CSE 182 L 5 Position specific scoring matrices







![From alignment to regular expressions * ALRDFATHDDF SMTAEATHDSI ECDQAATHEAS ATH-[DE] • Search Swissprot with From alignment to regular expressions * ALRDFATHDDF SMTAEATHDSI ECDQAATHEAS ATH-[DE] • Search Swissprot with](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-11.jpg)
![The sequence analysis perspective • • • Zinc Finger motif – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H The sequence analysis perspective • • • Zinc Finger motif – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-12.jpg)

![The sequence analysis perspective • Zinc Finger motif • – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H – The sequence analysis perspective • Zinc Finger motif • – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H –](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-15.jpg)




![Alg. For matching R. E. • If D[1. . c] is accepted by the Alg. For matching R. E. • If D[1. . c] is accepted by the](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-22.jpg)
![Alg. For matching R. E. • If D[1. . c] is accepted by the Alg. For matching R. E. • If D[1. . c] is accepted by the](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-23.jpg)
![D. P. to match regular expression u • Define: – A[u, ] = Automaton D. P. to match regular expression u • Define: – A[u, ] = Automaton](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-24.jpg)
![D. P. to match regular expression • Q: when is v N[c]? • A: D. P. to match regular expression • Q: when is v N[c]? • A:](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-25.jpg)
![The final step • We have answered the question: – Is D[1. . c] The final step • We have answered the question: – Is D[1. . c]](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-27.jpg)




- Slides: 39

CSE 182 -L 5: Position specific scoring matrices Regular Expression Matching Protein Domains Fa 05 CSE 182

Class Mailing List • fa 05_182@cs. ucsd. edu • To subscribe, send email to – fa 05_182 -subscribe@cs. ucsd. edu • You can subscribe from the course web page • Please subscribe with a UCSD email address if possible. Fa 05 CSE 182

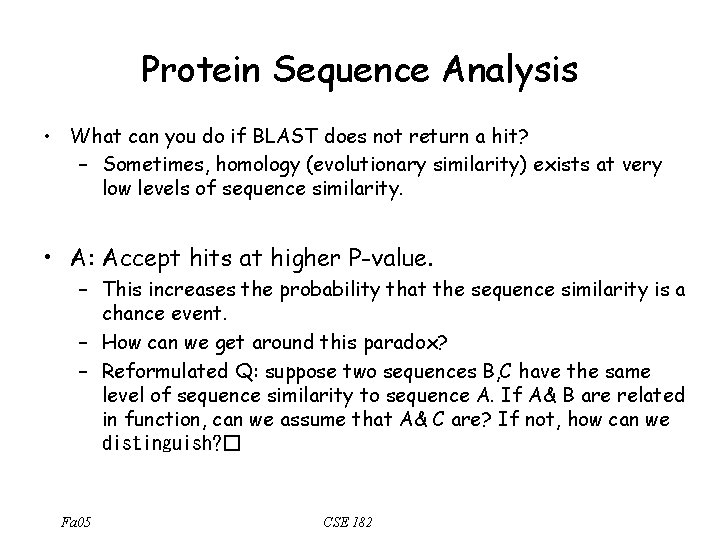
Protein Sequence Analysis • What can you do if BLAST does not return a hit? – Sometimes, homology (evolutionary similarity) exists at very low levels of sequence similarity. • A: Accept hits at higher P-value. – This increases the probability that the sequence similarity is a chance event. – How can we get around this paradox? – Reformulated Q: suppose two sequences B, C have the same level of sequence similarity to sequence A. If A& B are related in function, can we assume that A& C are? If not, how can we distinguish? � Fa 05 CSE 182

Silly Quiz Fa 05 CSE 182

Silly Quiz Fa 05 CSE 182

Protein sequence motifs • Premise: • The sequence of a protein sequence gives clues about its structure and function. • Not all residues are equally important in determining function. • How can we identify these key residues? Fa 05 CSE 182

Prosite • In some cases the sequence of an unknown protein is too distantly related to any protein of known structure to detect its resemblance by overall sequence alignment. However, relationships can be revealed by the occurrence in its sequence of a particular cluster of residue types, which is variously known as a pattern, motif, signature or fingerprint. These motifs arise because specific region(s) of a protein which may be important, for example, for their binding properties or for their enzymatic activity are conserved in both structure and sequence. These structural requirements impose very tight constraints on the evolution of this small but important portion(s) of a protein sequence. The use of protein sequence patterns or profiles to determine the function of proteins is becoming very rapidly one of the essential tools of sequence analysis. Many authors ( 3, 4) have recognized this reality. Based on these observations, we decided in 1988, to actively pursue the development of a database of regular expression-like patterns, which would be used to search against sequences of unknown function. Kay Hofmann , Philipp Bucher, Laurent Falquet and Amos Bairoch The PROSITE database, its status in 1999 Fa 05 CSE 182

Basic idea • It is a heuristic approach. Start with the following: – A collection of sequences with the same function. – Region/residues known to be significant for maintaining structure and function. • Develop a pattern of conserved residues around the residues of interest • Iterate for appropriate sensitivity and specificity Fa 05 CSE 182

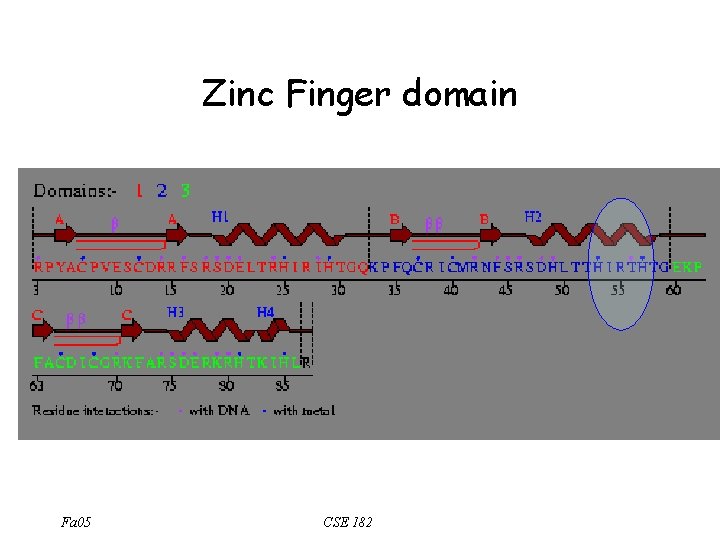
Zinc Finger domain Fa 05 CSE 182

Proteins containing zf domains How can we find a motif corresponding to a zf domain Fa 05 CSE 182
![From alignment to regular expressions ALRDFATHDDF SMTAEATHDSI ECDQAATHEAS ATHDE Search Swissprot with From alignment to regular expressions * ALRDFATHDDF SMTAEATHDSI ECDQAATHEAS ATH-[DE] • Search Swissprot with](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-11.jpg)
From alignment to regular expressions * ALRDFATHDDF SMTAEATHDSI ECDQAATHEAS ATH-[DE] • Search Swissprot with the resulting pattern • Refine pattern to eliminate false positives • Iterate Fa 05 CSE 182
![The sequence analysis perspective Zinc Finger motif Cx2 4Cx3LIVMFYWCx8Hx3 5H The sequence analysis perspective • • • Zinc Finger motif – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-12.jpg)
The sequence analysis perspective • • • Zinc Finger motif – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H – 2 conserved C, and 2 conserved H How can we search a database using these motifs? – The motif is described using a regular expression. What is a regular expression? – How can we search for a match to a regular expression? Not allowed to use Perl : -) The ‘regular expression’ motif is weak. How can we make it stronger Fa 05 CSE 182

Regular Expression Matching Protein structure basics Fa 05 CSE 182

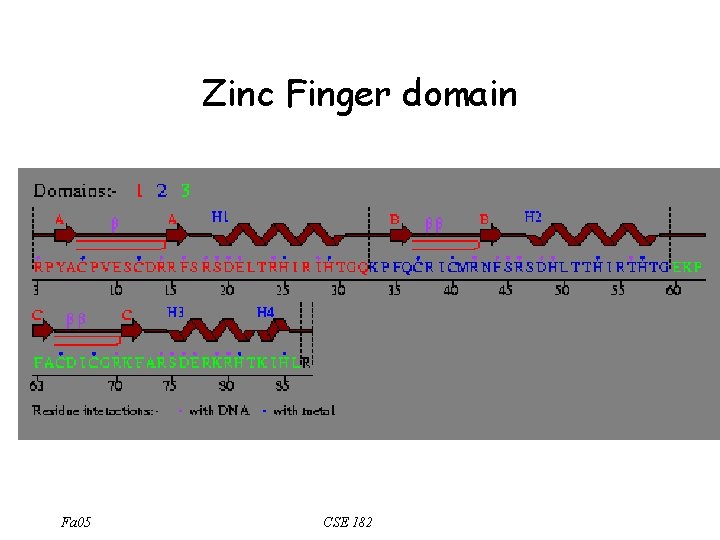
Zinc Finger domain Fa 05 CSE 182
![The sequence analysis perspective Zinc Finger motif Cx2 4Cx3LIVMFYWCx8Hx3 5H The sequence analysis perspective • Zinc Finger motif • – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H –](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-15.jpg)
The sequence analysis perspective • Zinc Finger motif • – C-x(2, 4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3, 5)-H – 2 conserved C, and 2 conserved H How can we search a database using these motifs? – The motif is described using a regular expression. What is a regular expression? Fa 05 CSE 182

Regular Expressions • Concise representation of a set of strings over alphabet . • Described by a string over • R is a r. e. if and only if Fa 05 CSE 182

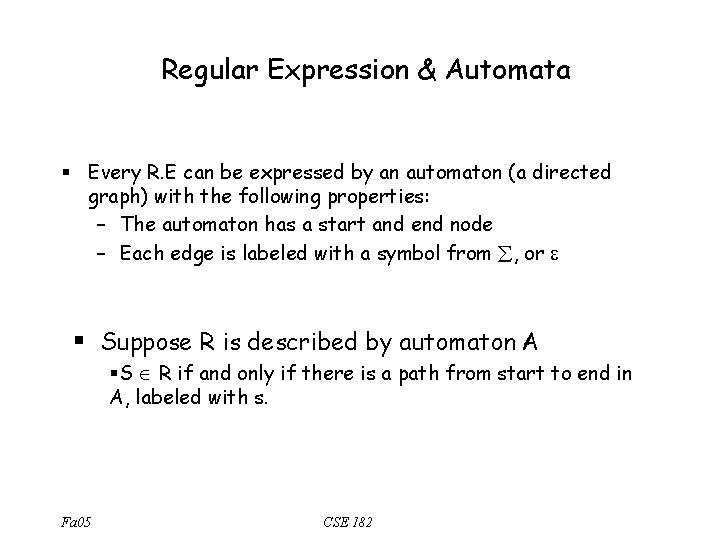
Regular Expression • Q: Let ={A, C, E} – Is (A+C)*EEC* a regular expression? – *(A+C)? – AC*. . E? • Q: When is a string s in a regular expression? – R =(A+C)*EEC* – Is CEEC in R? – AEC? – ACEE? Fa 05 CSE 182

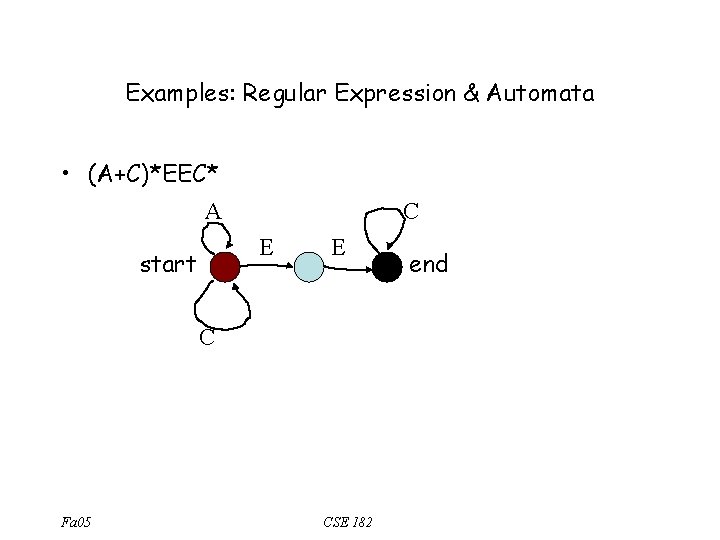
Regular Expression & Automata § Every R. E can be expressed by an automaton (a directed graph) with the following properties: – The automaton has a start and end node – Each edge is labeled with a symbol from , or § Suppose R is described by automaton A §S R if and only if there is a path from start to end in A, labeled with s. Fa 05 CSE 182

Examples: Regular Expression & Automata • (A+C)*EEC* A C E start E C Fa 05 CSE 182 end

Constructing automata from R. E • • • R = { }, R = R 1 + R 2 R = R 1 · R 2 R = R 1* Fa 05 CSE 182

Regular Expression Matching • Given a database D, and a regular expression R, is a substring of D in R? • Is there a string D[l. . c] that is accepted by the automaton of R? • Simpler Q: Is D[1. . c] accepted by the automaton of R? Fa 05 CSE 182
![Alg For matching R E If D1 c is accepted by the Alg. For matching R. E. • If D[1. . c] is accepted by the](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-22.jpg)
Alg. For matching R. E. • If D[1. . c] is accepted by the automaton RA – There is a path labeled D[1]…D[c] that goes from START to END in RA D[1] Fa 05 D[2] CSE 182 D[c]
![Alg For matching R E If D1 c is accepted by the Alg. For matching R. E. • If D[1. . c] is accepted by the](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-23.jpg)
Alg. For matching R. E. • If D[1. . c] is accepted by the automaton RA – There is a path labeled D[1]…D[c] that goes from START to END in RA – There is a path labeled D[1]. . D[c-1] from START to node u, and a path labeled D[c] from u to the END D[1]. . D[c-1] u D[c] Fa 05 CSE 182
![D P to match regular expression u Define Au Automaton D. P. to match regular expression u • Define: – A[u, ] = Automaton](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-24.jpg)
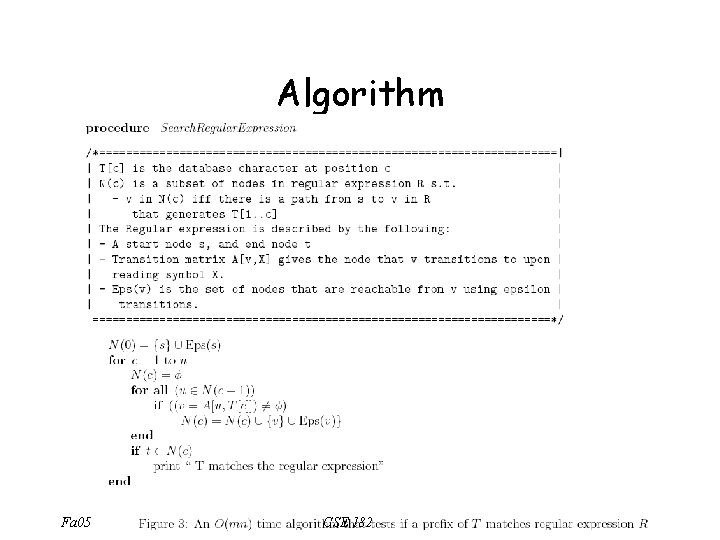
D. P. to match regular expression u • Define: – A[u, ] = Automaton node reached from u after reading – Eps(u): set of all nodes reachable from node u using epsilon transitions. – N[c] = subset of nodes reachable from START node after reading D[1. . c] – Q: when is v N[c] Fa 05 CSE 182 u v Eps(u)
![D P to match regular expression Q when is v Nc A D. P. to match regular expression • Q: when is v N[c]? • A:](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-25.jpg)
D. P. to match regular expression • Q: when is v N[c]? • A: If for some u N[c-1], w = A[u, D[c]], • v {w}+ Eps(w) Fa 05 CSE 182

Algorithm Fa 05 CSE 182
![The final step We have answered the question Is D1 c The final step • We have answered the question: – Is D[1. . c]](https://slidetodoc.com/presentation_image_h2/3c7397fdeea83ac3fd98655505001152/image-27.jpg)
The final step • We have answered the question: – Is D[1. . c] accepted by R? – Yes, if END N[c] • We need to answer – Is D[l. . c] (for some l, and some c) accepted by R Fa 05 CSE 182

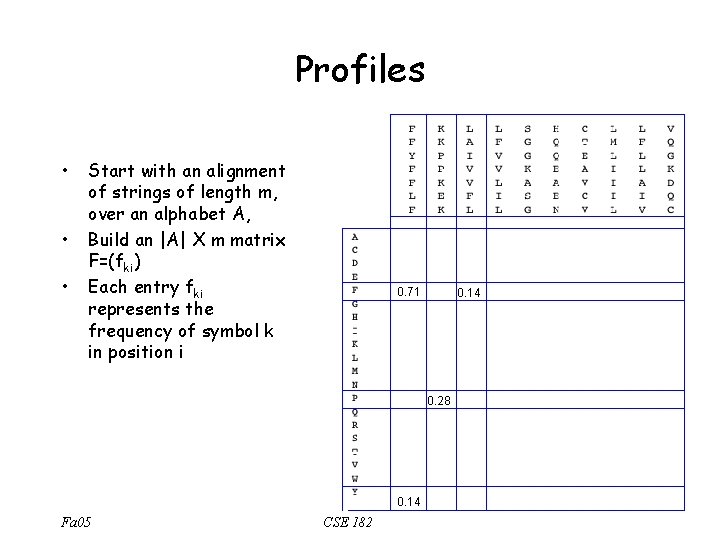
Profiles versus regular expressions • Regular expressions are intolerant to an occasional mis-match. • The Union operation (I+V+L) does not quantify the relative importance of I, V, L. It could be that V occurs in 80% of the family members. • Profiles capture some of these ideas. Fa 05 CSE 182

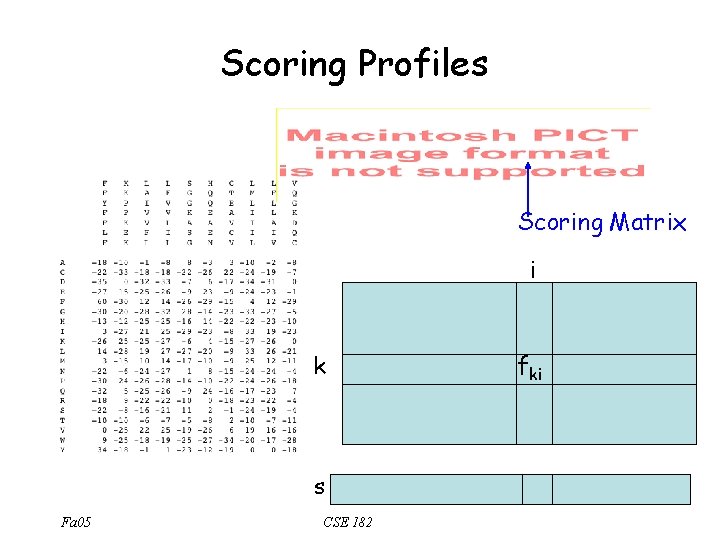
Profiles • • • Start with an alignment of strings of length m, over an alphabet A, Build an |A| X m matrix F=(fki) Each entry fki represents the frequency of symbol k in position i 0. 71 0. 14 0. 28 0. 14 Fa 05 CSE 182

Scoring Profiles Scoring Matrix i k s Fa 05 CSE 182 fki

Psi-BLAST idea • Multiple alignments are important for capturing remote homology. • Profile based scores are a natural way to handle this. • Q: What if the query is a single sequence. • A: Iterate: – Find homologs using Blast on query – Discard very similar homologs – Align, make a profile, search with profile. Fa 05 CSE 182

Psi-BLAST speed • Two time consuming steps. 1. Multiple alignment of homologs 2. Searching with Profiles. 1. Does the keyword search idea work? • • Multiple alignment: – Use ungapped multiple alignments only Fa 05 Pigeonhole principle again: – If profile of length m must score >= T – Then, a sub-profile of length l must score >= l. T/m – Generate all l-mers that score at least l. T|/M – Search using an automaton CSE 182

Databases of Motifs • Functionally related proteins have sequence motifs. • The sequence motifs can be represented in many ways, and different biological databases capture these representations – – Collection of sequences (SMART) Multiple alignments (BLOCKS) Profiles (Pfam (HMMs)/Impala)) Regular Expressions (Prosite) • Different representations must be queried in different ways Fa 05 CSE 182

Databases of protein domains Fa 05 CSE 182

Pfam http: //pfam. wustl. edu/ Also at Sanger Fa 05 CSE 182

PROSITE http: //us. expasy. org/prosite/ Fa 05 CSE 182

Fa 05 CSE 182

BLOCKS Fa 05 CSE 182
Fa 05 CSE 182