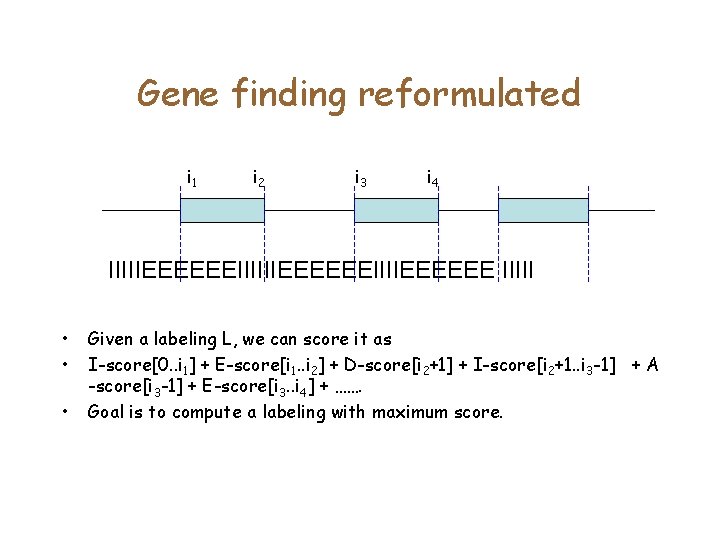
Gene finding reformulated IIIIIEEEEEEIIIIEEEEEEEIIIII • Recall that our goal was to identify the coordinates of the exons. • Instead, we label every nucleotide as I (Intron/Intergenic) or E (Exon). For simplicity, we treat intergenic and introns as identical.

Gene finding reformulated i 1 i 2 i 3 i 4 IIIIIEEEEEEIIIIEEEEEE IIIII • • • Given a labeling L, we can score it as I-score[0. . i 1] + E-score[i 1. . i 2] + D-score[i 2+1] + I-score[i 2+1. . i 3 -1] + A -score[i 3 -1] + E-score[i 3. . i 4] + ……. Goal is to compute a labeling with maximum score.

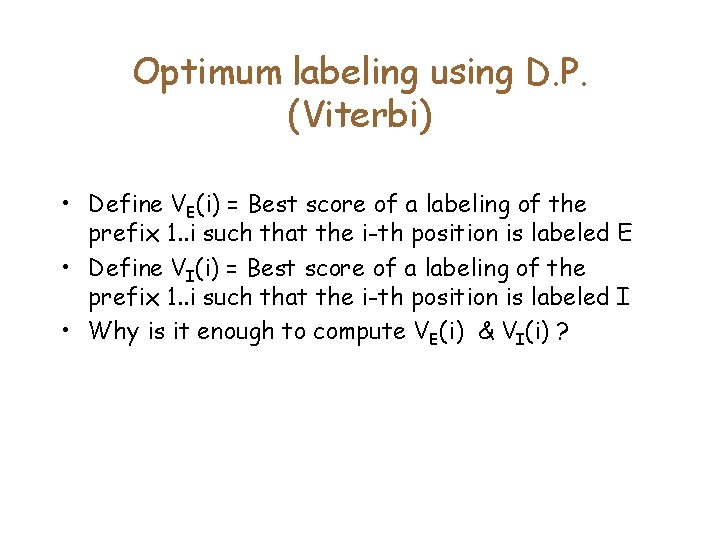
Optimum labeling using D. P. (Viterbi) • Define VE(i) = Best score of a labeling of the prefix 1. . i such that the i-th position is labeled E • Define VI(i) = Best score of a labeling of the prefix 1. . i such that the i-th position is labeled I • Why is it enough to compute VE(i) & VI(i) ?

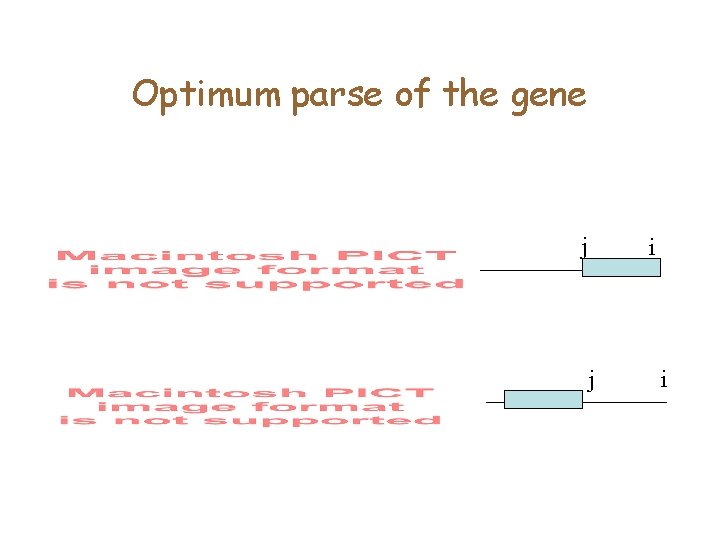
Optimum parse of the gene j i

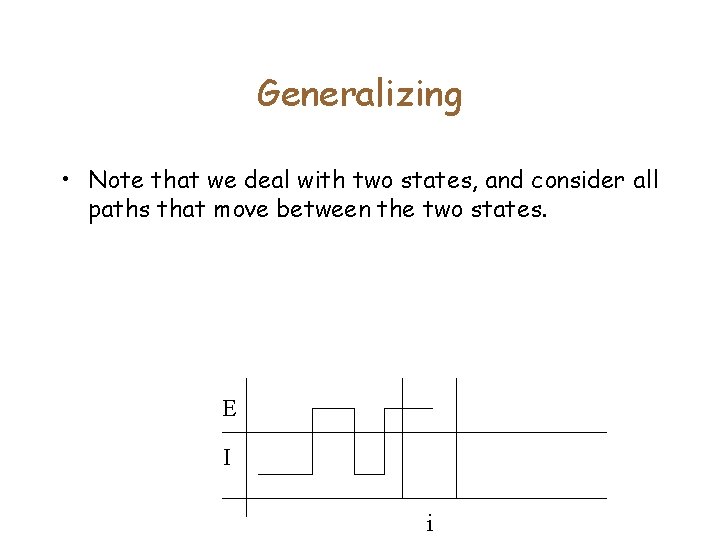
Generalizing • Note that we deal with two states, and consider all paths that move between the two states. E I i

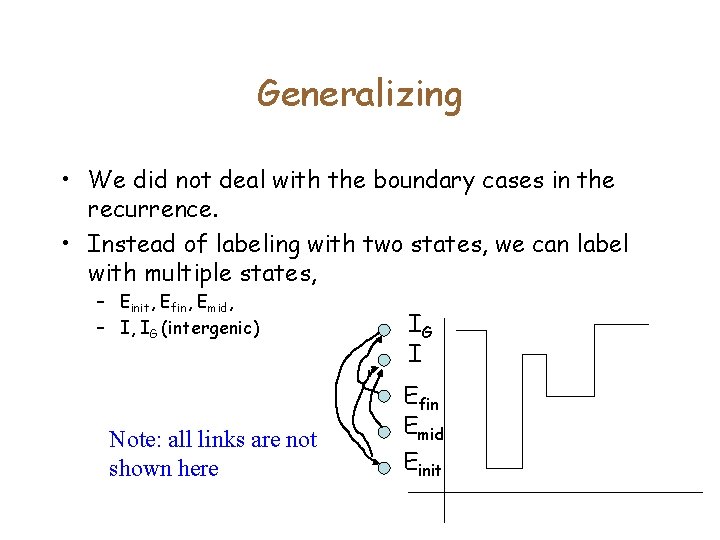
Generalizing • We did not deal with the boundary cases in the recurrence. • Instead of labeling with two states, we can label with multiple states, – Einit, Efin, Emid, – I, IG (intergenic) Note: all links are not shown here IG I Efin Emid Einit

HMMs and gene finding • HMMs allow for a systematic approach to merging many signals. • They can model multiple genes, partial genes in a genomic region, as also genes on both strands. • They allow an automated approach to weighting features.

An HMM for Gene structure

Generalized HMMs, and other refinements • A probabilistic model for each of the states (ex: Exon, Splice site) needs to be described • In standard HMMs, there is an exponential distribution on the duration of time spent in a state. • This is violated by many states of the gene structure HMM. Solution is to model these using generalized HMMs.

Length distributions of Introns & Exons

Generalized HMM for gene finding • Each state also emits a ‘duration’ for which it will cycle in the same state. The time is generated according to a random process that depends on the state.

Forward algorithm for gene finding qk j i Duration Prob. : Probability that you stayed in state qk for j-i+1 steps Emission Prob. : Probability that you emitted Xi. . Xj in state qk (given by the 5 th order markov model) Forward Prob: Probability that you emitted I symbols and ended up in state qk

HMMs and Gene finding • Generalized HMMs are an attractive model for computational gene finding – Allow incorporation of various signals – Quality of gene finding depends upon quality of signals.

DNA Signals • Coding versus non-coding • Splice Signals • Translation start

Splice signals • GT is a Donor signal, and AG is the acceptor signal GT AG

PWMs • • • Fixed length for the splice signal. Each position is generated independently according to a distribution Figure shows data from > 1200 donor sites 321123456 AAGGTGAGT CCGGTAAGT GAGGTGAGG TAGGTAAGG

MDD • PWMs do not capture correlations between positions • Many position pairs in the Donor signal are correlated

• Choose the position which has the highest correlation score. • Split sequences into two: those which have the consensus at position I, and the remaining. • Recurse until <Terminating conditions>

MDD for Donor sites

Gene prediction: Summary • Various signals distinguish coding regions from non-coding • HMMs are a reasonable model for Gene structures, and provide a uniform method for combining various signals. • Further improvement may come from improved signal detection

How many genes do we have? Nature Science

Alternative splicing

Comparative methods • Gene prediction is harder with alternative splicing. • One approach might be to use comparative methods to detect genes • Given a similar m. RNA/protein (from another species, perhaps? ), can you find the best parse of a genomic sequence that matches that target sequence • Yes, with a variant on alignment algorithms that penalize separately for introns, versus other gaps.

Comparative gene finding tools • • Procrustes/Sim 4: m. RNA vs. genomic Genewise: proteins versus genomic CEM: genomic versus genomic Twinscan: Combines comparative and de novo approach.

Databases • Ref. Seq and other databases maintain sequences of full -length transcripts. • We can query using sequence.