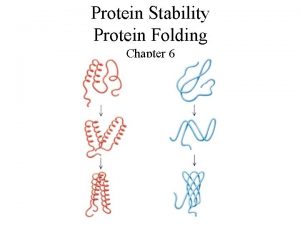
Protein Folding Energetics Kinetics and Models Oznur Tastan





- Slides: 50

Protein Folding Energetics, Kinetics and Models Oznur Tastan oznur@cs. cmu. edu Graduate Student Carnegie Mellon University 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan

Lecture Outline • Introduction: What is protein folding and why it is a problem? • Globular Protein Folding Models • Detection and characterization of denatured states, intermediates, such as the molten globule and comparison to folded states • Kinetics and pathways • Membrane Protein Folding Models • 2 -stage and 3 -stage hypothesis • New long range interaction hypothesis • Summary 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 2

Lecture Outline • Introduction: What is protein folding and why it is a problem? • Globular Protein Folding Models • Detection and characterization of denatured states, intermediates, such as the molten globule and comparison to folded states • Kinetics and pathways • Membrane Protein Folding Models • 2 -stage and 3 -stage hypothesis • New long range interaction hypothesis • Summary 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 3

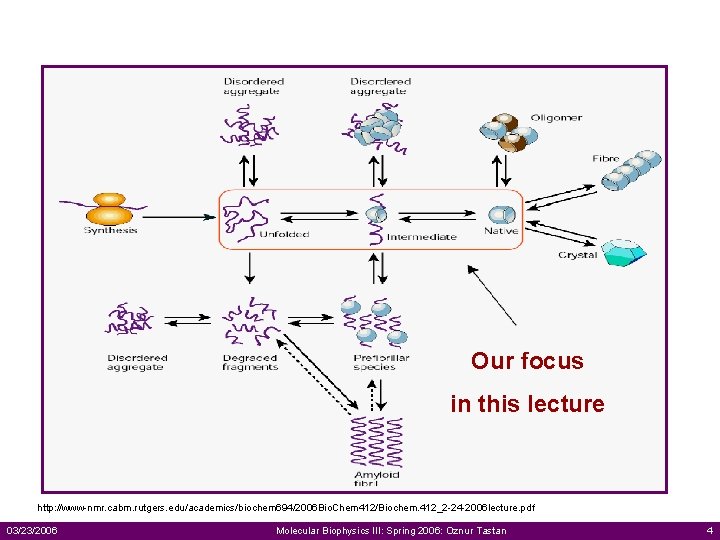
Our focus in this lecture http: //www-nmr. cabm. rutgers. edu/academics/biochem 694/2006 Bio. Chem 412/Biochem. 412_2 -24 -2006 lecture. pdf 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 4

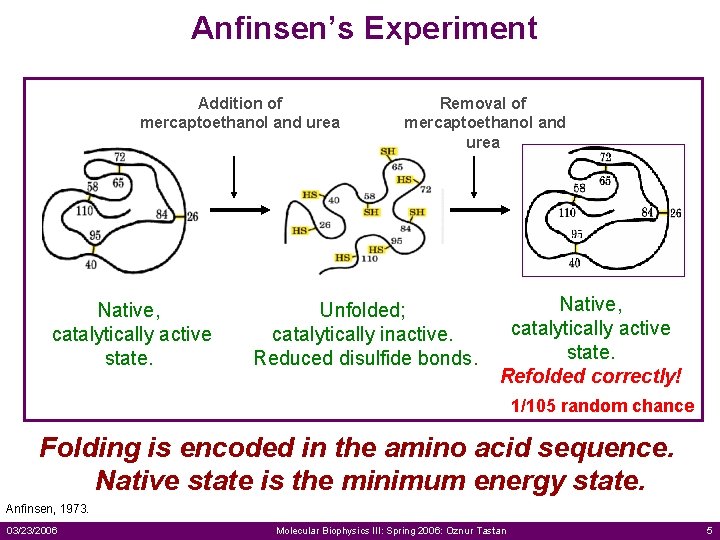
Anfinsen’s Experiment Addition of mercaptoethanol and urea Native, catalytically active state. Removal of mercaptoethanol and urea Unfolded; catalytically inactive. Reduced disulfide bonds. Native, catalytically active state. Refolded correctly! 1/105 random chance Folding is encoded in the amino acid sequence. Native state is the minimum energy state. Anfinsen, 1973. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 5

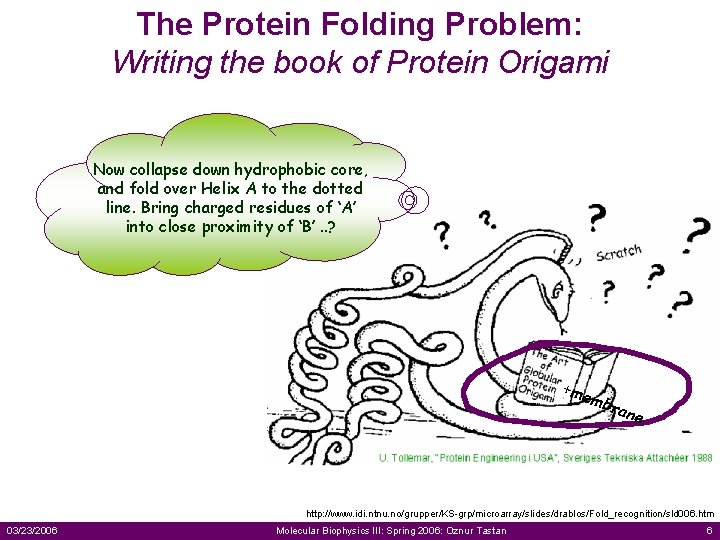
The Protein Folding Problem: Writing the book of Protein Origami Now collapse down hydrophobic core, and fold over Helix A to the dotted line. Bring charged residues of ‘A’ into close proximity of ‘B’. . ? +me mbr ane http: //www. idi. ntnu. no/grupper/KS-grp/microarray/slides/drablos/Fold_recognition/sld 006. htm 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 6

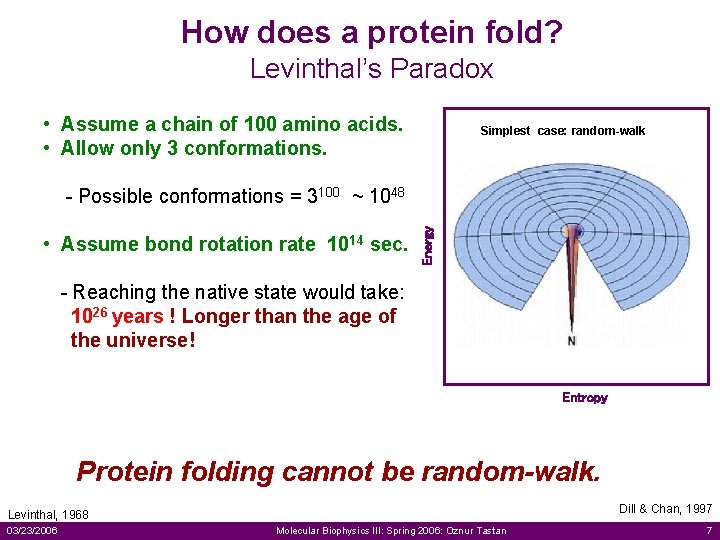
How does a protein fold? Levinthal’s Paradox • Assume a chain of 100 amino acids. • Allow only 3 conformations. Simplest case: random-walk • Assume bond rotation rate 1014 sec. Energy - Possible conformations = 3100 ~ 1048 - Reaching the native state would take: 1026 years ! Longer than the age of the universe! Entropy Protein folding cannot be random-walk. Dill & Chan, 1997 Levinthal, 1968 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 7

Why is protein folding problem difficult? • Folding can be very fast, millisecond to second (slow folding is easier) • Small energy changes between the denatured state to the native state ( 1 -15 kcal/mol) - equivalent to the strength of a few hydrogen bonds • The states populated along pathway are ensembles of structures Comparison from multiple complementary techniques are required. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 8

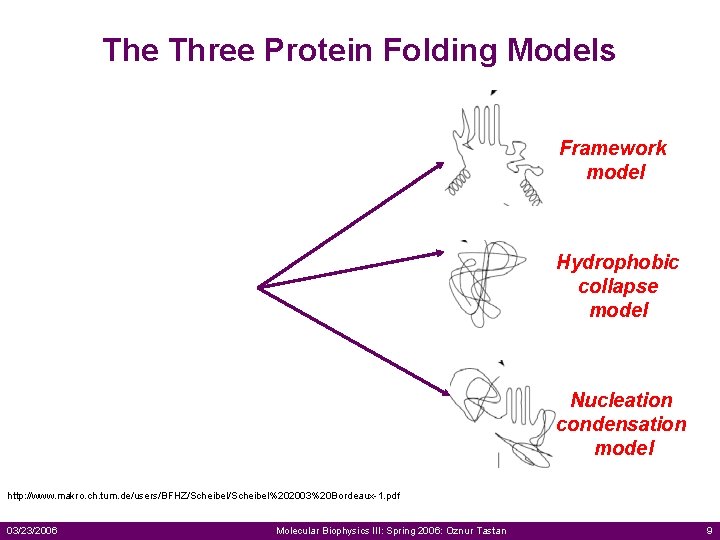
The Three Protein Folding Models Framework model Hydrophobic collapse model Nucleation condensation model http: //www. makro. ch. tum. de/users/BFHZ/Scheibel%202003%20 Bordeaux-1. pdf 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 9

Lecture Outline • Introduction: What is protein folding and why it is a problem? • Globular Protein Folding Models • Detection and characterization of denatured states, intermediates, such as the molten globule and comparison to folded states • Kinetics and pathways • Membrane Protein Folding Models • 2 -stage and 3 -stage hypothesis • New long range interaction hypothesis • Summary 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 10

The Native State • A complex balance between: 1) Short-range local interactions -intrinsic conformational preferences of the amino acids 2) Medium-range interactions -stabilizing regions of secondary structure 3) Long-range interactions - tertiary interactions determining the global fold • Generally single conformation (with small fluctuations around the mean torsion angles). 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 11

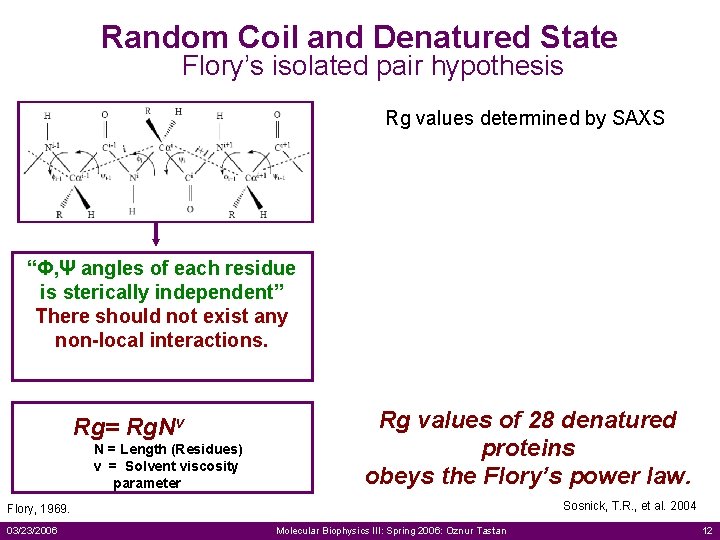
Random Coil and Denatured State Flory’s isolated pair hypothesis Rg values determined by SAXS “Φ, Ψ angles of each residue is sterically independent” There should not exist any non-local interactions. Rg= Rg. Nv N = Length (Residues) v = Solvent viscosity parameter Rg values of 28 denatured proteins obeys the Flory’s power law. Sosnick, T. R. , et al. 2004 Flory, 1969. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 12

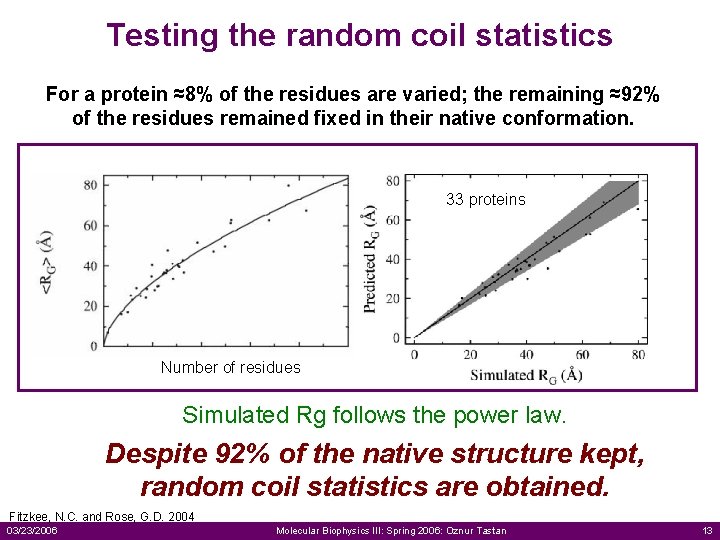
Testing the random coil statistics For a protein ≈8% of the residues are varied; the remaining ≈92% of the residues remained fixed in their native conformation. 33 proteins Number of residues Simulated Rg follows the power law. Despite 92% of the native structure kept, random coil statistics are obtained. Fitzkee, N. C. and Rose, G. D. 2004 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 13

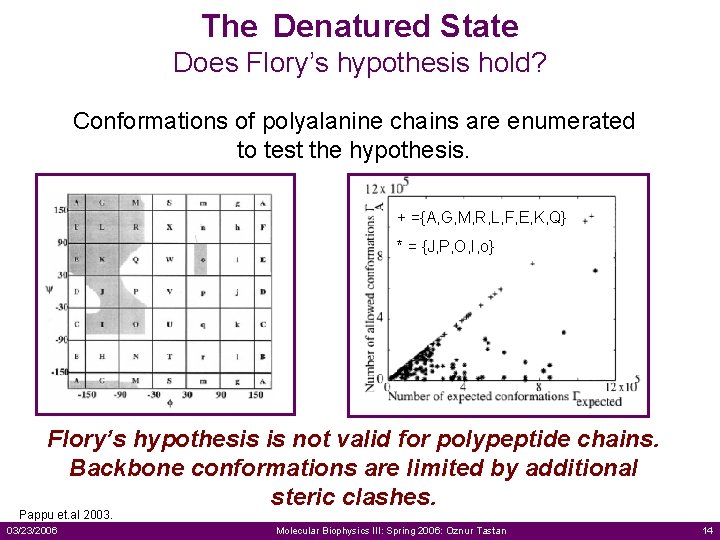
The Denatured State Does Flory’s hypothesis hold? Conformations of polyalanine chains are enumerated to test the hypothesis. + ={A, G, M, R, L, F, E, K, Q} * = {J, P, O, I, o} Flory’s hypothesis is not valid for polypeptide chains. Backbone conformations are limited by additional steric clashes. Pappu et. al 2003. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 14

Can we get a structure of the “denatured state”? When the folded state breaks down: 1. The dispersion of all resonances decreases: 2. - Extensive overlap of peaks. 2. Greater dynamic motions between residues: - Weak or eliminated NOEs between protons. 3. Ensemble of conformations: - Each NMR parameter reflects an average over a dynamic ensemble of conformations. Attainment of a high-resolution structure is not possible in the non-native state. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 15

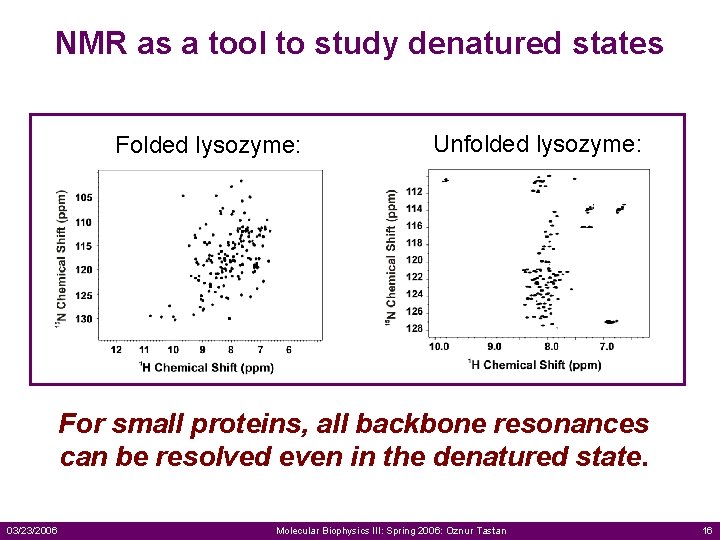
NMR as a tool to study denatured states Folded lysozyme: Unfolded lysozyme: For small proteins, all backbone resonances can be resolved even in the denatured state. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 16

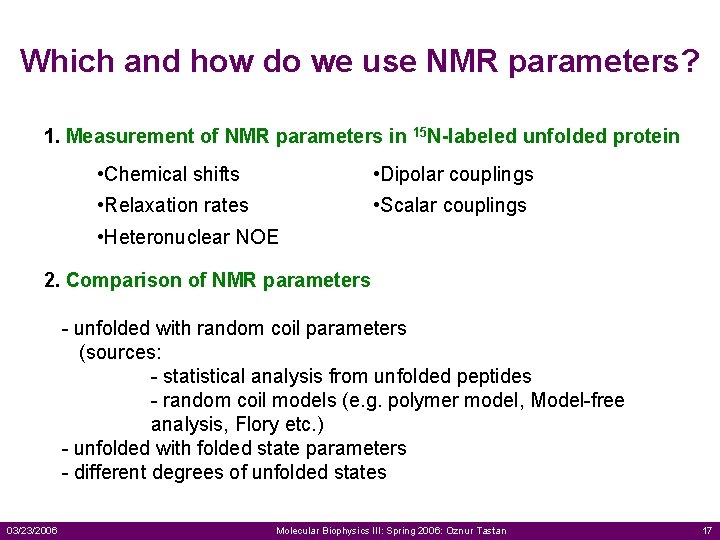
Which and how do we use NMR parameters? 1. Measurement of NMR parameters in 15 N-labeled unfolded protein • Chemical shifts • Dipolar couplings • Relaxation rates • Scalar couplings • Heteronuclear NOE 2. Comparison of NMR parameters - unfolded with random coil parameters (sources: - statistical analysis from unfolded peptides - random coil models (e. g. polymer model, Model-free analysis, Flory etc. ) - unfolded with folded state parameters - different degrees of unfolded states 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 17

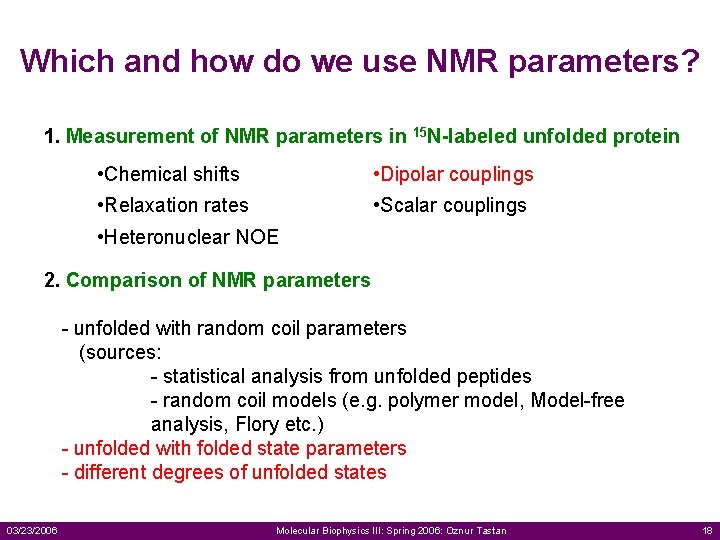
Which and how do we use NMR parameters? 1. Measurement of NMR parameters in 15 N-labeled unfolded protein • Chemical shifts • Dipolar couplings • Relaxation rates • Scalar couplings • Heteronuclear NOE 2. Comparison of NMR parameters - unfolded with random coil parameters (sources: - statistical analysis from unfolded peptides - random coil models (e. g. polymer model, Model-free analysis, Flory etc. ) - unfolded with folded state parameters - different degrees of unfolded states 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 18

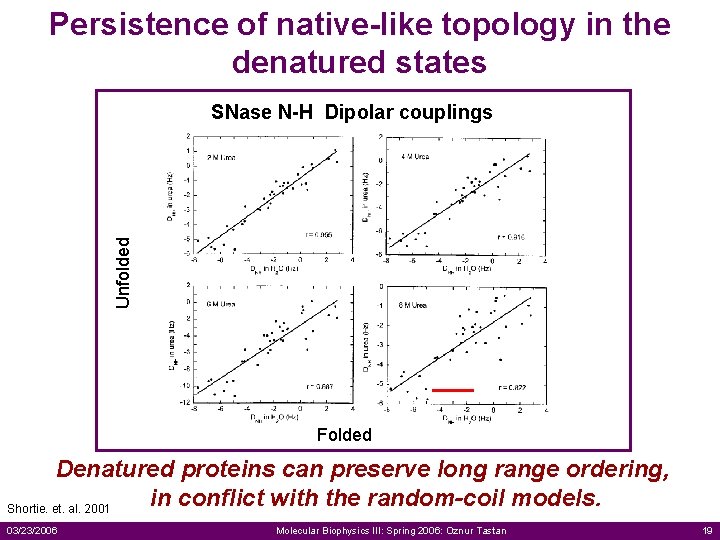
Persistence of native-like topology in the denatured states Unfolded SNase N-H Dipolar couplings Folded Denatured proteins can preserve long range ordering, in conflict with the random-coil models. Shortie. et. al. 2001 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 19

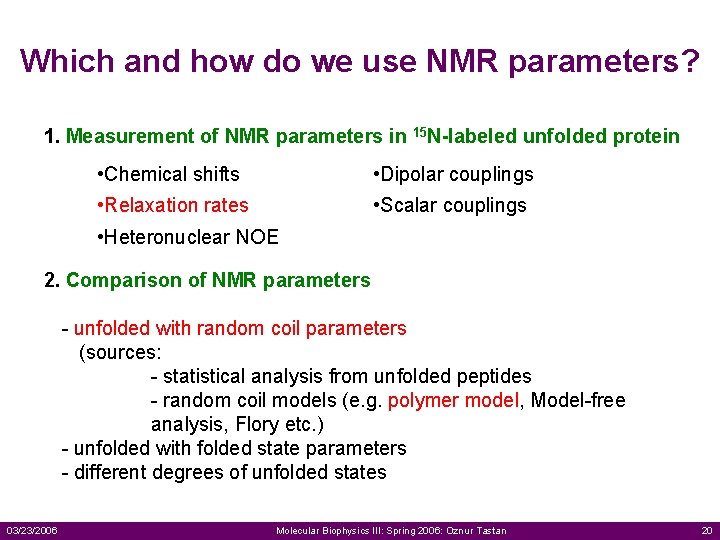
Which and how do we use NMR parameters? 1. Measurement of NMR parameters in 15 N-labeled unfolded protein • Chemical shifts • Dipolar couplings • Relaxation rates • Scalar couplings • Heteronuclear NOE 2. Comparison of NMR parameters - unfolded with random coil parameters (sources: - statistical analysis from unfolded peptides - random coil models (e. g. polymer model, Model-free analysis, Flory etc. ) - unfolded with folded state parameters - different degrees of unfolded states 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 20

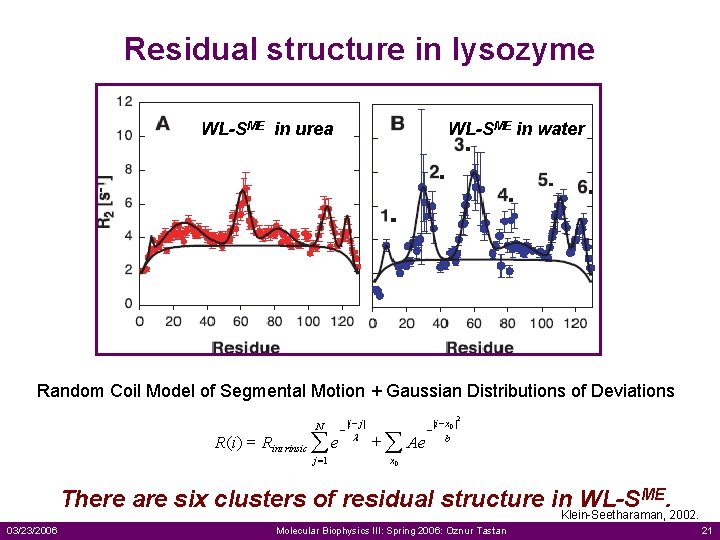
Residual structure in lysozyme WL-SME in urea WL-SME in water 3. 2. 4. 1. 5. 6. Random Coil Model of Segmental Motion + Gaussian Distributions of Deviations N R (i ) = Rint rinsic å e j =1 - |i - j | l + å Ae - |i - x 0 | b 2 x 0 There are six clusters of residual structure in WL-SME. Klein-Seetharaman, 2002. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 21

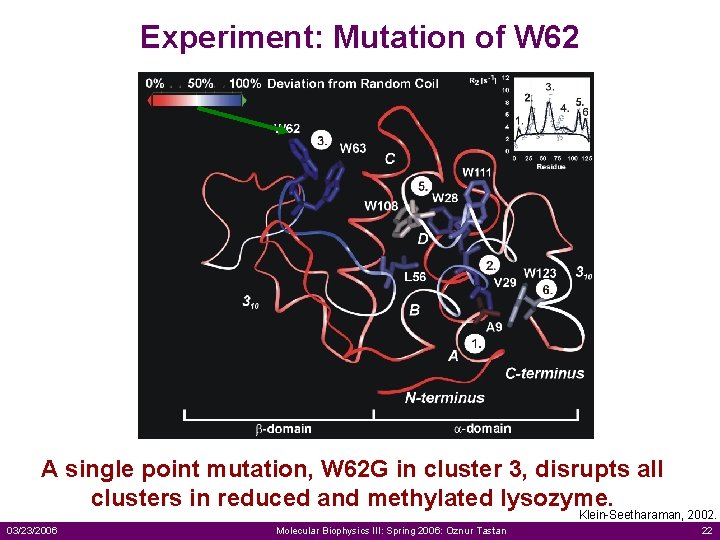
Experiment: Mutation of W 62 A single point mutation, W 62 G in cluster 3, disrupts all clusters in reduced and methylated lysozyme. Klein-Seetharaman, 2002. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 22

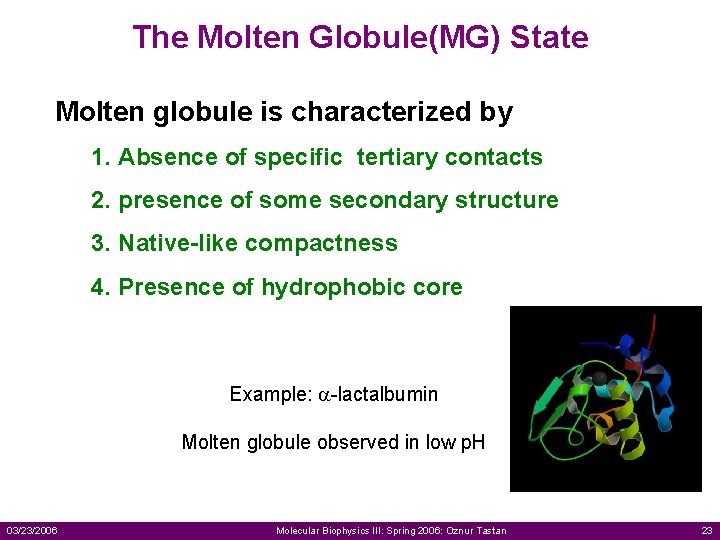
The Molten Globule(MG) State Molten globule is characterized by 1. Absence of specific tertiary contacts 2. presence of some secondary structure 3. Native-like compactness 4. Presence of hydrophobic core Example: a-lactalbumin Molten globule observed in low p. H 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 23

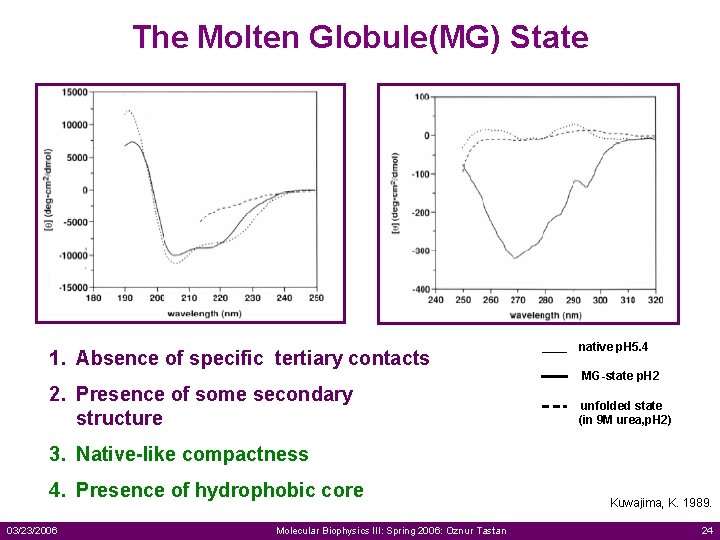
The Molten Globule(MG) State 1. Absence of specific tertiary contacts 2. Presence of some secondary structure native p. H 5. 4 MG-state p. H 2 unfolded state (in 9 M urea, p. H 2) 3. Native-like compactness 4. Presence of hydrophobic core 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan Kuwajima, K. 1989. 24

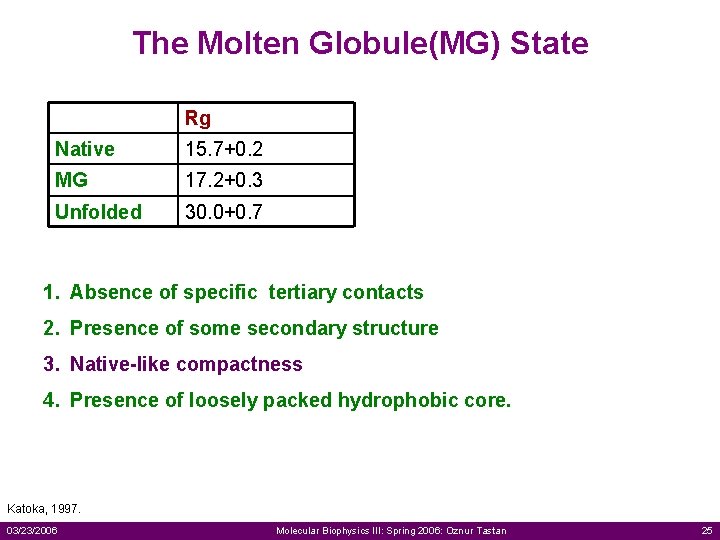
The Molten Globule(MG) State Rg Native 15. 7+0. 2 MG 17. 2+0. 3 Unfolded 30. 0+0. 7 1. Absence of specific tertiary contacts 2. Presence of some secondary structure 3. Native-like compactness 4. Presence of loosely packed hydrophobic core. Katoka, 1997. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 25

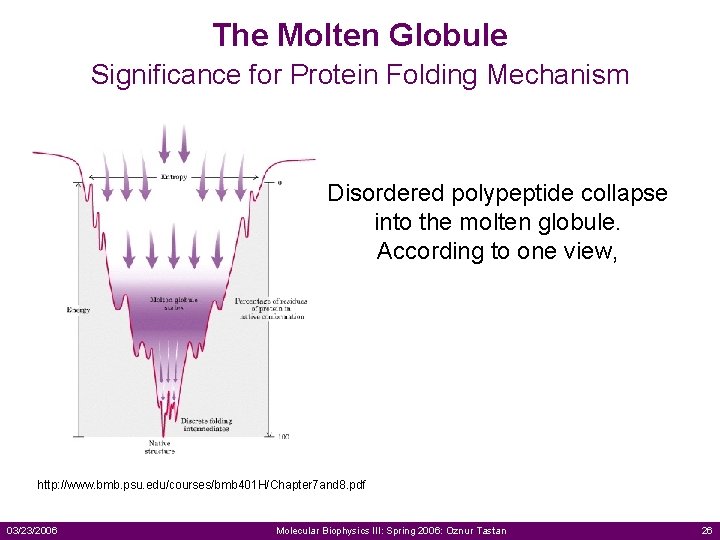
The Molten Globule Significance for Protein Folding Mechanism Disordered polypeptide collapse into the molten globule. According to one view, http: //www. bmb. psu. edu/courses/bmb 401 H/Chapter 7 and 8. pdf 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 26

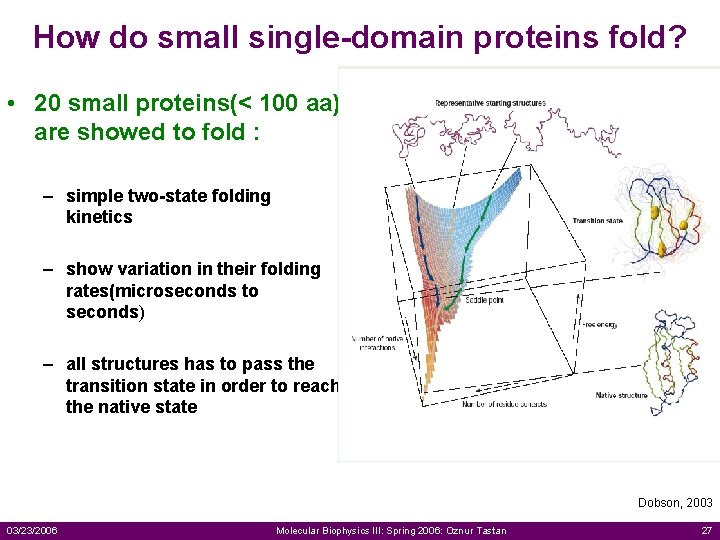
How do small single-domain proteins fold? • 20 small proteins(< 100 aa) are showed to fold : – simple two-state folding kinetics – show variation in their folding rates(microseconds to seconds) – all structures has to pass the transition state in order to reach the native state Dobson, 2003 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 27

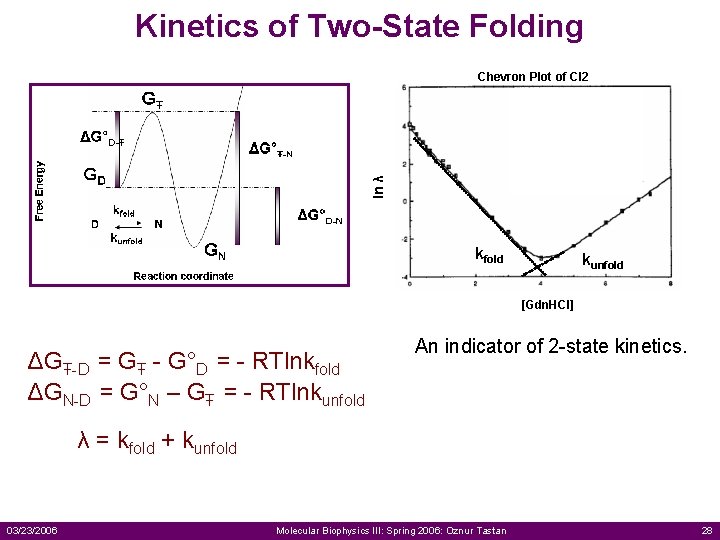
Kinetics of Two-State Folding ln λ Chevron Plot of CI 2 kfold kunfold [Gdn. HCl] ΔGŦ-D = GŦ - G°D = - RTlnkfold ΔGN-D = G°N – GŦ = - RTlnkunfold An indicator of 2 -state kinetics. λ = kfold + kunfold 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 28

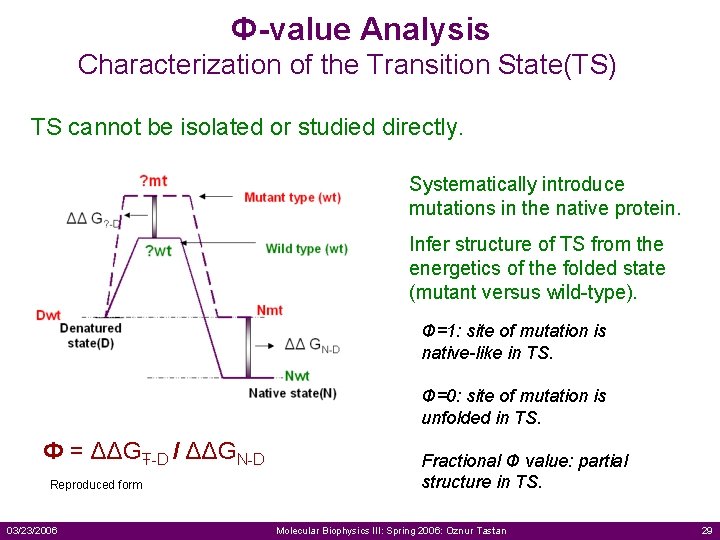
Ф-value Analysis Characterization of the Transition State(TS) TS cannot be isolated or studied directly. Systematically introduce mutations in the native protein. Infer structure of TS from the energetics of the folded state (mutant versus wild-type). Ф=1: site of mutation is native-like in TS. Ф=0: site of mutation is unfolded in TS. Ф = ΔΔGŦ-D / ΔΔGN-D Reproduced form 03/23/2006 Fractional Ф value: partial structure in TS. Molecular Biophysics III: Spring 2006: Oznur Tastan 29

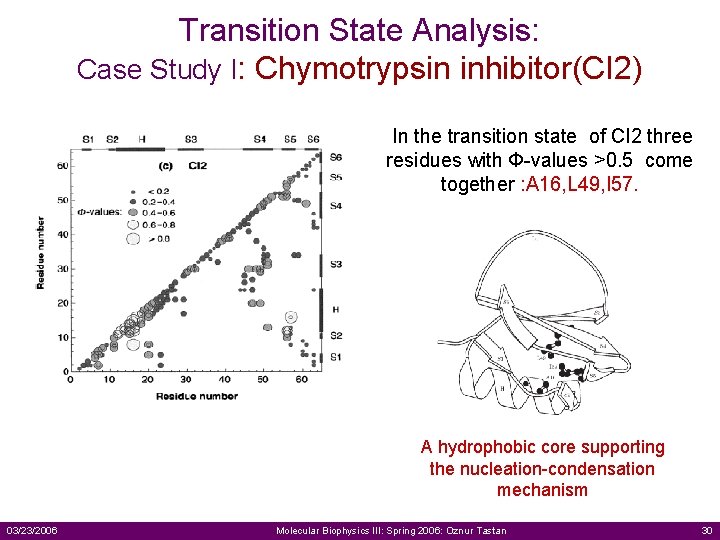
Transition State Analysis: Case Study I: Chymotrypsin inhibitor(CI 2) In the transition state of CI 2 three residues with Ф-values >0. 5 come together : A 16, L 49, I 57. A hydrophobic core supporting the nucleation-condensation mechanism 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 30

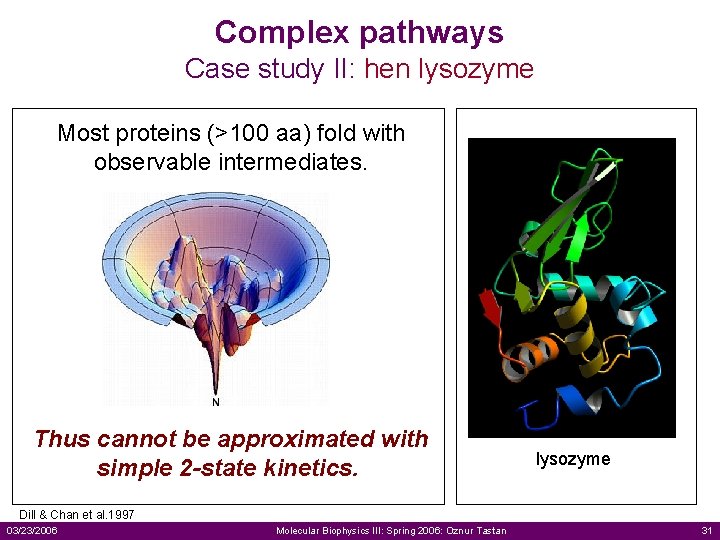
Complex pathways Case study II: hen lysozyme Most proteins (>100 aa) fold with observable intermediates. Thus cannot be approximated with simple 2 -state kinetics. lysozyme Dill & Chan et al. 1997 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 31

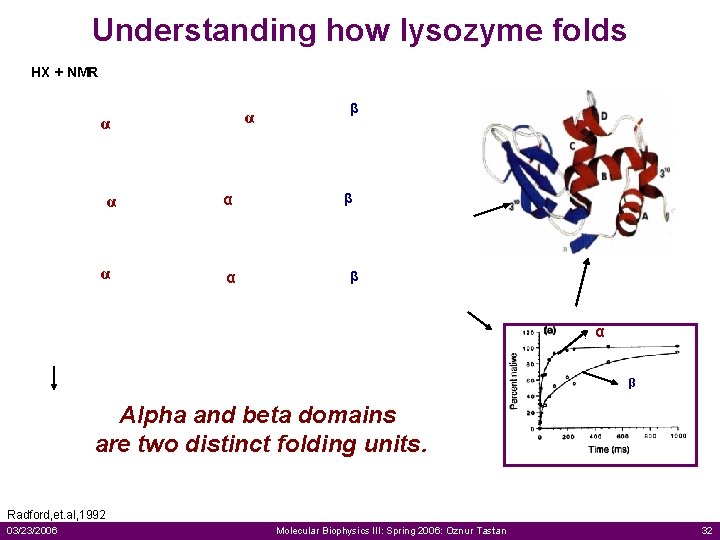
Understanding how lysozyme folds HX + NMR α α α β α β α α β Alpha and beta domains are two distinct folding units. Radford, et. al, 1992 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 32

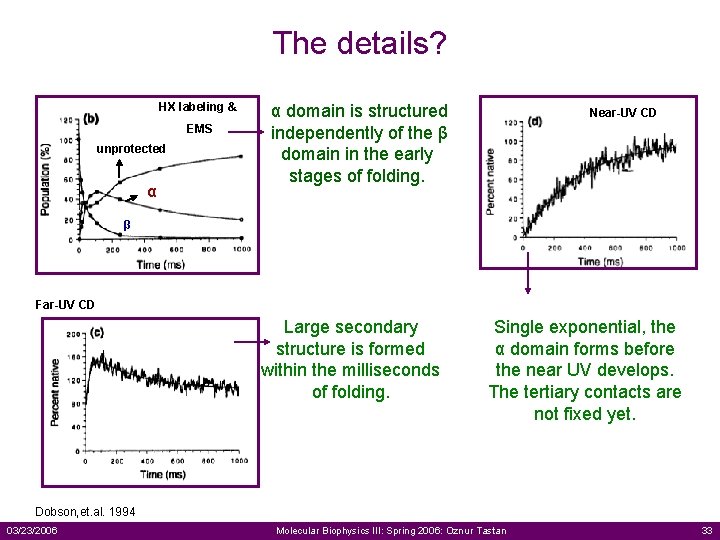
The details? HX labeling & EMS unprotected α α domain is structured independently of the β domain in the early stages of folding. Near-UV CD β Far-UV CD Large secondary structure is formed within the milliseconds of folding. Single exponential, the α domain forms before the near UV develops. The tertiary contacts are not fixed yet. Dobson, et. al. 1994 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 33

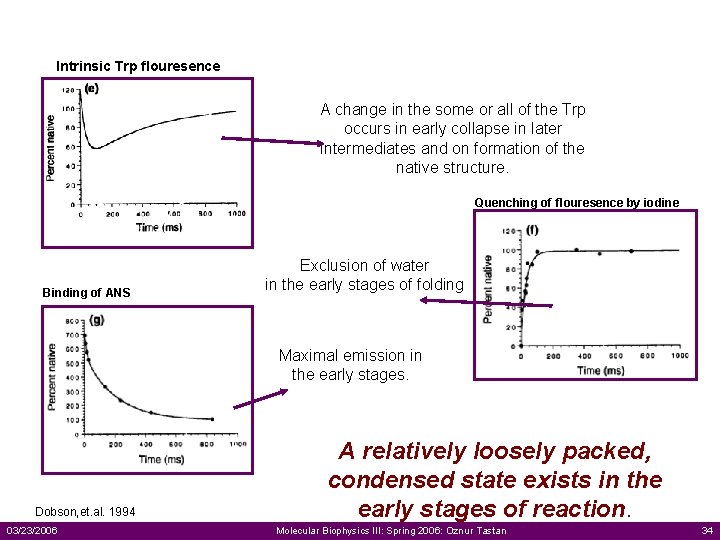
Intrinsic Trp flouresence A change in the some or all of the Trp occurs in early collapse in later intermediates and on formation of the native structure. Quenching of flouresence by iodine Binding of ANS Exclusion of water in the early stages of folding Maximal emission in the early stages. Dobson, et. al. 1994 03/23/2006 A relatively loosely packed, condensed state exists in the early stages of reaction. Molecular Biophysics III: Spring 2006: Oznur Tastan 34

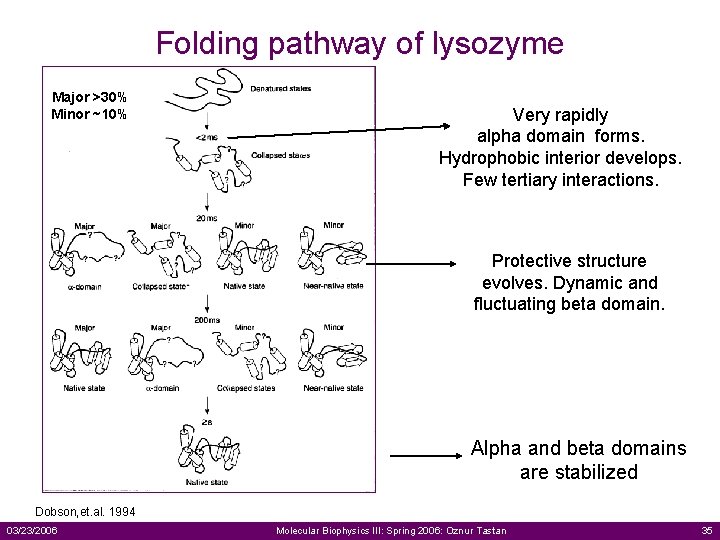
Folding pathway of lysozyme Major >30% Minor ~10% Very rapidly alpha domain forms. Hydrophobic interior develops. Few tertiary interactions. Protective structure evolves. Dynamic and fluctuating beta domain. Alpha and beta domains are stabilized Dobson, et. al. 1994 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 35

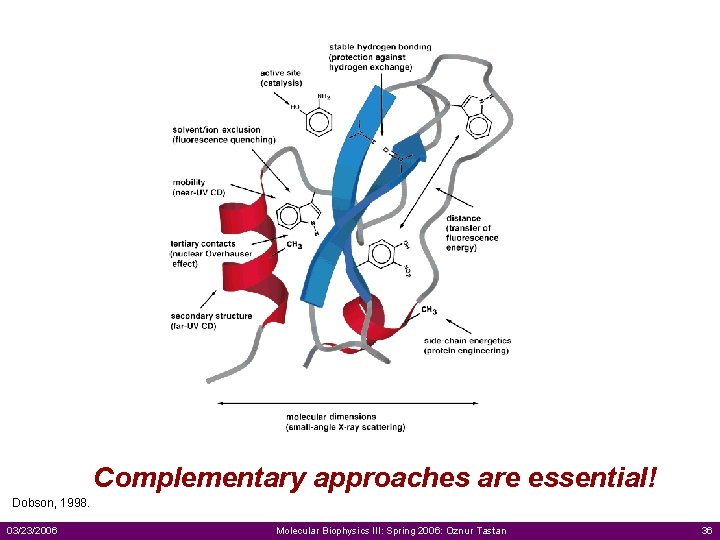
Complementary approaches are essential! Dobson, 1998. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 36

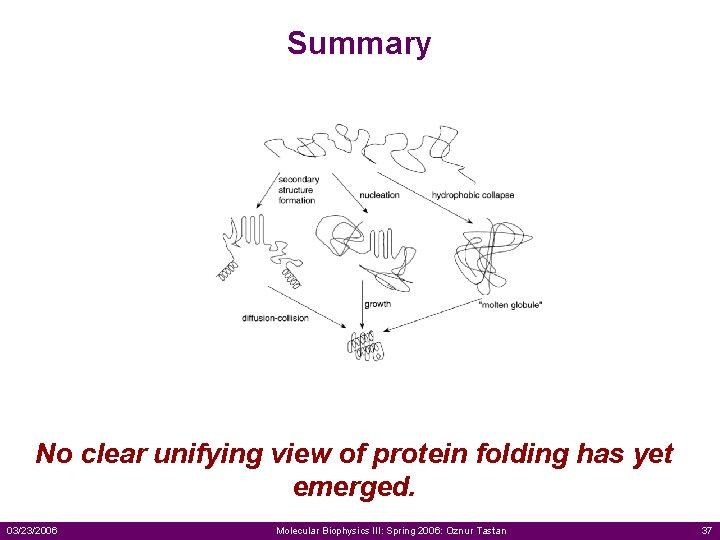
Summary No clear unifying view of protein folding has yet emerged. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 37

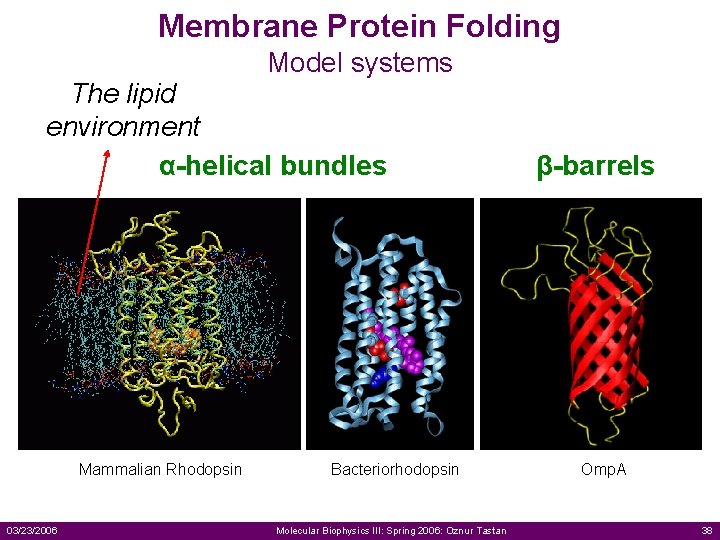
Membrane Protein Folding Model systems The lipid environment α-helical bundles Mammalian Rhodopsin 03/23/2006 Bacteriorhodopsin Molecular Biophysics III: Spring 2006: Oznur Tastan β-barrels Omp. A 38

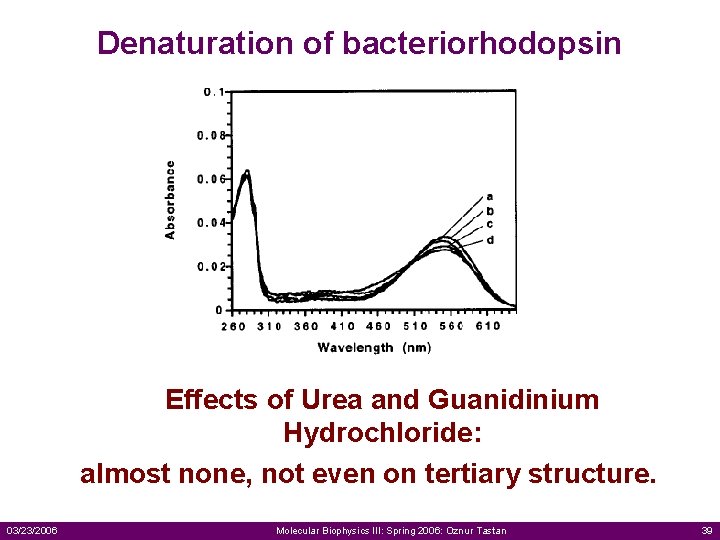
Denaturation of bacteriorhodopsin Effects of Urea and Guanidinium Hydrochloride: almost none, not even on tertiary structure. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 39

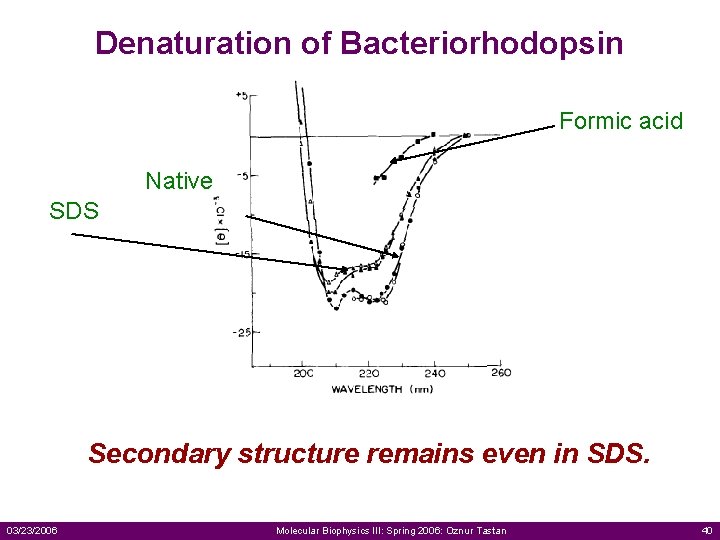
Denaturation of Bacteriorhodopsin Formic acid Native SDS Secondary structure remains even in SDS. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 40

Why is it so difficult to disrupt secondary structure in membrane proteins? 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 41

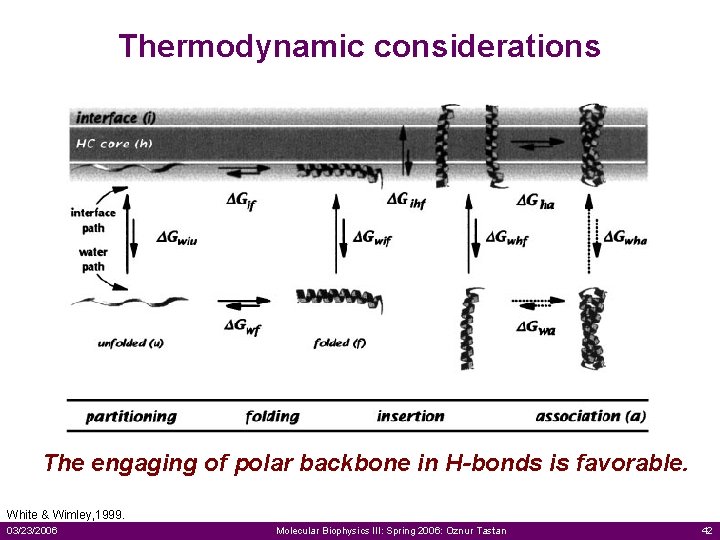
Thermodynamic considerations The engaging of polar backbone in H-bonds is favorable. White & Wimley, 1999. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 42

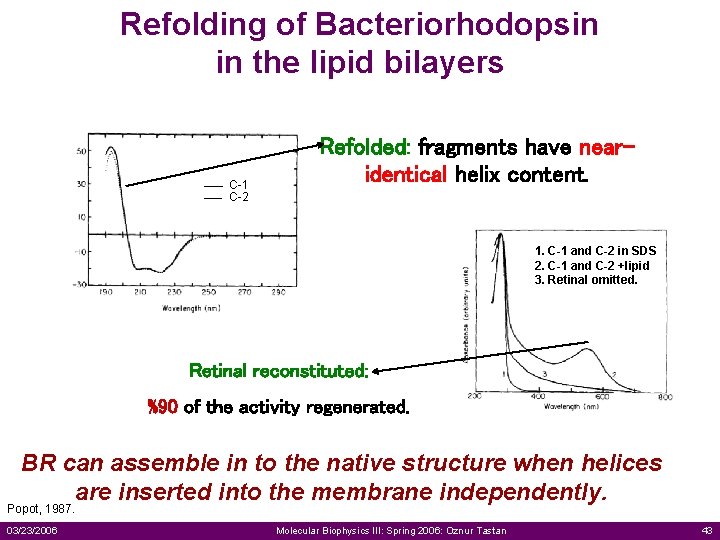
Refolding of Bacteriorhodopsin in the lipid bilayers C-1 C-2 Refolded: fragments have nearidentical helix content. 1. C-1 and C-2 in SDS 2. C-1 and C-2 +lipid 3. Retinal omitted. Retinal reconstituted: %90 of the activity regenerated. BR can assemble in to the native structure when helices are inserted into the membrane independently. Popot, 1987. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 43

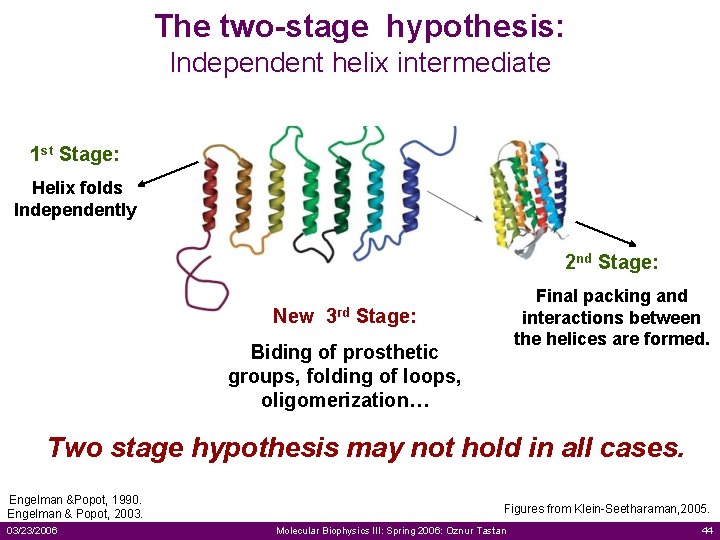
The two-stage hypothesis: Independent helix intermediate 1 st Stage: Helix folds Independently 2 nd Stage: Final packing and interactions between the helices are formed. New 3 rd Stage: Biding of prosthetic groups, folding of loops, oligomerization… Two stage hypothesis may not hold in all cases. Engelman &Popot, 1990. Engelman & Popot, 2003. 03/23/2006 Figures from Klein-Seetharaman, 2005. Molecular Biophysics III: Spring 2006: Oznur Tastan 44

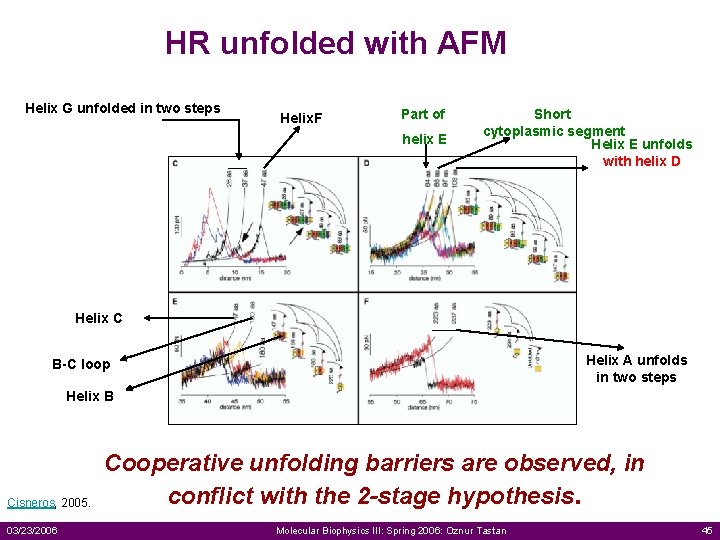
HR unfolded with AFM Helix G unfolded in two steps Helix. F Part of helix E Short cytoplasmic segment Helix E unfolds with helix D Helix C Helix A unfolds in two steps B-C loop Helix B Cisneros, 2005. 03/23/2006 Cooperative unfolding barriers are observed, in conflict with the 2 -stage hypothesis. Molecular Biophysics III: Spring 2006: Oznur Tastan 45

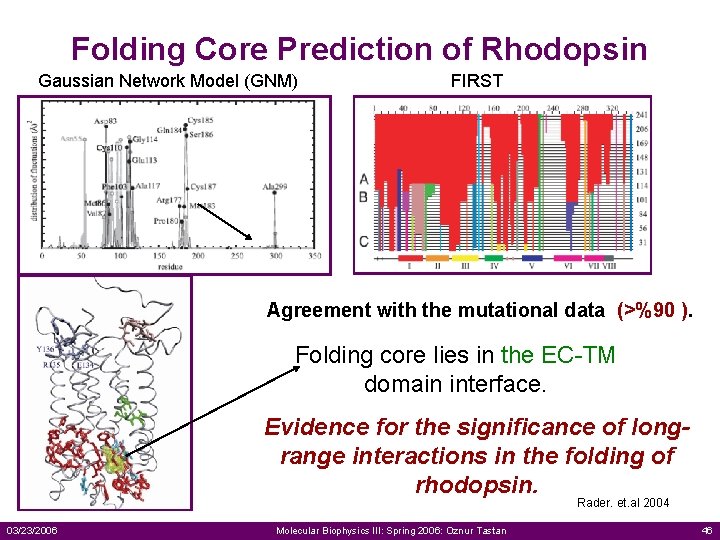
Folding Core Prediction of Rhodopsin Gaussian Network Model (GNM) FIRST Agreement with the mutational data (>%90 ). Folding core lies in the EC-TM domain interface. Evidence for the significance of longrange interactions in the folding of rhodopsin. Rader. et. al 2004 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 46

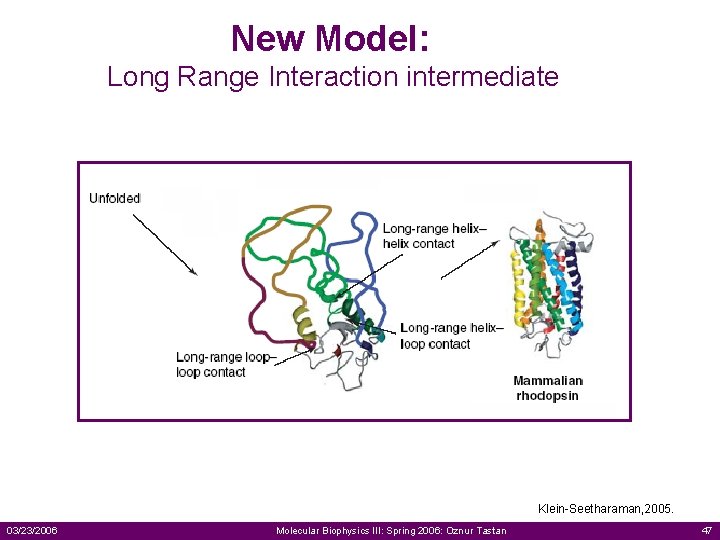
New Model: Long Range Interaction intermediate Klein-Seetharaman, 2005. 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 47

References • • • • • • 03/23/2006 Anfinsen, C. B. (1973) "Principles that govern the folding of protein chains. " Science 181 223 -230. Cisneros, D. A. , D. Oesterhelt, and D. J. Muller, Probing origins of molecular interactions stabilizing the membrane proteins halorhodopsin and bacteriorhodopsin. Structure, 2005. 13(2): p. 235 -42. Dobson, C. M. Sali A. , and Karplus, M. , "Protein Folding: A Perspective from Theory and Experiment", Angew. Chem. Int. Ed. Eng. 37, 868 -893 ( 1998). Dobson, C. M. "Protein Folding and Misfolding", Nature 426, 884 -890 ( 2003). Dobson, C. M. , P. A. Evans, and S. E. Radford, Understanding how proteins fold: the lysozyme story so far. Trends Biochem Sci, 1994. 19(1): p. 31 -7. Engelman, D. M. , Chen, Y. , Chin, C. N. , Curran, R. , Dixon, A. M. , Dupuy, A, Lee, A. , Lehnert, U. , Matthews, E. , Reshetnyak, Y. , Senes, A. , Popot, J-L. “Membrane Protein Folding: Beyond the Two Stage Model” FEBS Lett. (2003) 555: 122 -5. Flory, P. J. (1969) Statistical Mechanics of Chain Molecules (Wiley, New York). Fitzkee, N. C. and Rose, G. D. (2004). Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. 101: 12497– 12502. Klein-Seetharaman, J. , Oikawa, M. , Wirmer, J. , Duchardt, E. , Ueda, T. , Imoto, T. , Smith, L. J. , Dobson, C. and Schwalbe, H. (2002) Long-Range Interactions within a Non-Native Protein. Science 295, 1719 -1722. Klein-Seetharaman, J. (2005) Dual role of interactions between membranous and soluble portions of helical membrane receptors for folding and signaling. Trends in Pharmacological Science 26(4), 183 -189 Kuwajima, K. (1989). The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins: Struct. Funct. Genet. 6: 87– 103. Kataoka, M. , Y. Hagihara, K. Mihara, and Y. Goto. (1993). Molten globule of cytochrome c studied by the small angle X-ray scattering. J. Mol. Biol. 229: 591 -596. Radford, S. E. , Dobson, C. M. & Evans, P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature 358, 302 -307 (1992). Pappu, R. V. , Srinivasan, R. & Rose, G. D. (2000) Proc. Natl. Acad. Sci. USA 97, 12565 -12570. Popot, J-L and Engelman D. M. "Membrane Protein Folding and Oligomerization: The Two-Stage Model“ Biochemistry (1990), 29 (17), 4031 -7. Popot J. L. , Gerchman S. E. , Engelman D. M. (1987) Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J. Mol. Biol. 198: 655 -76 Shortle D, Ackerman MS. (2001) Persistence of native-like topology in a denatured protein in 8 M urea. Science. Jul 20; 293(5529): 487 -9. White S. H. and Wimley, W. C. (1999). Membrane protein folding and stability: Physical principles. Annu. Rev. Biophys. Biomol. Struct. 28: 319365. http: //www. otago. ac. nz/humannutrition/dietetics/gfx/philosophy. jpg, March 22, 2006. http: //www-nmr. cabm. rutgers. edu/academics/biochem 694/2006 Bio. Chem 412/Biochem. 412_2 -24 -2006 lecture. pdf, March 22, 2006. http: //www. makro. ch. tum. de/users/BFHZ/Scheibel%202003%20 Bordeaux-1. pdf, March 22, 2006. Molecular Biophysics III: Spring 2006: Oznur Tastan 48

Acknowledgements Dr. Judith Klein-Seetharaman Dr. Sanford Leuba & The class of MB 3 (Spring 2006) 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 49

QUESTIONS? http: //www. otago. ac. nz/humannutrition/dietetics/gfx/philosophy. jpg 03/23/2006 Molecular Biophysics III: Spring 2006: Oznur Tastan 50
 Protein denaturation egg white
Protein denaturation egg white Hydrophobic collapse in protein folding
Hydrophobic collapse in protein folding Sefer baday
Sefer baday Role of chaperones in protein folding ppt
Role of chaperones in protein folding ppt Garland science
Garland science Protein folding
Protein folding Protein folding
Protein folding Hmp significance
Hmp significance Glycolysis energetics
Glycolysis energetics Significance of hmp shunt in biochemistry
Significance of hmp shunt in biochemistry Sodium azide electron transport chain
Sodium azide electron transport chain Energetics power tower 180
Energetics power tower 180 Gluconeogenesis from lactate
Gluconeogenesis from lactate Building energetics
Building energetics Ib energetics
Ib energetics Energetics janesville
Energetics janesville Calorimetry 2 chemsheets answers 2015
Calorimetry 2 chemsheets answers 2015 Carrier vs channel proteins
Carrier vs channel proteins Protein-protein docking
Protein-protein docking What is modals and semi modals
What is modals and semi modals Kinetics and equilibrium
Kinetics and equilibrium Explain law of mass action
Explain law of mass action Metaboloism
Metaboloism Kinetics of rigid bodies
Kinetics of rigid bodies Kinetics of a particle: force and acceleration
Kinetics of a particle: force and acceleration Kinetics of a particle: impulse and momentum
Kinetics of a particle: impulse and momentum Hemotology
Hemotology Kinematics of rigid bodies problems and solutions
Kinematics of rigid bodies problems and solutions Planar kinetics of a rigid body work and energy
Planar kinetics of a rigid body work and energy Cell kinetics and fermenter design
Cell kinetics and fermenter design Define folding faulting and volcanic activity
Define folding faulting and volcanic activity Types of folds
Types of folds Overfold diagram
Overfold diagram At a country concert the ratio of the number
At a country concert the ratio of the number Regional metamorphism
Regional metamorphism How are fold mountains formed
How are fold mountains formed Molekülerite nedir
Molekülerite nedir Zeroth order kinetics
Zeroth order kinetics Rigid body kinetics
Rigid body kinetics Collision theory of kinetics
Collision theory of kinetics Grade 11 chemistry unit 4
Grade 11 chemistry unit 4 Kinetics flotation reagents
Kinetics flotation reagents Kinetics order
Kinetics order Chemical kinetics definition
Chemical kinetics definition Kinetics of rigid bodies engineering mechanics
Kinetics of rigid bodies engineering mechanics Kinetics ap chemistry
Kinetics ap chemistry Kinetics of crystal violet fading
Kinetics of crystal violet fading Octet kinetics
Octet kinetics Km in enzyme kinetics
Km in enzyme kinetics Kcat equation mcat
Kcat equation mcat Kinetics of particles newton's second law
Kinetics of particles newton's second law