CSE 182 L 11 Protein sequencing and Mass






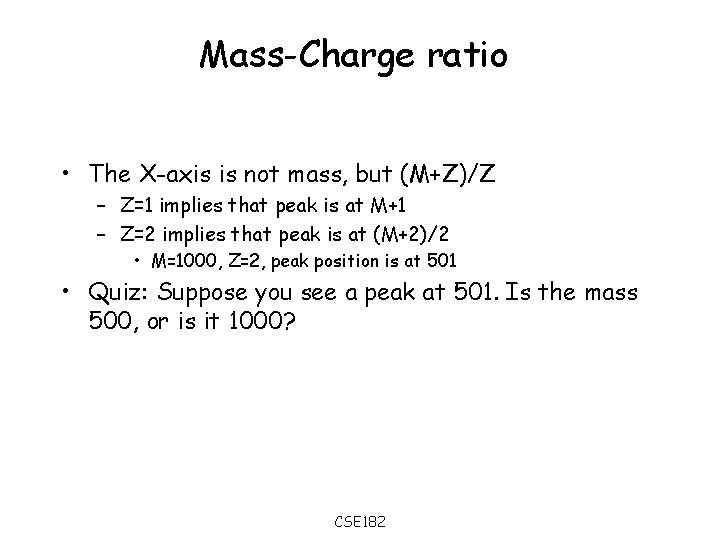





- Slides: 31

CSE 182 -L 11 Protein sequencing and Mass Spectrometry CSE 182

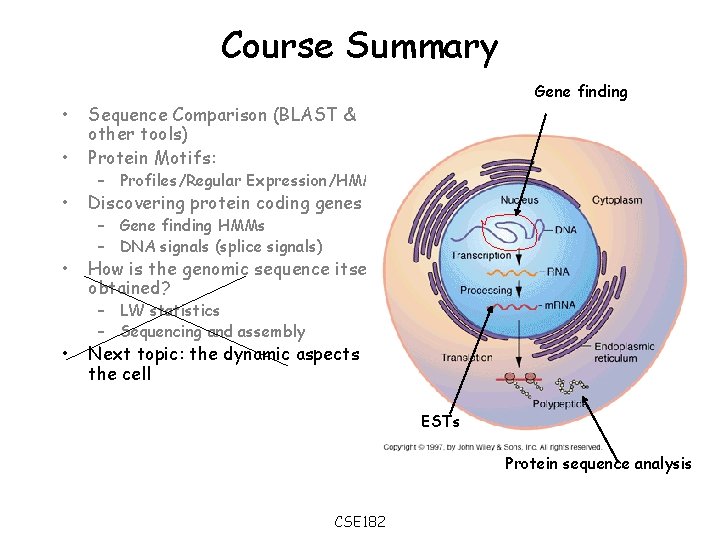
Course Summary • • Sequence Comparison (BLAST & other tools) Protein Motifs: • Discovering protein coding genes • How is the genomic sequence itself obtained? • Gene finding – Profiles/Regular Expression/HMMs – Gene finding HMMs – DNA signals (splice signals) – LW statistics – Sequencing and assembly Next topic: the dynamic aspects of the cell ESTs Protein sequence analysis CSE 182

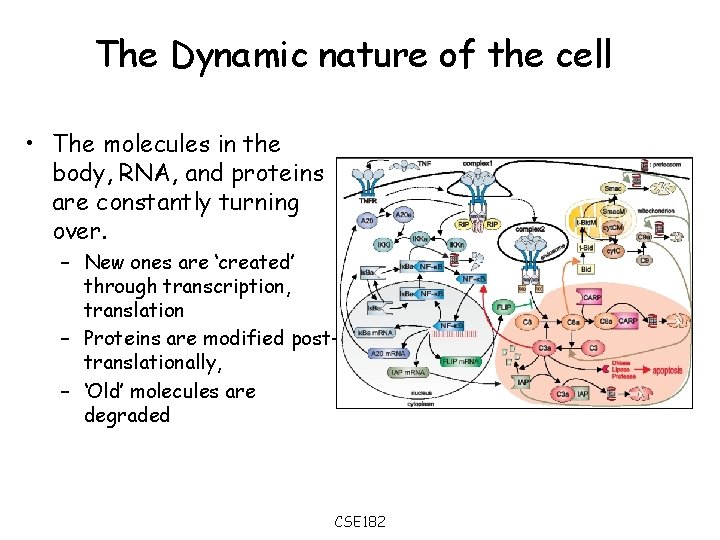
The Dynamic nature of the cell • The molecules in the body, RNA, and proteins are constantly turning over. – New ones are ‘created’ through transcription, translation – Proteins are modified posttranslationally, – ‘Old’ molecules are degraded CSE 182

Dynamic aspects of cellular function • Expressed transcripts – Microarrays to ‘count’ the number of copies of RNA • Expressed proteins – Mass spectrometry is used to ‘count’ the number of copies of a protein sequence. • Protein-protein interactions (protein networks) • Protein-DNA interactions • Population studies CSE 182

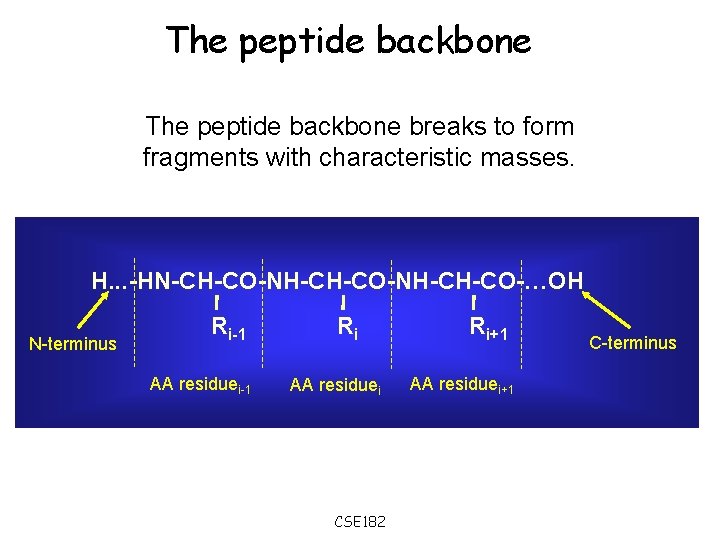
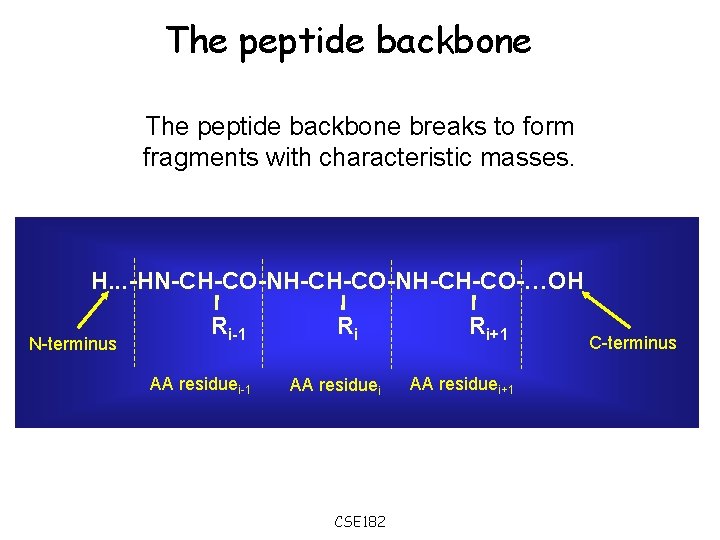
The peptide backbone breaks to form fragments with characteristic masses. H. . . -HN-CH-CO-NH-CH-CO-…OH N-terminus Ri-1 AA residuei-1 Ri AA residuei CSE 182 Ri+1 AA residuei+1 C-terminus

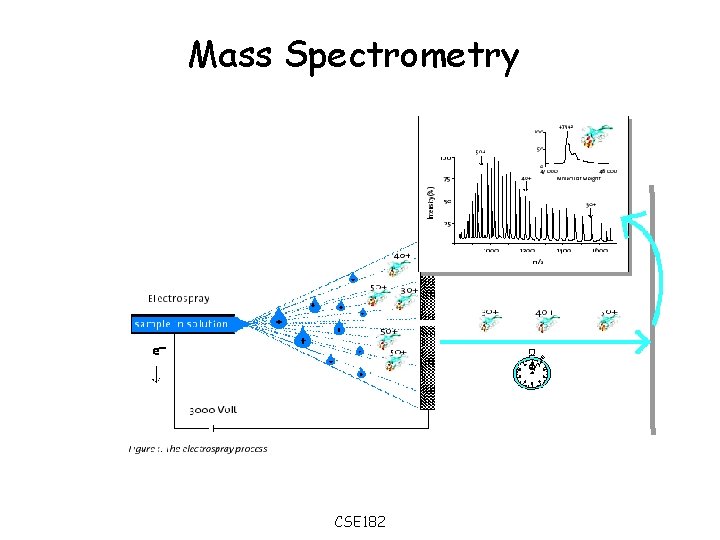
Mass Spectrometry CSE 182

Nobel citation ’ 02 CSE 182

The promise of mass spectrometry • Mass spectrometry is coming of age as the tool of choice for proteomics – Protein sequencing, networks, quantitation, interactions, structure…. • Computation has a big role to play in the interpretation of MS data. • We will discuss algorithms for – Sequencing, Modifications, Interactions. . CSE 182

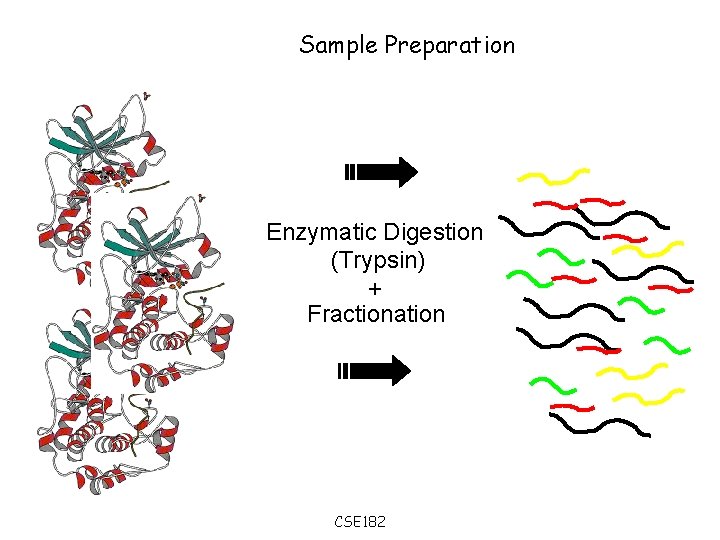
Sample Preparation Enzymatic Digestion (Trypsin) + Fractionation CSE 182

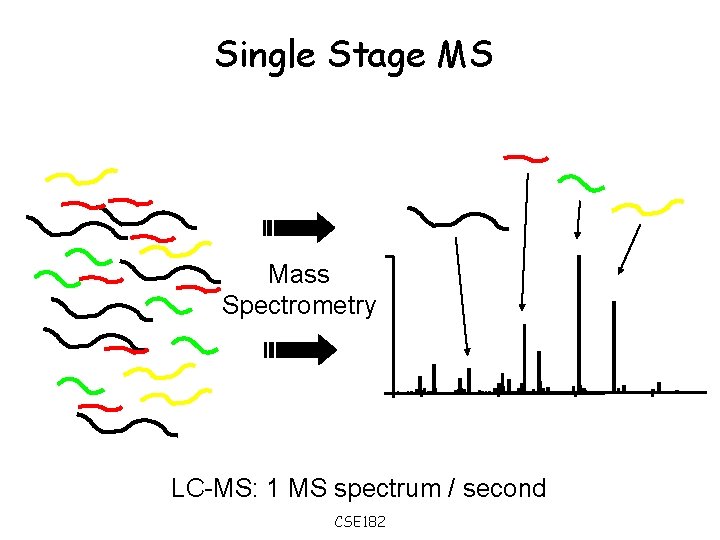
Single Stage MS Mass Spectrometry LC-MS: 1 MS spectrum / second CSE 182

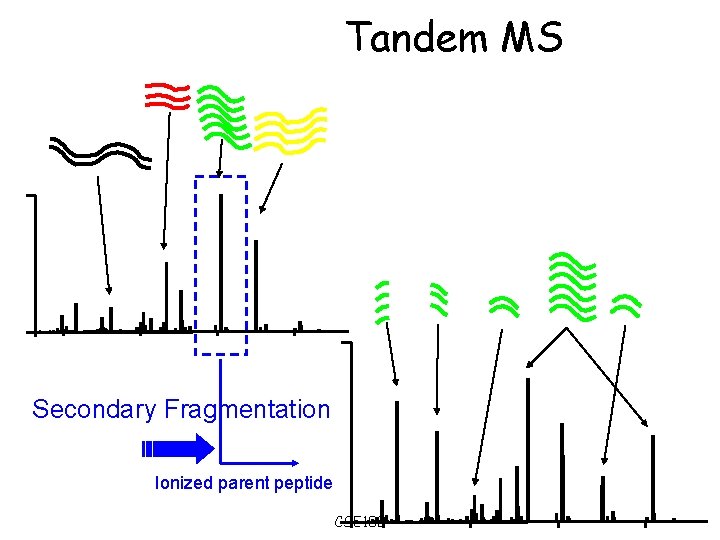
Tandem MS Secondary Fragmentation Ionized parent peptide CSE 182

The peptide backbone breaks to form fragments with characteristic masses. H. . . -HN-CH-CO-NH-CH-CO-…OH N-terminus Ri-1 AA residuei-1 Ri AA residuei CSE 182 Ri+1 AA residuei+1 C-terminus

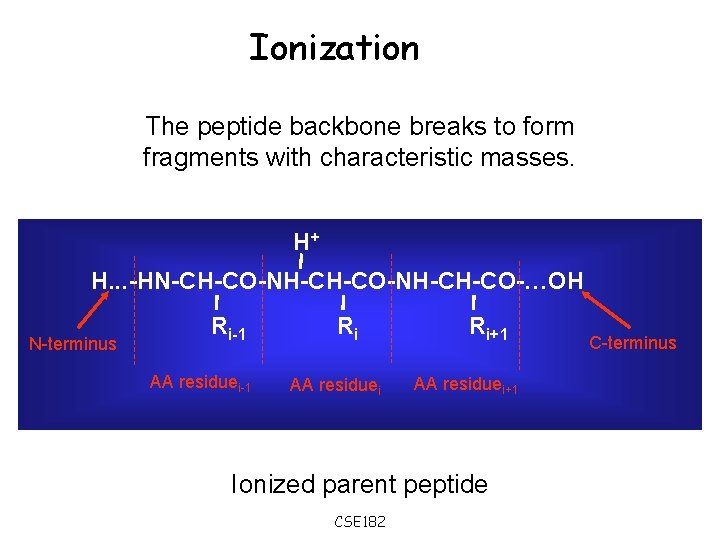
Ionization The peptide backbone breaks to form fragments with characteristic masses. H+ H. . . -HN-CH-CO-NH-CH-CO-…OH N-terminus Ri-1 AA residuei-1 Ri AA residuei Ri+1 AA residuei+1 Ionized parent peptide CSE 182 C-terminus

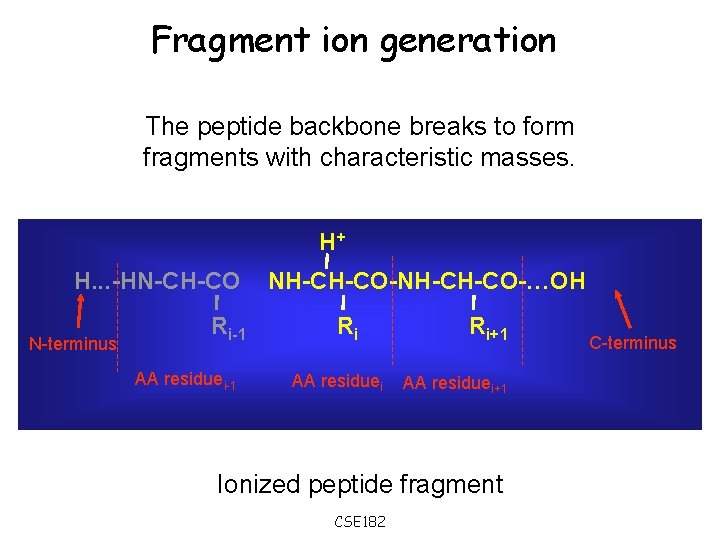
Fragment ion generation The peptide backbone breaks to form fragments with characteristic masses. H+ H. . . -HN-CH-CO N-terminus Ri-1 AA residuei-1 NH-CH-CO-…OH Ri AA residuei Ri+1 AA residuei+1 Ionized peptide fragment CSE 182 C-terminus

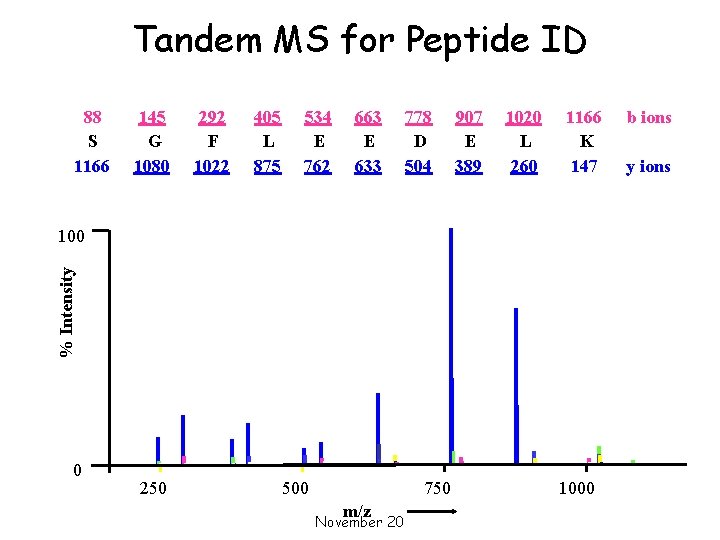
Tandem MS for Peptide ID 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 907 E 389 1020 L 260 1166 K 147 % Intensity 100 0 [M+2 H]2+ 250 500 750 m/z November 20 1000 b ions y ions

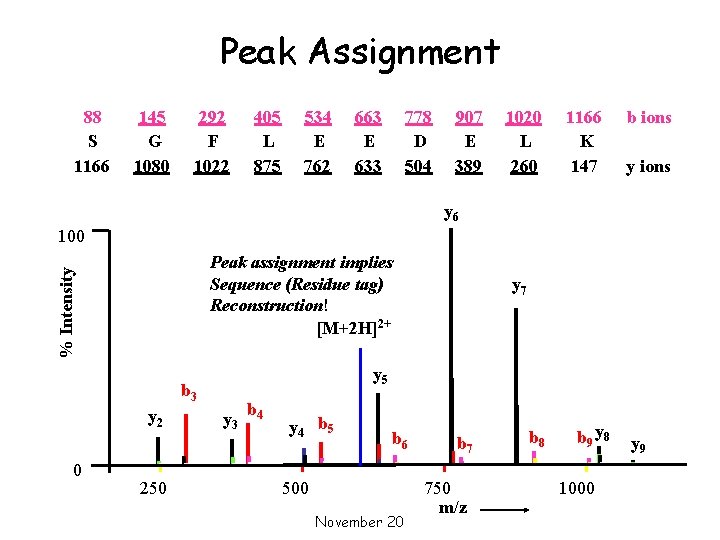
Peak Assignment 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 907 E 389 1020 L 260 1166 K 147 b ions y 6 100 % Intensity Peak assignment implies Sequence (Residue tag) Reconstruction! [M+2 H]2+ y 5 b 3 y 2 0 250 y 7 y 3 b 4 y 4 b 5 b 6 500 November 20 b 7 750 m/z b 8 b 9 y 8 1000 y 9

Database Searching for peptide ID • For every peptide from a database – Generate a hypothetical spectrum – Compute a correlation between observed and experimental spectra – Choose the best • Database searching is very powerful and is the de facto standard for MS. – Sequest, Mascot, and many others CSE 182

Spectra: the real story • Noise Peaks • Ions, not prefixes & suffixes • Mass to charge ratio, and not mass – Multiply charged ions • Isotope patterns, not single peaks CSE 182

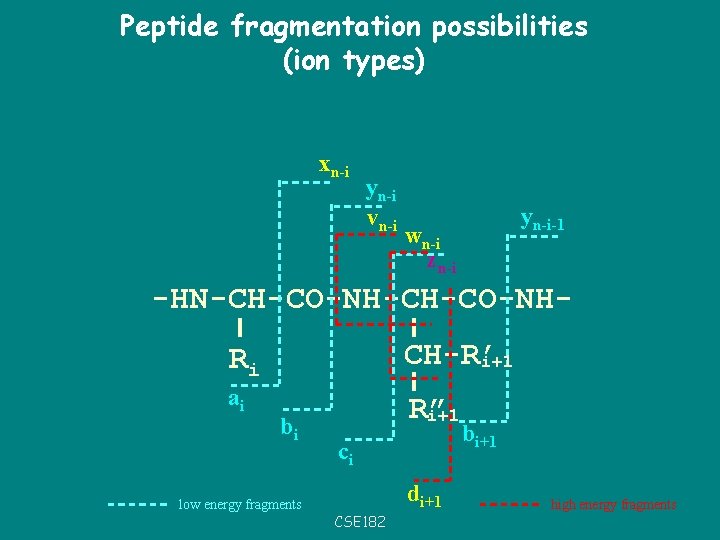
Peptide fragmentation possibilities (ion types) xn-i yn-i vn-i yn-i-1 wn-i zn-i -HN-CH-CO-NHRi CH-R’ i+1 ai R” i+1 bi low energy fragments ci di+1 CSE 182 bi+1 high energy fragments

Ion types, and offsets • • • P = prefix residue mass S = Suffix residue mass b-ions = P+1 y-ions = S+19 a-ions = P-27 CSE 182
Mass-Charge ratio • The X-axis is not mass, but (M+Z)/Z – Z=1 implies that peak is at M+1 – Z=2 implies that peak is at (M+2)/2 • M=1000, Z=2, peak position is at 501 • Quiz: Suppose you see a peak at 501. Is the mass 500, or is it 1000? CSE 182

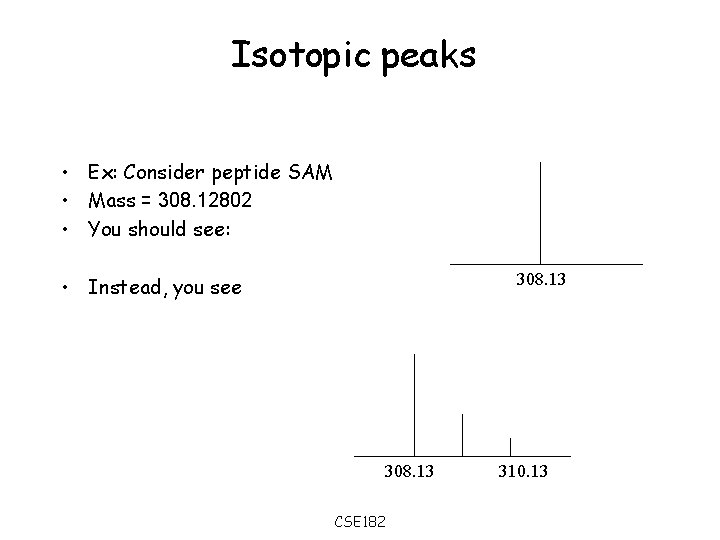
Isotopic peaks • Ex: Consider peptide SAM • Mass = 308. 12802 • You should see: 308. 13 • Instead, you see 308. 13 CSE 182 310. 13

Isotopes • C-12 is the most common. Suppose C-13 occurs with probability 1% • EX: SAM – Composition: C 11 H 22 N 3 O 5 S 1 • What is the probability that you will see a single C-13? • Note that C, S, O, N all have isotopes. Can you compute the isotopic distribution? CSE 182

All atoms have isotopes • Isotopes of atoms – O 16, 18, C-12, 13, S 32, 34…. – Each isotope has a frequency of occurrence • If a molecule (peptide) has a single copy of C-13, that will shift its peak by 1 Da • With multiple copies of a peptide, we have a distribution of intensities over a range of masses (Isotopic profile). • How can you compute the isotopic profile of a peak? CSE 182

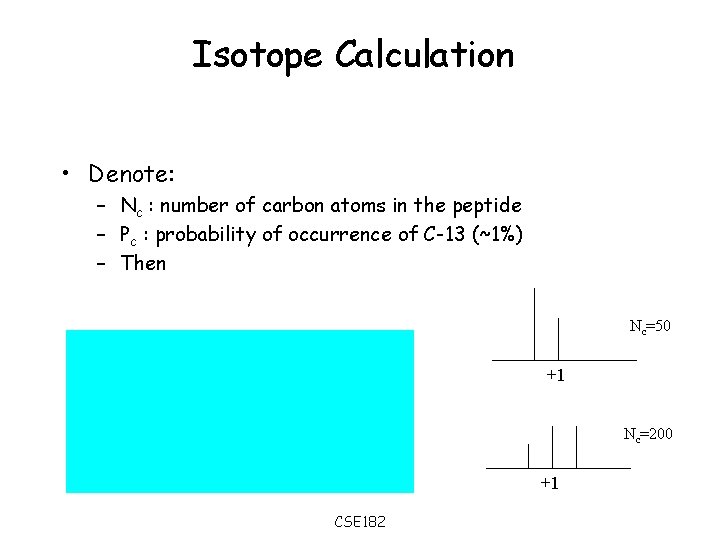
Isotope Calculation • Denote: – Nc : number of carbon atoms in the peptide – Pc : probability of occurrence of C-13 (~1%) – Then Nc=50 +1 Nc=200 +1 CSE 182

Isotope Calculation Example • • • Suppose we consider Nitrogen, and Carbon NN: number of Nitrogen atoms PN: probability of occurrence of N-15 Pr(peak at M) Pr(peak at M+1)? Pr(peak at M+2)? How do we generalize? How can we handle Oxygen (O-16, 18)? CSE 182

General isotope computation • Definition: – Let pi, a be the abundance of the isotope with mass i Da above the least mass – Ex: P 0, C : abundance of C-12, P 2, O: O-18 etc. • Characteristic polynomial • Prob{M+i}: coefficient of xi in (x) (a binomial convolution) CSE 182

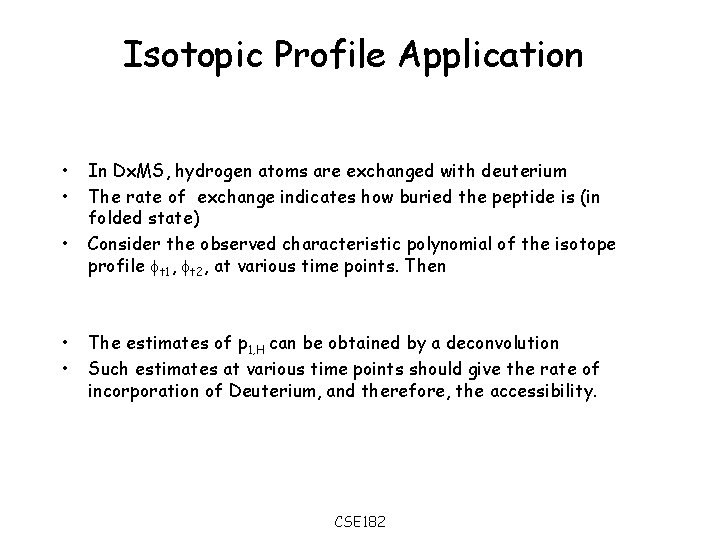
Isotopic Profile Application • • • In Dx. MS, hydrogen atoms are exchanged with deuterium The rate of exchange indicates how buried the peptide is (in folded state) Consider the observed characteristic polynomial of the isotope profile t 1, t 2, at various time points. Then The estimates of p 1, H can be obtained by a deconvolution Such estimates at various time points should give the rate of incorporation of Deuterium, and therefore, the accessibility. CSE 182

Quiz § How can you determine the charge on a peptide? § Difference between the first and second isotope peak is 1/Z § Proposal: §Given a mass, predict a composition, and the isotopic profile § Do a ‘goodness of fit’ test to isolate the peaks corresponding to the isotope § Compute the difference CSE 182

Tandem MS summary • The basics of peptide ID using tandem MS is simple. – Correlate experimental with theoretical spectra • In practice, there might be many confounding problems. – Isotope peaks, noise peaks, varying charges, post-translational modifications, no database. • Recall that we discussed how peptides could be identified by scanning a database. • What if the database did not contain the peptide of interest? CSE 182

De novo analysis basics • Suppose all ions were prefix ions? Could you tell what the peptide was? • Can post-translational modifications help? CSE 182
 Cse 182
Cse 182 Cse 182 ucsd
Cse 182 ucsd Cse 182
Cse 182 Channel vs carrier proteins
Channel vs carrier proteins Protein-protein docking
Protein-protein docking Tungsten-186
Tungsten-186 Mandatos con nosotros (p. 182)
Mandatos con nosotros (p. 182) Department order 182
Department order 182 Flam 182
Flam 182 Engine on half power ahead - steer 182 degrees port side.
Engine on half power ahead - steer 182 degrees port side. Pg 167
Pg 167 Cessna 182 diesel
Cessna 182 diesel Cs 182
Cs 182 Love 182
Love 182 Berkeley cs 182
Berkeley cs 182 Is atomic mass and relative atomic mass the same
Is atomic mass and relative atomic mass the same Atomic
Atomic Mass of oxygen
Mass of oxygen Atomic number mass
Atomic number mass Sequence selection and iteration
Sequence selection and iteration Binary microprogram
Binary microprogram Loading sequencing and scheduling
Loading sequencing and scheduling Difference between ngs and sanger sequencing
Difference between ngs and sanger sequencing Sequential conditional and iterative
Sequential conditional and iterative Example of selection in pseudocode
Example of selection in pseudocode Sequencing strategies and tactics
Sequencing strategies and tactics Cloning and sequencing explorer series
Cloning and sequencing explorer series Sequencing batch reactor advantages and disadvantages
Sequencing batch reactor advantages and disadvantages Stoichiometry example
Stoichiometry example Percent composition of propane
Percent composition of propane Inertial mass vs gravitational mass
Inertial mass vs gravitational mass Grams to mols
Grams to mols