CSE 182 L 8 Protein domain analysis via

![Regular expressions as Protein sequence motifs C-X-[DE]-X{10, 12}-C-X-C--[STYLV] Fam(B) A C • E F Regular expressions as Protein sequence motifs C-X-[DE]-X{10, 12}-C-X-C--[STYLV] Fam(B) A C • E F](https://slidetodoc.com/presentation_image_h2/1937e99e82495f4ba97bbf35da3f523f/image-2.jpg)



![HMM T[j, k] j k i • • Q is a set of states HMM T[j, k] j k i • • Q is a set of states](https://slidetodoc.com/presentation_image_h2/1937e99e82495f4ba97bbf35da3f523f/image-19.jpg)






- Slides: 41

CSE 182 -L 8 Protein domain analysis via HMMs Gene finding December 21
![Regular expressions as Protein sequence motifs CXDEX10 12CXCSTYLV FamB A C E F Regular expressions as Protein sequence motifs C-X-[DE]-X{10, 12}-C-X-C--[STYLV] Fam(B) A C • E F](https://slidetodoc.com/presentation_image_h2/1937e99e82495f4ba97bbf35da3f523f/image-2.jpg)
Regular expressions as Protein sequence motifs C-X-[DE]-X{10, 12}-C-X-C--[STYLV] Fam(B) A C • E F Problem: if there is a mis-match, the sequence is not accepted. December 18, 2021

Representation 2: Profiles • Profiles versus regular expressions – – – Regular expressions are intolerant to an occasional mis-match. The Union operation (I+V+L) does not quantify the relative importance of I, V, L. It could be that V occurs in 80% of the family members. Profiles capture some of these ideas. December 21

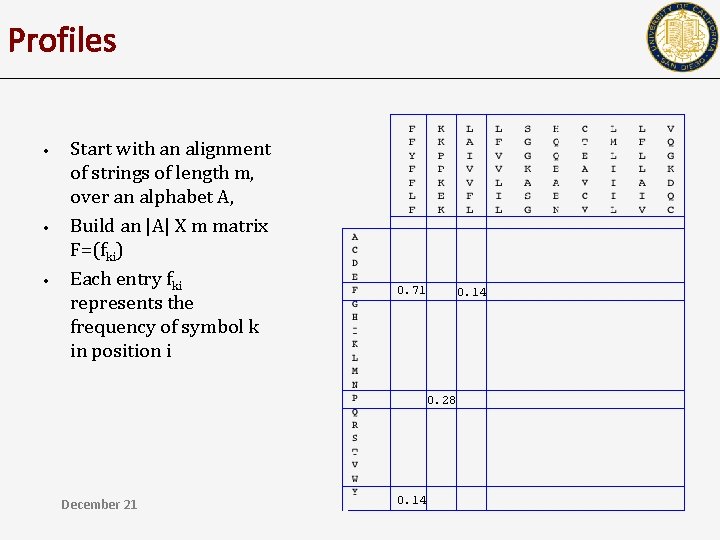
Profiles • • • Start with an alignment of strings of length m, over an alphabet A, Build an |A| X m matrix F=(fki) Each entry fki represents the frequency of symbol k in position i 0. 71 0. 14 0. 28 December 21 0. 14

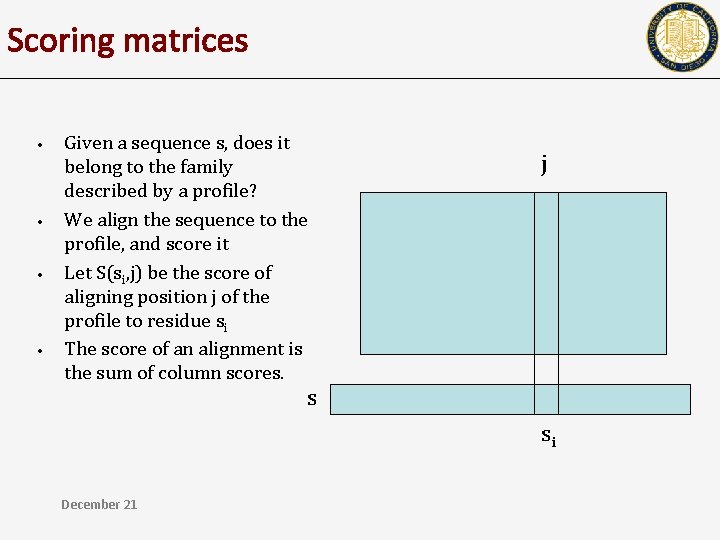
Scoring matrices • • Given a sequence s, does it belong to the family described by a profile? We align the sequence to the profile, and score it Let S(si, j) be the score of aligning position j of the profile to residue si The score of an alignment is the sum of column scores. j s si December 21

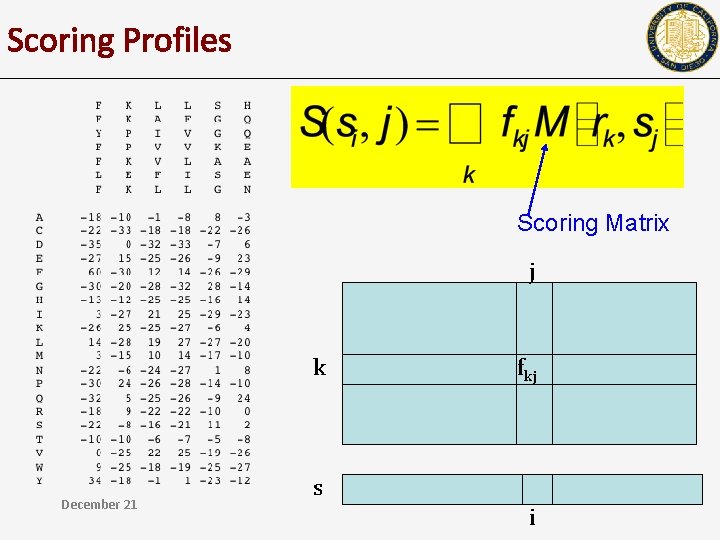
Scoring Profiles Scoring Matrix j k December 21 fkj s i

Domain analysis via profiles • • Given a database of profiles of known domains/families, we can query our sequence against each of them, and choose the high scoring ones to functionally characterize our sequences. What if the sequence matches some other sequences weakly (using BLAST), but does not match any known profile? December 21

Psi-BLAST idea Seq Db --In the next iteration, the red sequence will be thrown out. --It matches the query in non-essential residues • Iterate: – – Find homologs using Blast on query Discard very similar homologs Align, make a profile, search with profile. Why is this more sensitive? December 21

EXTENDING PROFILES USING HIDDEN MARKOV MODELS December 21

QUIZ! • • • Question: Your ‘friend’ likes to gamble. She tosses a coin: HEADS, she gives you a dollar. TAILS, you give her a dollar. Usually, she uses a fair coin, but ‘once in a while’, she uses a loaded coin. Can you say what fraction of the times she loads the coin? December 21

Representation 3: HMMs • Building good profiles relies upon good alignments. – – • • Difficult if there are gaps in the alignment. Psi-BLAST/BLOCKS etc. work with gapless alignments. An HMM representation of Profiles helps put the alignment construction/membership query in a uniform framework. Also allows for position specific gap scoring. December 21 V

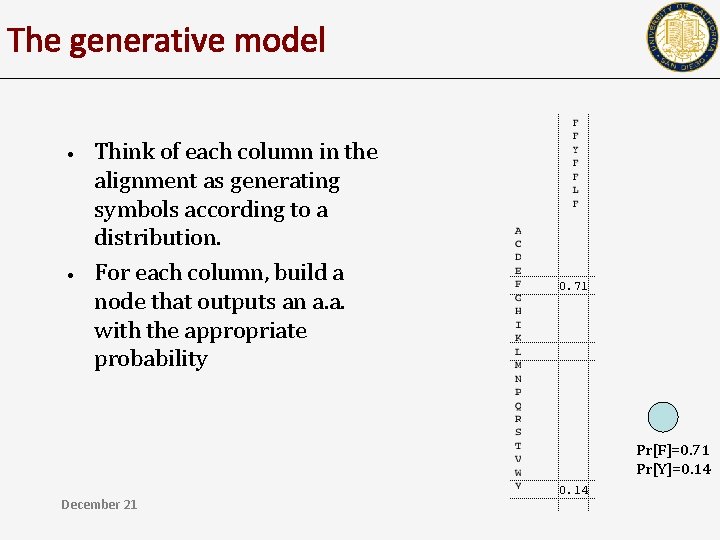
The generative model • • Think of each column in the alignment as generating symbols according to a distribution. For each column, build a node that outputs an a. a. with the appropriate probability 0. 71 Pr[F]=0. 71 Pr[Y]=0. 14 December 21 0. 14

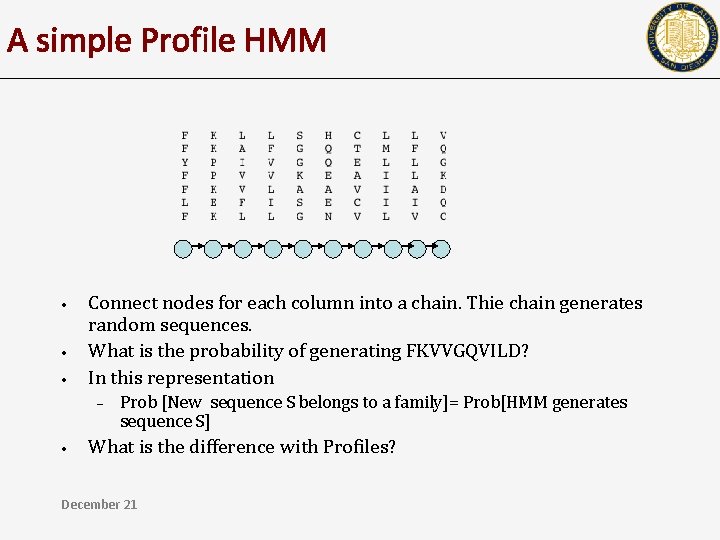
A simple Profile HMM • • • Connect nodes for each column into a chain. Thie chain generates random sequences. What is the probability of generating FKVVGQVILD? In this representation – • Prob [New sequence S belongs to a family]= Prob[HMM generates sequence S] What is the difference with Profiles? December 21

HMMs with indels December 21

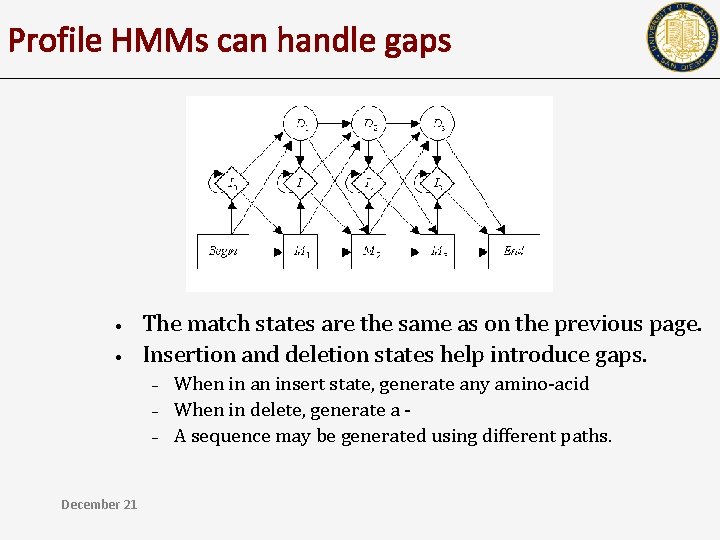
Profile HMMs can handle gaps • • The match states are the same as on the previous page. Insertion and deletion states help introduce gaps. – – – December 21 When in an insert state, generate any amino-acid When in delete, generate a A sequence may be generated using different paths.

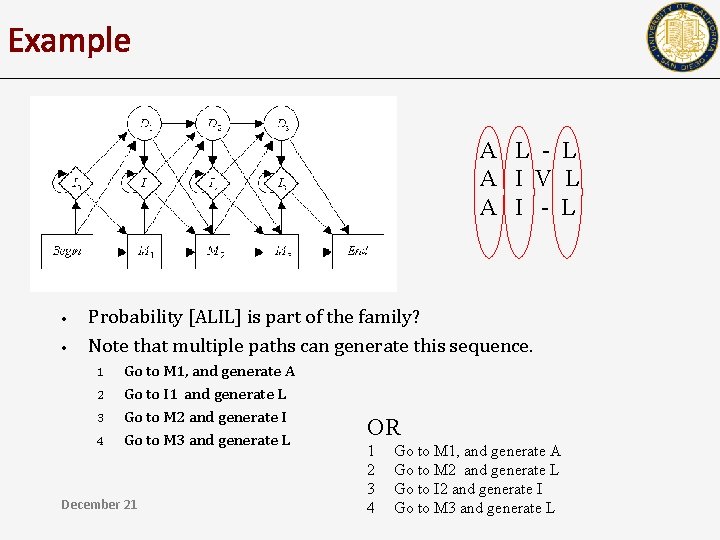
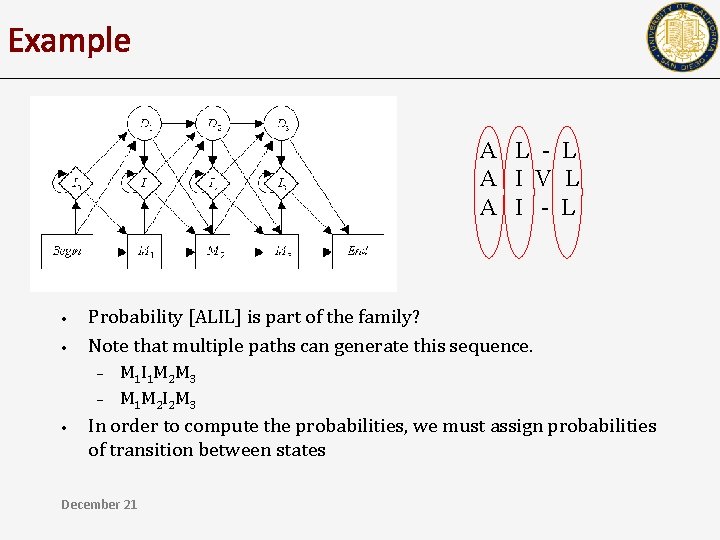
Example A L - L A I V L A I - L • • Probability [ALIL] is part of the family? Note that multiple paths can generate this sequence. 1 2 3 4 Go to M 1, and generate A Go to I 1 and generate L Go to M 2 and generate I Go to M 3 and generate L December 21 OR 1 2 3 4 Go to M 1, and generate A Go to M 2 and generate L Go to I 2 and generate I Go to M 3 and generate L

Example A L - L A I V L A I - L • • Probability [ALIL] is part of the family? Note that multiple paths can generate this sequence. – – • M 1 I 1 M 2 M 3 M 1 M 2 I 2 M 3 In order to compute the probabilities, we must assign probabilities of transition between states December 21

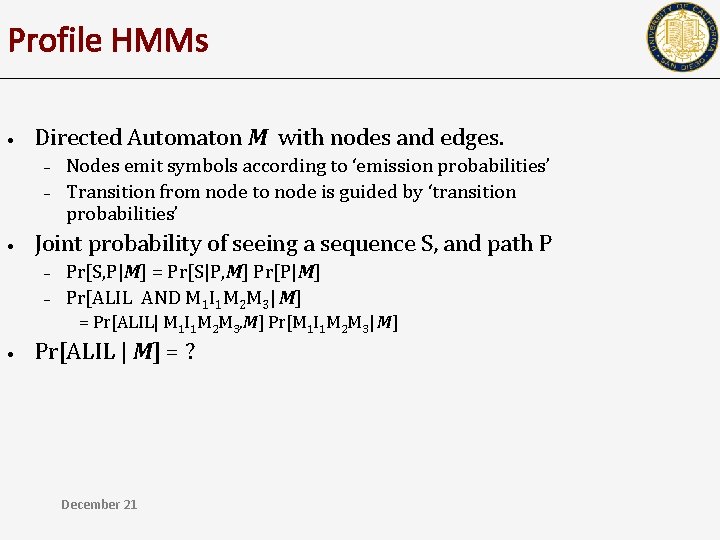
Profile HMMs • Directed Automaton M with nodes and edges. – – • Nodes emit symbols according to ‘emission probabilities’ Transition from node to node is guided by ‘transition probabilities’ Joint probability of seeing a sequence S, and path P – – Pr[S, P|M] = Pr[S|P, M] Pr[P|M] Pr[ALIL AND M 1 I 1 M 2 M 3| M] = Pr[ALIL| M 1 I 1 M 2 M 3, M] Pr[M 1 I 1 M 2 M 3| M] • Pr[ALIL | M] = ? December 21
![HMM Tj k j k i Q is a set of states HMM T[j, k] j k i • • Q is a set of states](https://slidetodoc.com/presentation_image_h2/1937e99e82495f4ba97bbf35da3f523f/image-19.jpg)
HMM T[j, k] j k i • • Q is a set of states • Π is the probability distribution on initial state • T is a matrix of transition probabilities • T[j, k]: probability of moving from state j to state k • Σ is a set of symbols • ej(S) is the probability of emitting S while in state j. Automaton M=(Q, T, π, Σ, e) At first, M goes to initial state j with probability πj In state j, M emits a symbol from Σ according to ej, and moves to state k with probability T[j, k]. December 18, 2021

Two solutions • • An unknown (hidden) path is traversed to produce (emit) the sequence S. The probability that M emits S can be either – The sum over the joint probabilities over all paths. • – OR, it is the probability of the most likely path • • Pr(S|M) = ∑P Pr(S, P|M) Pr(S|M) = max. P Pr(S, P|M) Both are appropriate ways to model, and have similar algorithms to solve them. December 21

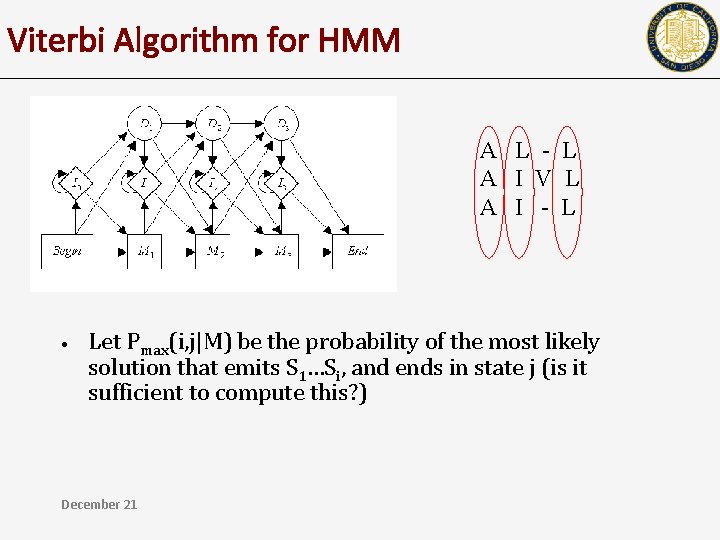
Viterbi Algorithm for HMM A L - L A I V L A I - L • Let Pmax(i, j|M) be the probability of the most likely solution that emits S 1…Si, and ends in state j (is it sufficient to compute this? ) December 21

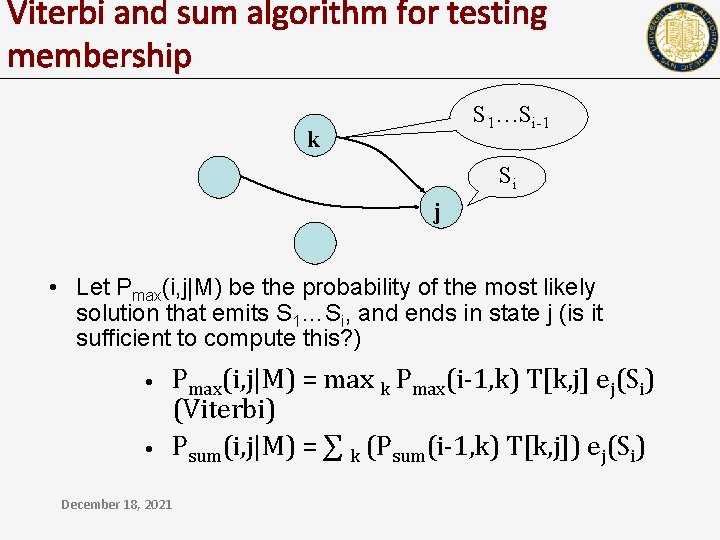
Viterbi and sum algorithm for testing membership S 1…Si-1 k Si j • Let Pmax(i, j|M) be the probability of the most likely solution that emits S 1…Si, and ends in state j (is it sufficient to compute this? ) • • Pmax(i, j|M) = max k Pmax(i-1, k) T[k, j] ej(Si) (Viterbi) Psum(i, j|M) = ∑ k (Psum(i-1, k) T[k, j]) ej(Si) December 18, 2021

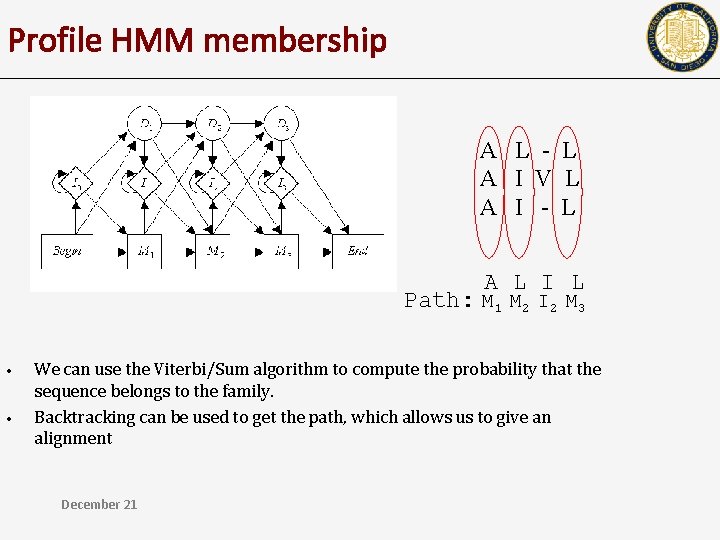
Profile HMM membership A L - L A I V L A I - L A L I L Path: M 1 M 2 I 2 M 3 • • We can use the Viterbi/Sum algorithm to compute the probability that the sequence belongs to the family. Backtracking can be used to get the path, which allows us to give an alignment December 21

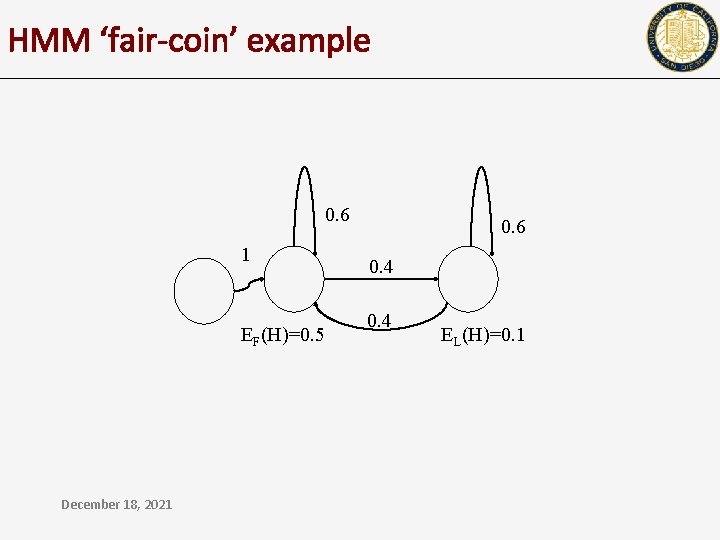
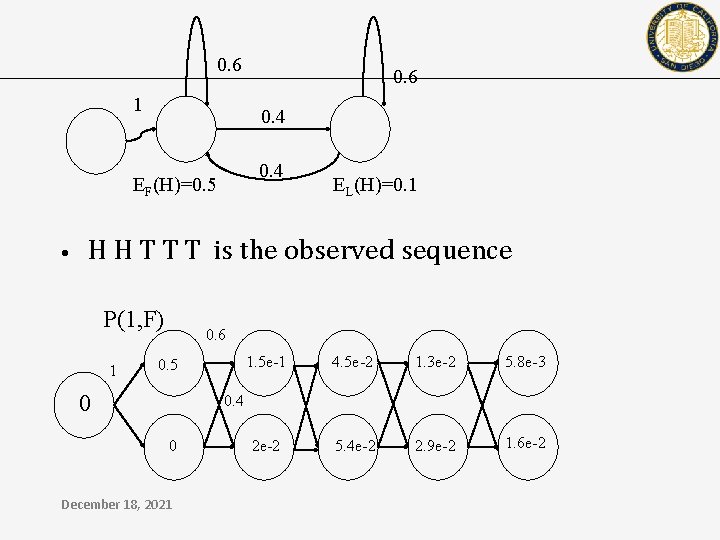
HMM ‘fair-coin’ example 0. 6 1 EF(H)=0. 5 December 18, 2021 0. 6 0. 4 EL(H)=0. 1

0. 6 1 0. 4 EF(H)=0. 5 • 0. 6 EL(H)=0. 1 H H T T T is the observed sequence P(1, F) 1 0. 6 0. 5 0 1. 5 e-1 4. 5 e-2 1. 3 e-2 5. 8 e-3 2 e-2 5. 4 e-2 2. 9 e-2 1. 6 e-2 0. 4 0 December 18, 2021

Summary • • • HMMs allow us to model position specific gap penalties, and allow for automated training to get a good alignment. Patterns/Profiles/HMMs allow us to represent families and foucs on key residues Each has its advantages and disadvantages, and needs special algorithms to query efficiently. December 21

Protein Domain databases HMM • • • A number of databases capture proteins (domains) using various representations Each domain is also associated with structure/function information, parsed from the literature. Each database has specific query mechanisms that allow us to compare our sequences against them, and assign function 3 D December 21

Biology • In our discussion of BLAST, we alternated between looking at DNA, and protein sequences, treating them as strings. – • DNA, RNA, and proteins are the 3 important molecules What is the relation between the three? December 21

December 21

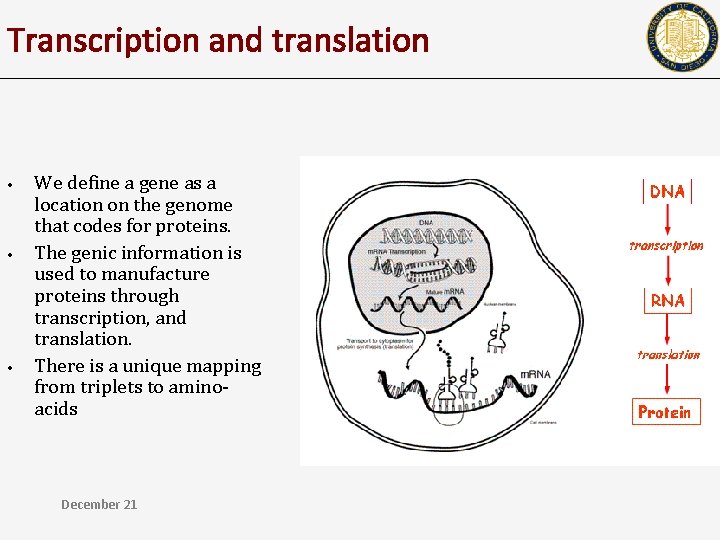
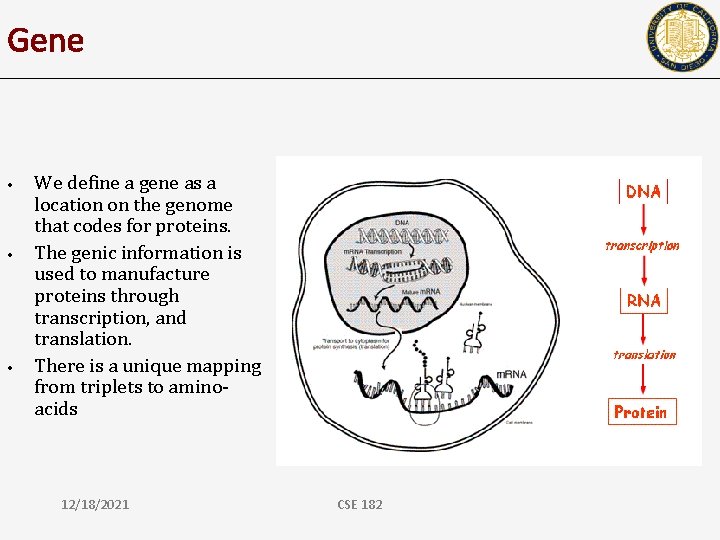
Transcription and translation • • • We define a gene as a location on the genome that codes for proteins. The genic information is used to manufacture proteins through transcription, and translation. There is a unique mapping from triplets to aminoacids December 21

Gene • • • We define a gene as a location on the genome that codes for proteins. The genic information is used to manufacture proteins through transcription, and translation. There is a unique mapping from triplets to aminoacids 12/18/2021 CSE 182

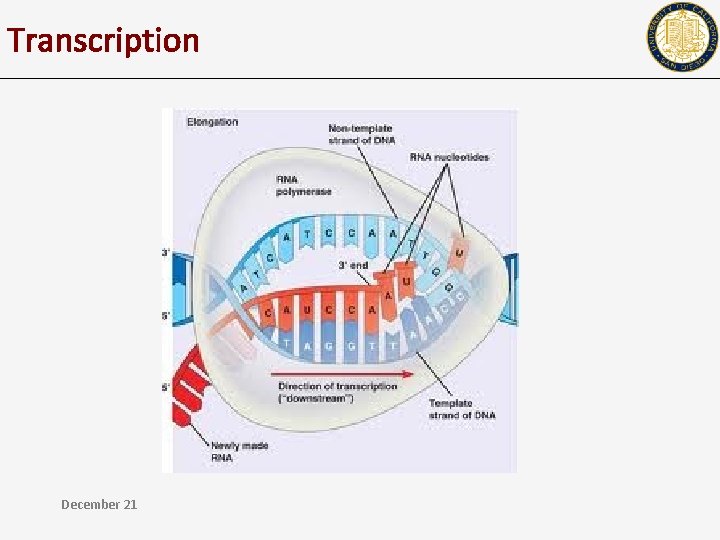
Transcription December 21

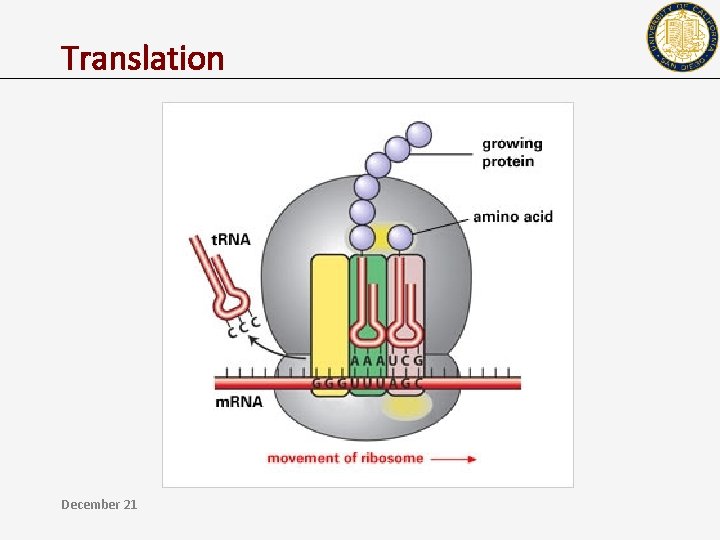
Translation December 21

Transcription start ATAGATGATGTACGATGAGAATGTGATTAATG Translation start Donor Acceptor

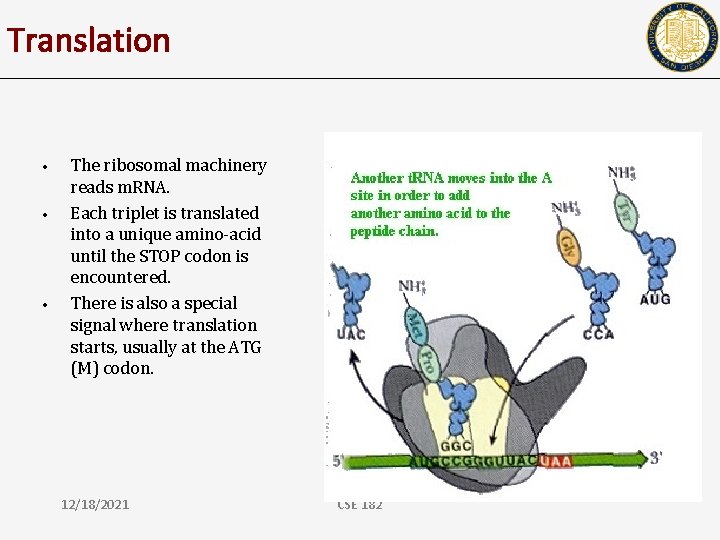
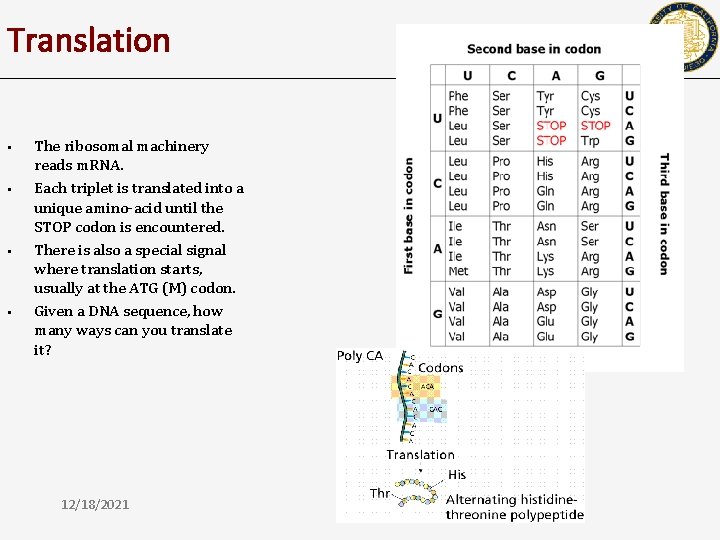
Translation • • • The ribosomal machinery reads m. RNA. Each triplet is translated into a unique amino-acid until the STOP codon is encountered. There is also a special signal where translation starts, usually at the ATG (M) codon. 12/18/2021 CSE 182

Translation • • The ribosomal machinery reads m. RNA. Each triplet is translated into a unique amino-acid until the STOP codon is encountered. There is also a special signal where translation starts, usually at the ATG (M) codon. Given a DNA sequence, how many ways can you translate it? 12/18/2021 CSE 182

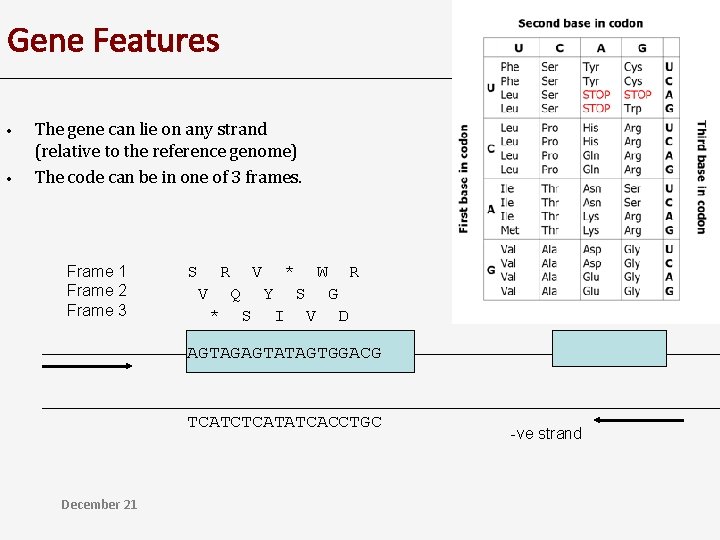
Gene Features • • The gene can lie on any strand (relative to the reference genome) The code can be in one of 3 frames. Frame 1 Frame 2 Frame 3 S R V * W R V Q Y S G * S I V D AGTAGAGTATAGTGGACG TCATCTCATATCACCTGC December 21 -ve strand

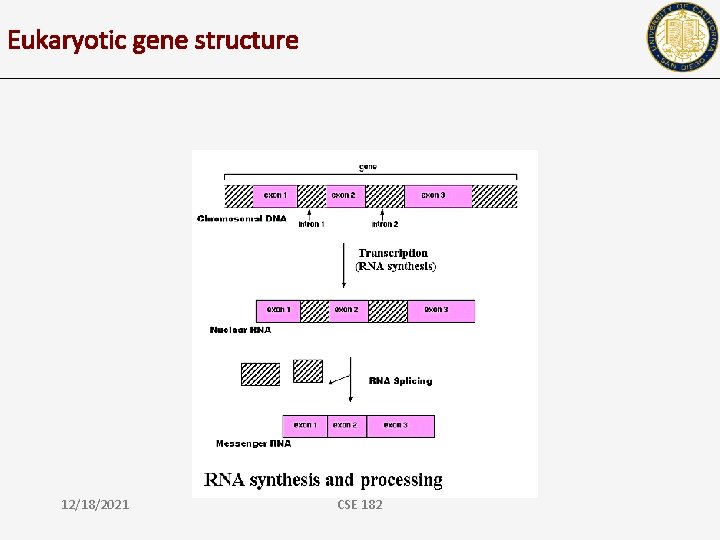
Eukaryotic gene structure 12/18/2021 CSE 182

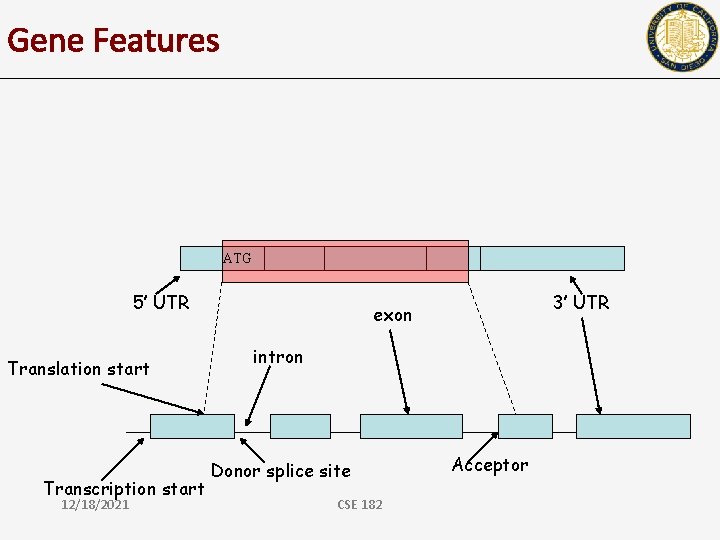
Gene Features ATG 5’ UTR Translation start Transcription start 12/18/2021 3’ UTR exon intron Donor splice site CSE 182 Acceptor

Gene identification • Eukaryotic gene definitions: – – – • • • Location that codes for a protein The transcript sequence(s) that encodes the protein The protein sequence(s) Suppose you want to know all of the genes in an organism. This was a major problem in the 70 s. Ph. Ds, and careers were spent isolating a single gene sequence. All of that changed with better reagents and the development of high throughput methods like EST sequencing 12/18/2021 CSE 182

End of L 9 December 21