Protein Synthesis How cells follow DNA directions on










- Slides: 36

Protein Synthesis How cells follow DNA directions on how to make proteins

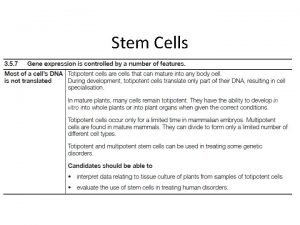
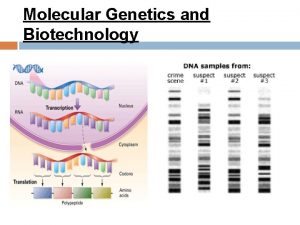
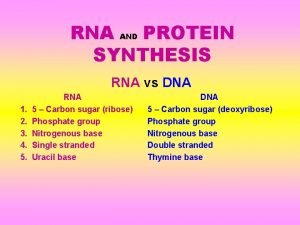
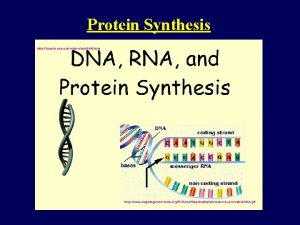
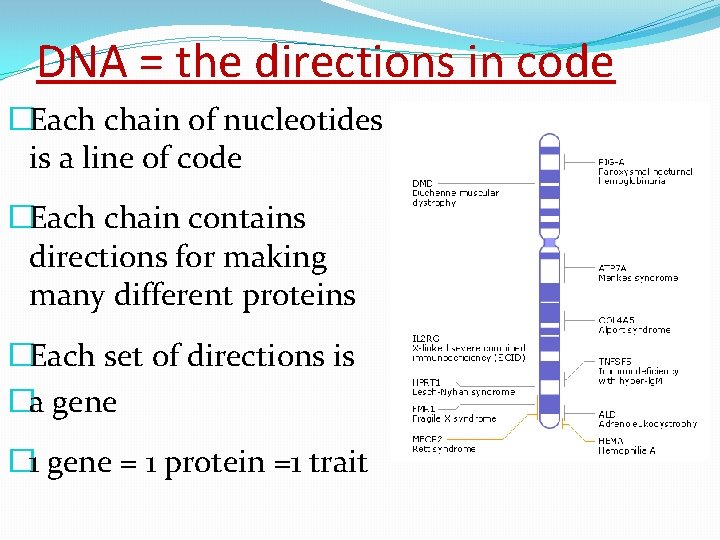
DNA = the directions in code �Each chain of nucleotides is a line of code �Each chain contains directions for making many different proteins �Each set of directions is �a gene � 1 gene = 1 protein =1 trait

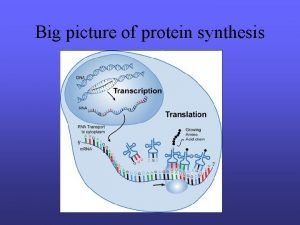
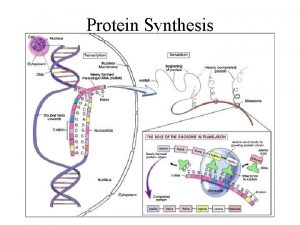
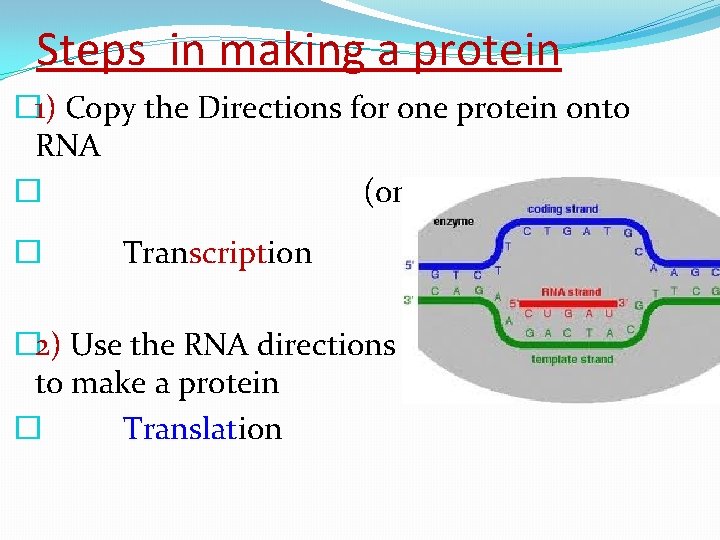
Steps in making a protein � 1) Copy the Directions for one protein onto RNA � (one gene) � Transcription � 2) Use the RNA directions to make a protein � Translation

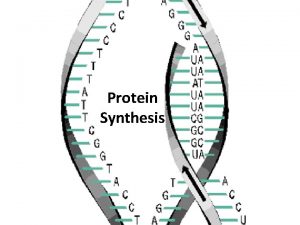
1) Transcription �A) RNA polymerase scans the DNA to find the gene it needs to copy �B) RNA polymerase has 4 jobs � 1) unwind the DNA double helix � 2) breaks hydrogen bonds to separate chains � 3) match RNA nucleotides to the DNA gene � 4) fuse the RNA nucleotides together to make a chain of RNA �http: //www. youtube. com/watch? v=zt. Pkv 7 wc 3 y. U& feature=related �http: //www. youtube. com/watch? v=5 Mf. SYn. It. Yvg& feature=related

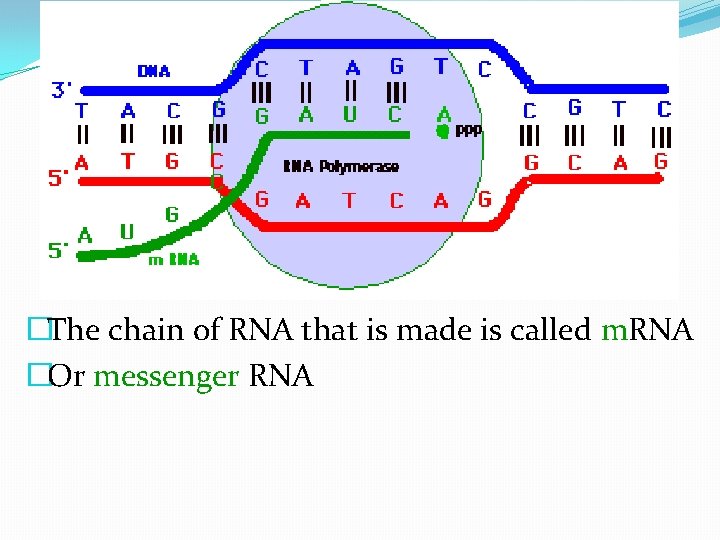
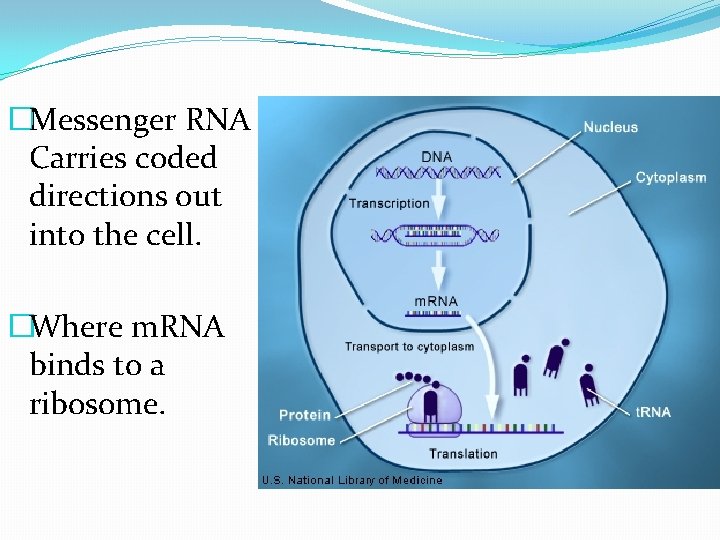
�The chain of RNA that is made is called m. RNA �Or messenger RNA

�Messenger RNA Carries coded directions out into the cell. �Where m. RNA binds to a ribosome.

2) Translation �A) Translates nucleotide code of RNA into � the amino acid code of a protein �B) Takes place in the cytoplasm �C) Is done by a ribosome �http: //www. biostudio. com/demo_freeman_ protein_synthesis. htm

m. RNA = chain of nuclotides that make up the coded directions for making the protein � Every 3 nuclotides of m. RNA is a codon � Each codon is the code for 1 amino acid


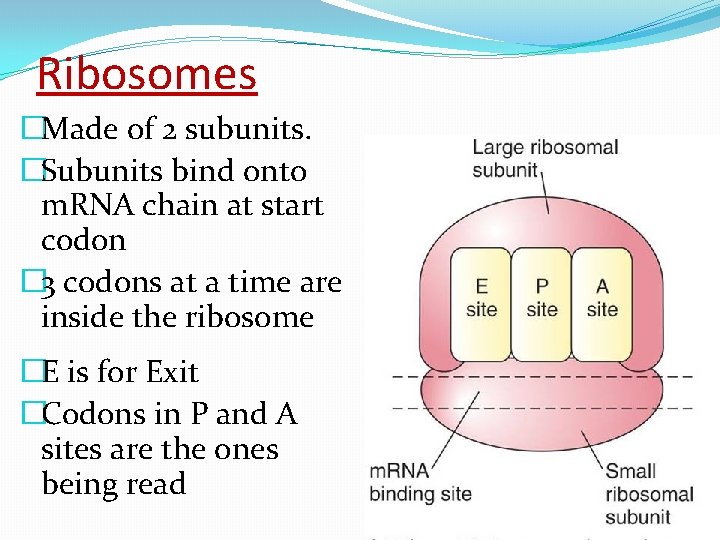
Ribosomes �Made of 2 subunits. �Subunits bind onto m. RNA chain at start codon � 3 codons at a time are inside the ribosome �E is for Exit �Codons in P and A sites are the ones being read

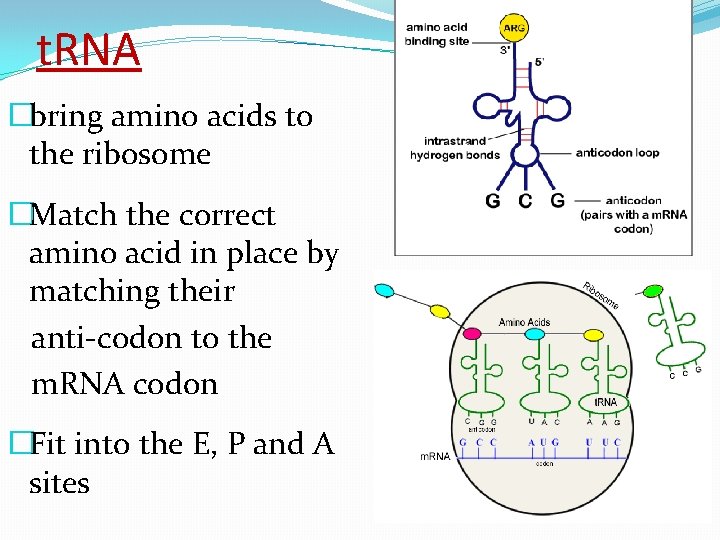
t. RNA �bring amino acids to the ribosome �Match the correct amino acid in place by matching their anti-codon to the m. RNA codon �Fit into the E, P and A sites

t. RNA �Each t. RNA only picks up ONE kind of amino acid �t. RNA drops off amino acids at the ribosome then moves back into the cytoplasm to pick up another amino acid(but always the same kind) � aminoacyl-t. RNA synthetase enzymes bind aa to

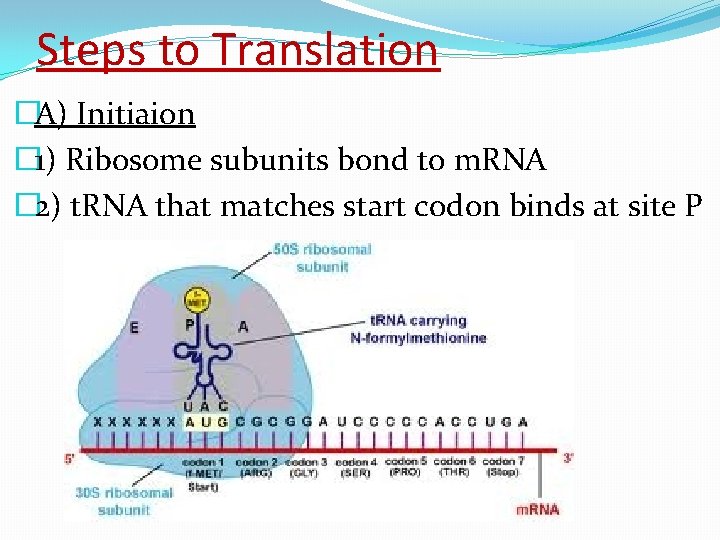
Steps to Translation �A) Initiaion � 1) Ribosome subunits bond to m. RNA � 2) t. RNA that matches start codon binds at site P

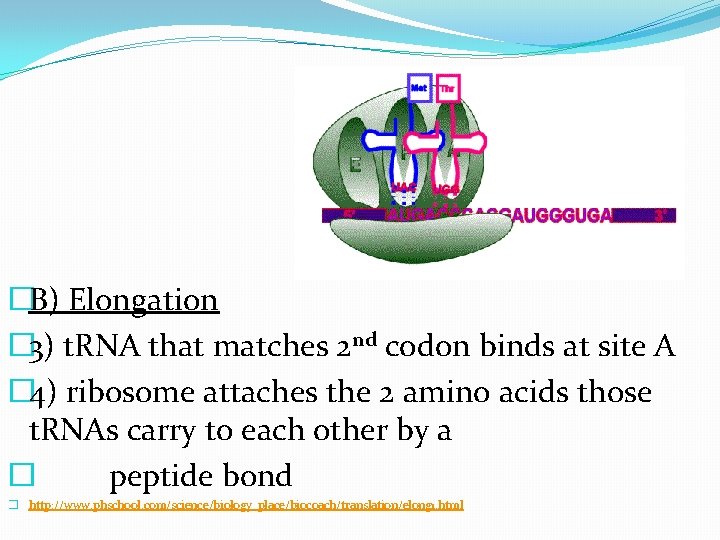
�B) Elongation � 3) t. RNA that matches 2 nd codon binds at site A � 4) ribosome attaches the 2 amino acids those t. RNAs carry to each other by a � peptide bond � http: //www. phschool. com/science/biology_place/biocoach/translation/elong 1. html

wobble effect �The last base in the anti-codon has some flexibility in what it binds to (wobble room) �So anticodon AGU could bind to UCA or UCG

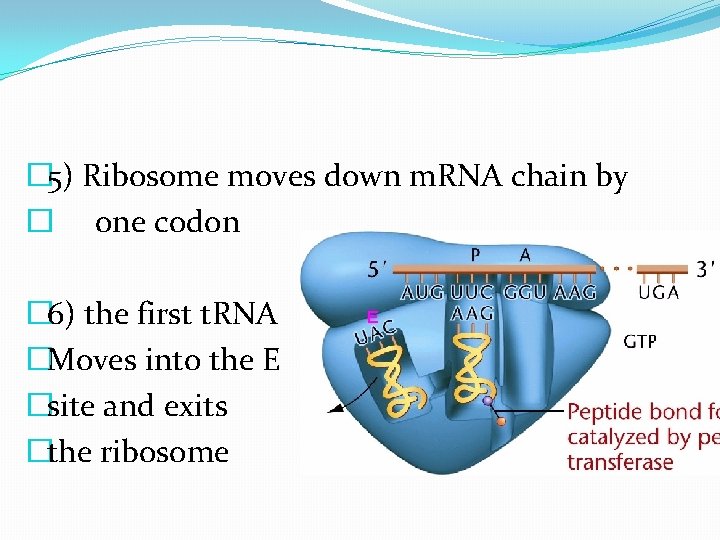
� 5) Ribosome moves down m. RNA chain by � one codon � 6) the first t. RNA �Moves into the E �site and exits �the ribosome

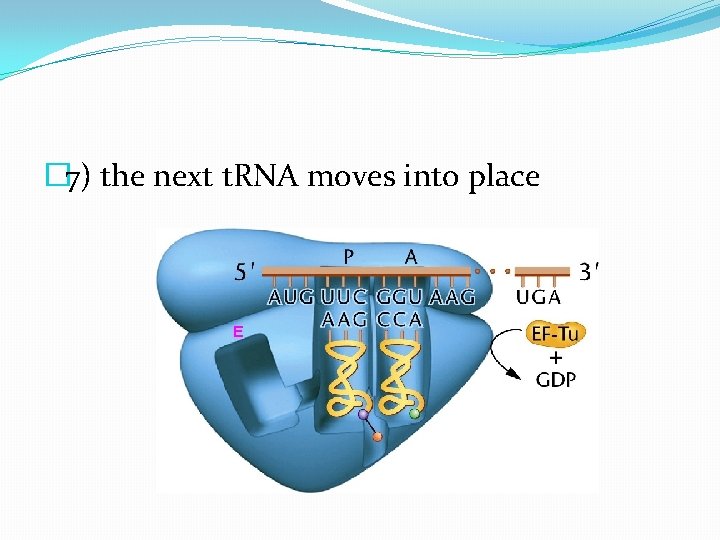
� 7) the next t. RNA moves into place

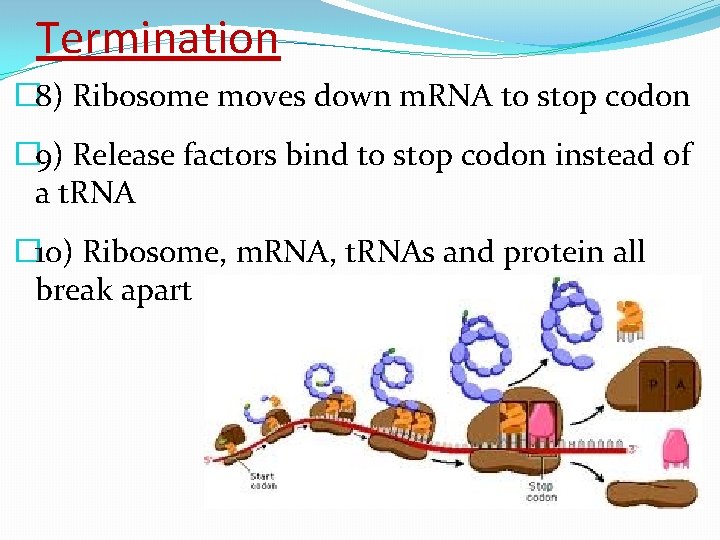
Termination � 8) Ribosome moves down m. RNA to stop codon � 9) Release factors bind to stop codon instead of a t. RNA � 10) Ribosome, m. RNA, t. RNAs and protein all break apart

Mutations= changes in a cell’s DNA �Caused by: � 1) spontaneous errors � 2) mutagens: chemicals/radiation �Can happen in body cells (somatic cells) or �In reproductive cells (egg & sperm)

Somatic cell mutations �May cause no change = if mutated gene is one that is not use by that particular cell �May cause cell to die �May cause cancer �Not passed on to children

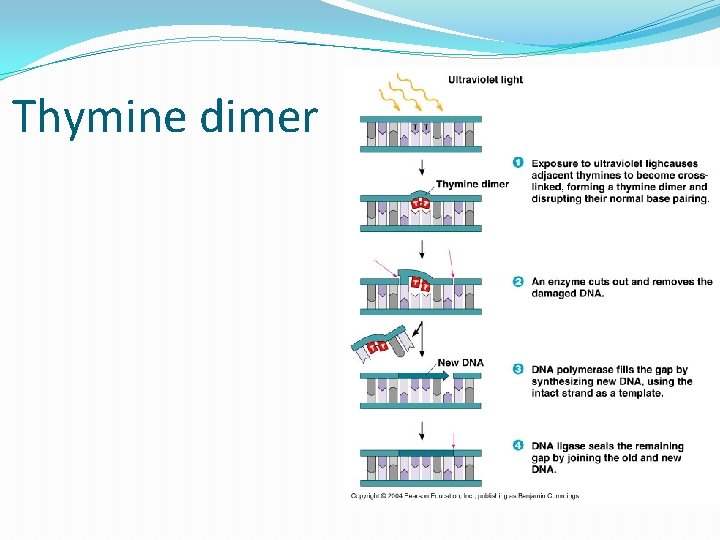
Thymine dimer

Reproductive Cell or Early Embryo Cell Mutations �Child with mutation �Every cell of the child is mutated �May cause miscarriage �May cause a genetic disorder in child �May have no effect at all

2 types of mutation �Gene mutation = DNA coding error � may be a missense mutation where the codons code for the wrong aa …. �or nonsense where stop codon or partial codon �Chromosome Mutation = change in chromosome number, missing or extra chromosome pieces.

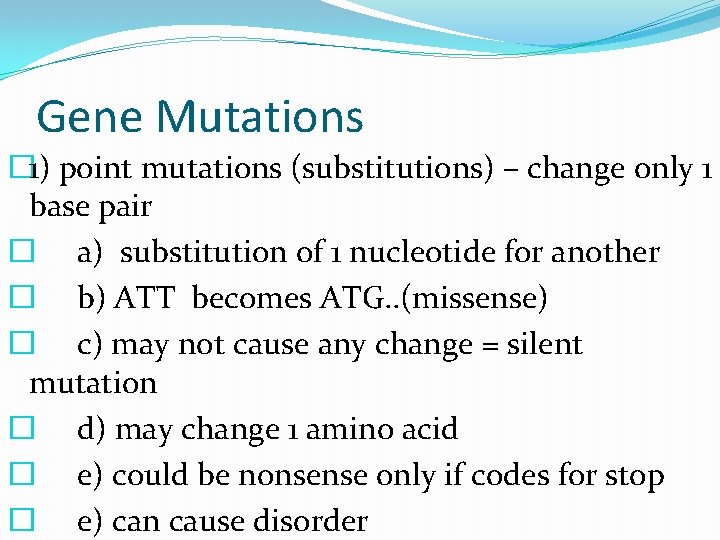
Gene Mutations � 1) point mutations (substitutions) – change only 1 base pair � a) substitution of 1 nucleotide for another � b) ATT becomes ATG. . (missense) � c) may not cause any change = silent mutation � d) may change 1 amino acid � e) could be nonsense only if codes for stop � e) can cause disorder

� 2) Frameshift mutations- change all the codons � a) insertion � b) deletion � c) THE FAT CAT = HEF ATC AT � d) most always ruins protein

� 3) tandem repeats – codons repeated over & over � most often ruins protein � more repeats = more problems �

Transcripts �Transcript = chain of RNA as copied from DNA template Pre-m. RNA = a transcript that will become m. RNA Other transcripts become t. RNA or r. RNA

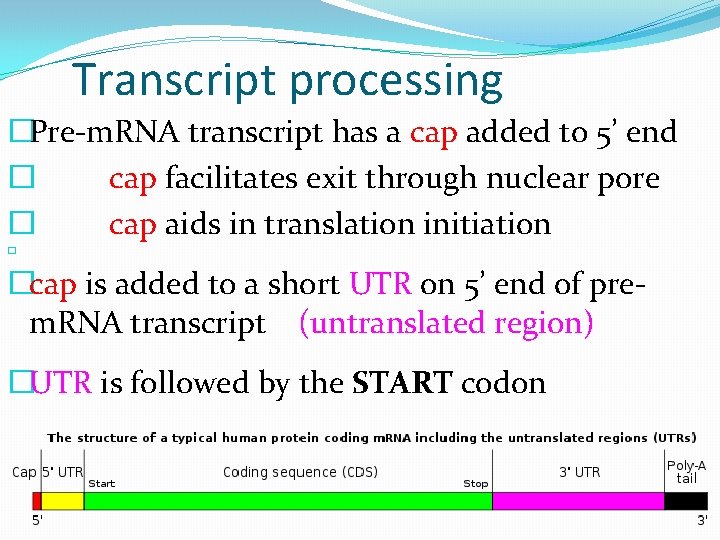
Transcript processing �Pre-m. RNA transcript has a cap added to 5’ end � cap facilitates exit through nuclear pore � cap aids in translation initiation � �cap is added to a short UTR on 5’ end of prem. RNA transcript (untranslated region) �UTR is followed by the START codon

Poly-A tail � 3’ end of transcript gets a poly-A tail added to it �Enzyme adds 50 -250 more A ribonucleotides �More As added make the m. RNA last longer �Fewer As make the m. RNA break down quicker �Breakdown starts immediately on entry to cytosol

RNA splicing �Introns = in between coding regions of RNA � do not code for a. a. s � are cut out before m. RNA leaves nucleus � bacteria do NOT have introns �Exons = have codons that are executed by translation…. code for a. a. s �Introns are cut out, exons fused together


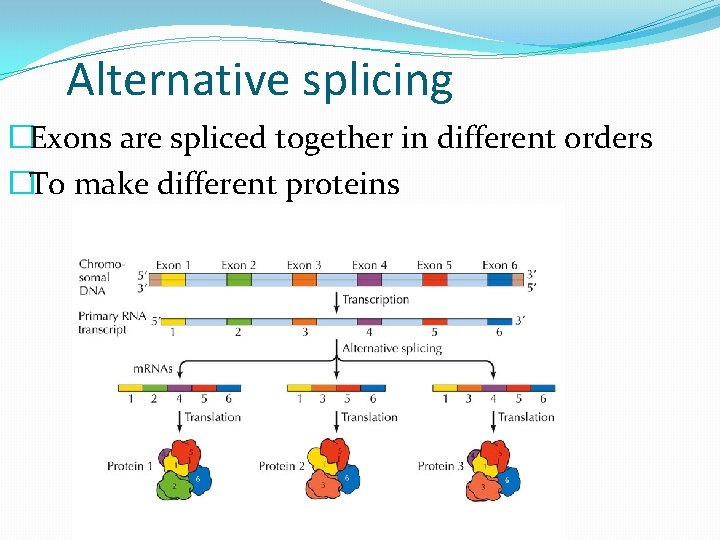
Alternative splicing �Exons are spliced together in different orders �To make different proteins

spliceosome �Complex of proteins and small RNAs �Remove introns �Joins exons in proper order �RNAs in spliceosome are Ribozymes… � RNA enzymes �Sometimes the intron being removed IS the ribozyme

Evidence for RNA before DNA �RNA can act as its own enzyme �RNA polymerase can initiate polymerization on its own �DNA polymerase can only start polymerization at a primer �That is created by RNA polymerase

Post-translational modification �Amino acid modification – add functional groups �Trim aa from cap end �Cleave polypeptide into pieces �Join 2 polypeptide into quaternary structure � with disulfide bonds

Protein location w/i the cell �All translation begins in cytoplasm �For proteins destined for endomembrane system � (ER, golgi, lysosome, plasma membrane) �OR proteins produced for secretion (insulin) �The first few aa at leading end of polypeptide = signal peptide �Signal peptide binds to a Signal Recognition Particle (SRP) �SRP escorts ribosome to receptor protein on ER
 Chapter 11 dna and genes
Chapter 11 dna and genes Rna and protein synthesis study guide
Rna and protein synthesis study guide I will follow you wherever you will go
I will follow you wherever you will go Totipotent cells
Totipotent cells Section 12 3 rna and protein synthesis
Section 12 3 rna and protein synthesis Protein synthesis
Protein synthesis Protein synthesis restaurant analogy
Protein synthesis restaurant analogy Transcription and translation
Transcription and translation Protein synthesis gcse
Protein synthesis gcse Protein synthesis and mutations
Protein synthesis and mutations Cookie monster analogy
Cookie monster analogy Whats translation in biology
Whats translation in biology Transfer rna
Transfer rna Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis Protein synthesis and mutations
Protein synthesis and mutations Protein synthesis story
Protein synthesis story Protein synthesis and mutations
Protein synthesis and mutations Protein synthesis animation mcgraw hill
Protein synthesis animation mcgraw hill Test cross definition
Test cross definition Transcription and translation
Transcription and translation Protein synthesis
Protein synthesis Ribosome
Ribosome Protein synthesis ppt
Protein synthesis ppt Picture transcription
Picture transcription Sulfonamides mechanism of action
Sulfonamides mechanism of action Which best summarizes the process of protein synthesis?
Which best summarizes the process of protein synthesis? Concept map of protein synthesis
Concept map of protein synthesis Protein synthesis
Protein synthesis Protein synthesis scramble
Protein synthesis scramble Fictional character μετάφραση
Fictional character μετάφραση Nucleic acids are made up of
Nucleic acids are made up of Teste de ames
Teste de ames Sintese de proteinas na celula
Sintese de proteinas na celula Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis Protein synthesis
Protein synthesis