DENATURATION OF PROTEIN DENATURATION Denaturation is a process


















- Slides: 32

DENATURATION OF PROTEIN

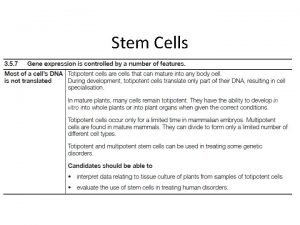
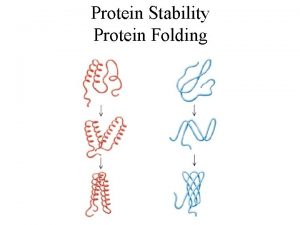
DENATURATION Denaturation is a process in which a protein loses its native shape due to the disruption of weak chemical bonds and interactions, thereby becoming biologically inactive. When proteins denature, the cells go through a series of changes, first loosening, then tightening. In case of proteins : • A loss of three-dimensional structure, sufficient to cause loss of function ⟰ Loss of secondary, tertiary and quaternary structure of proteins. ⟰ Change in physical, chemical and biological properties of protein molecules.

FOR EXAMPLE: i. Changing p. H denatures proteins because it changes the charges on many of the side chains. This disrupts electrostatic attractions and hydrogen bonds. ii. Certain reagents such as urea and guanidine hydrochloride denature proteins by forming hydrogen bonds to the protein groups that are stronger than the hydrogen bonds formed between the groups. iii. Detergents such as sodium dodecyl sulphate denature proteins by associating with the non-polar groups of protein, thus interfering with the normal hydrophobic interactions. iv. Organic solvents such as acetone alcohols denature proteins by disrupting hydrophobic interactions. v. Proteins can also be denatured by heat. Heat increase molecular motion which can disrupt the attractive forces.

• None of these agents breaks peptide bonds, so the primary structure of a protein remains intact when it is denatured. • When a protein is denatured, it loses its function. Example: • A denatured enzyme ceases to function. • A denatured antibody no longer can bind its antigen.

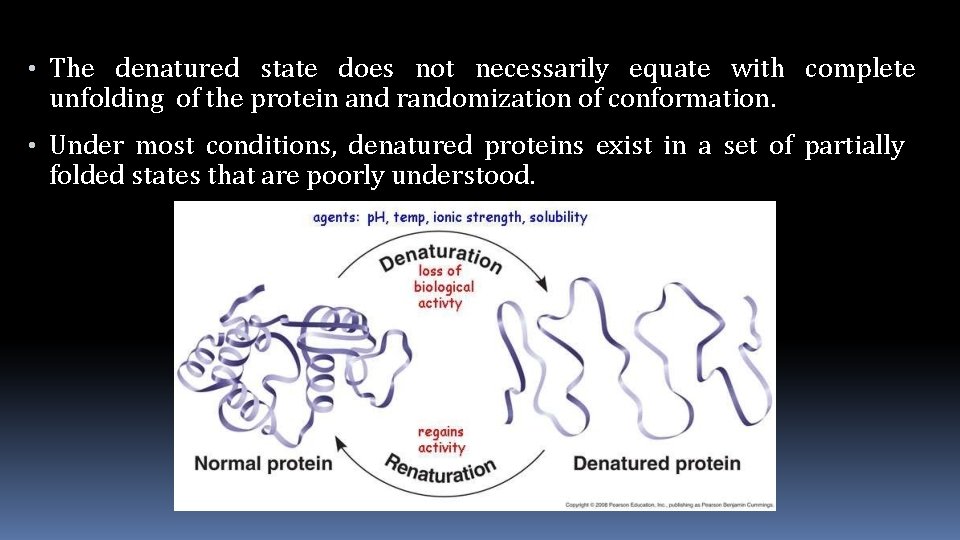
• The denatured state does not necessarily equate with complete unfolding of the protein and randomization of conformation. • Under most conditions, denatured proteins exist in a set of partially folded states that are poorly understood.

AGENTS OF DENATURATION Physical agents : 1. Heat, 2. Violent shaking, 3. X-rays, 4. Hydrostatic Pressure (5, 000 – 10, 000 atm) 5. UV radiation.

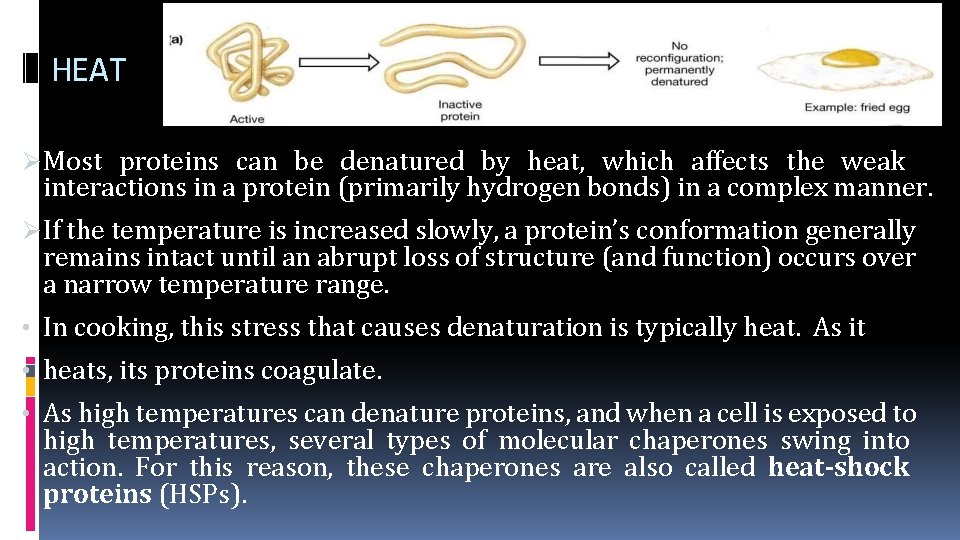
HEAT Most proteins can be denatured by heat, which affects the weak interactions in a protein (primarily hydrogen bonds) in a complex manner. If the temperature is increased slowly, a protein’s conformation generally remains intact until an abrupt loss of structure (and function) occurs over a narrow temperature range. • In cooking, this stress that causes denaturation is typically heat. As it • heats, its proteins coagulate. • As high temperatures can denature proteins, and when a cell is exposed to high temperatures, several types of molecular chaperones swing into action. For this reason, these chaperones are also called heat-shock proteins (HSPs).


VIOLENT SHAKING • Agitation also denatures protein. • We see this clearly in the whipping of egg whites. • In nature, you can see the denaturation of protein in the waves at the beach. The constant churning creates foam from various proteins in the sea water.

HYDROSTATIC PRESSURE (5, 000 – 10, 000 atm) • Pressure destabilization of hydrophobic aggregates by using an information theory model of hydrophobic interactions. • Pressure-denatured proteins, unlike heat-denatured proteins, retain a compact structure with water molecules penetrating their core.

UV RADIATION • UV radiation supplies kinetic energy to protein molecules, causing their atoms to vibrate more rapidly and disrupting relatively weak hydrogen bonding and dispersion forces. [http: //www. ncbi. nlm. nih. gov/pubmed/8933720]

CHEMICAL AGENTS : • Acids and alkalies • Organic solvents (ether, alcohol), • Salts of heavy metals (Pb, Hg), • Chaotropic agents • Detergents • Altered p. H

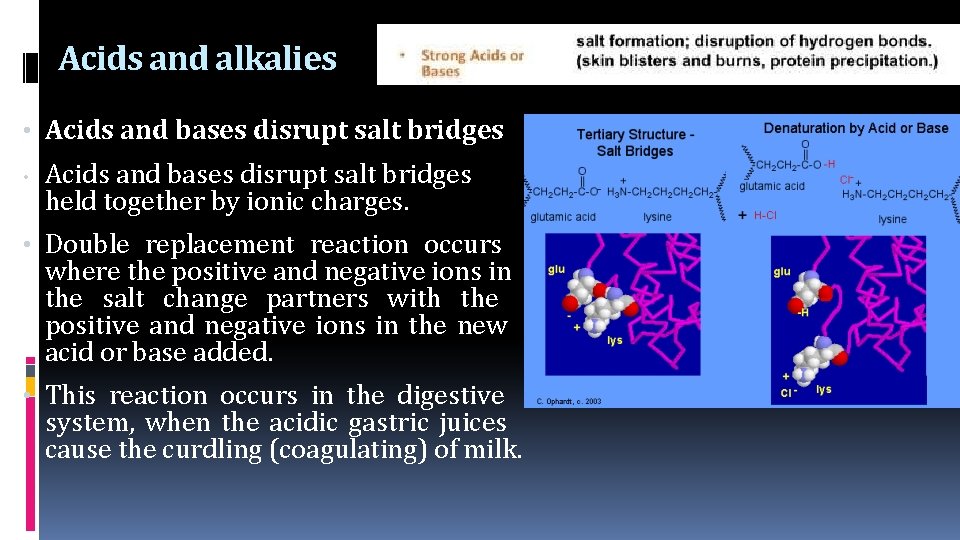
Acids and alkalies • Acids and bases disrupt salt bridges • Acids and bases disrupt salt bridges held together by ionic charges. • Double replacement reaction occurs where the positive and negative ions in the salt change partners with the positive and negative ions in the new acid or base added. • This reaction occurs in the digestive system, when the acidic gastric juices cause the curdling (coagulating) of milk.

ACIDIC PROTEIN DENATURANTS INCLUDE: • Acetic acid • Trichloroacetic acid 12% in water • Sulfosalicylic acid Bases work similarly to acids in denaturation. • Sodium bicarbonate

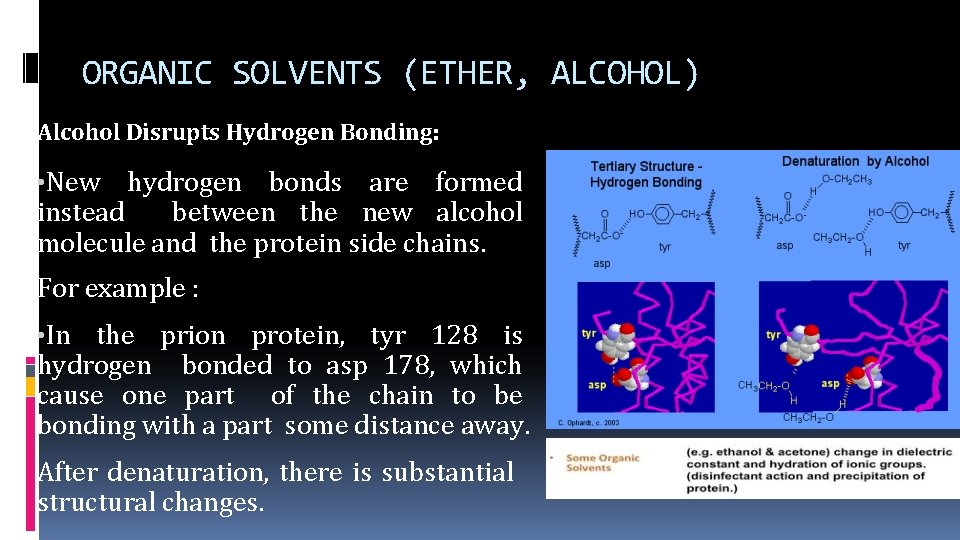
ORGANIC SOLVENTS (ETHER, ALCOHOL) Alcohol Disrupts Hydrogen Bonding: • New hydrogen bonds are formed instead between the new alcohol molecule and the protein side chains. For example : • In the prion protein, tyr 128 is hydrogen bonded to asp 178, which cause one part of the chain to be bonding with a part some distance away. After denaturation, there is substantial structural changes.

Salts of heavy metals (Pb, Hg), • Heavy metal salts usually contain Hg+2, Pb+2, Ag+1 Tl+1, Cd+2 and other metals with high atomic weights. • Since salts are ionic they disrupt salt bridges in proteins. • The reaction of a heavy metal salt with a protein usually leads to an insoluble metal protein salt • This reaction is used for its disinfectant properties in external applications. For example – ⎎ Ag. NO 3 is used to prevent gonorrhea infections in the eyes of new born infants. Silver nitrate is also used in the treatment of nose and throat infections, as well as to cauterize wounds. “Heavy metals may also disrupt disulfide bonds because of their high affinity and attraction for sulfur and will also lead to the denaturation of proteins. ”

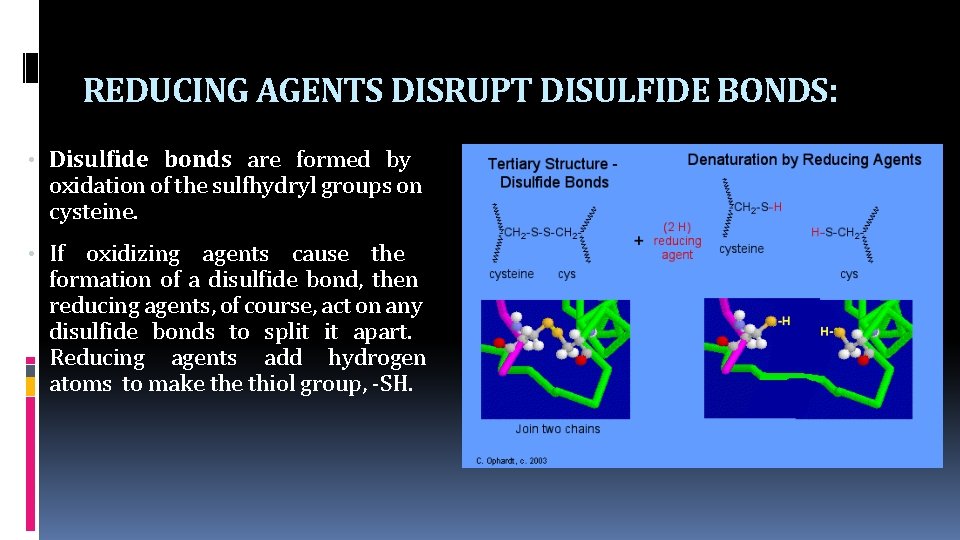
REDUCING AGENTS DISRUPT DISULFIDE BONDS: • Disulfide bonds are formed by oxidation of the sulfhydryl groups on cysteine. • If oxidizing agents cause the formation of a disulfide bond, then reducing agents, of course, act on any disulfide bonds to split it apart. Reducing agents add hydrogen atoms to make thiol group, -SH.

CHAOTROPIC AGENTS Chaotropic agents include: 1. Urea 6 – 8 mol/l 2. Guanidinium chloride 6 mol/l 3. Lithium perchlorate 4. 5 mol/l

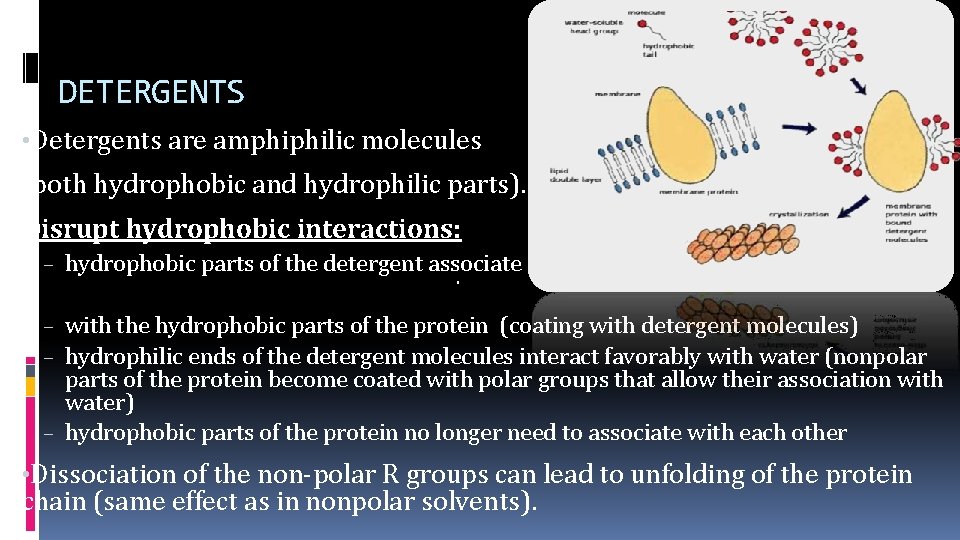
DETERGENTS • Detergents are amphiphilic molecules (both hydrophobic and hydrophilic parts). Example: Disrupt hydrophobic interactions: – hydrophobic parts of the detergent associate – with the hydrophobic parts of the protein (coating with detergent molecules) – hydrophilic ends of the detergent molecules interact favorably with water (nonpolar parts of the protein become coated with polar groups that allow their association with water) – hydrophobic parts of the protein no longer need to associate with each other • Dissociation of the non-polar R groups can lead to unfolding of the protein chain (same effect as in nonpolar solvents).

DISULFIDE BOND REDUCERS Agents that break disulfide bonds by reduction include: 1. 2 -Mercaptoethanol 2. Dithiothreitol 3. TCEP (tris(2 -carboxyethyl)phosphine)

CROSS-LINKING REAGENTS Cross-linking agents for proteins include: 1. Formaldehyde 2. Glutaraldehyde

CHARACTERISTICS OF DENATURATION ⧗ The native helical structure of protein is lost ⧗ The primary structure of a protein with peptide linkages remains intact i. e. , peptide bonds are not hydrolyzed. ⧗ The protein loses its biological activity. ⧗ Denatured protein becomes insoluble in the solvent in which it was originally soluble. ⧗ The viscosity of denatured protein (solution) increases while its surface tension decreases. ⧗ Denaturation is associated with increase in ionizable and sulfhydryl groups of protein. This is due to loss of hydrogen and disulfide bonds.

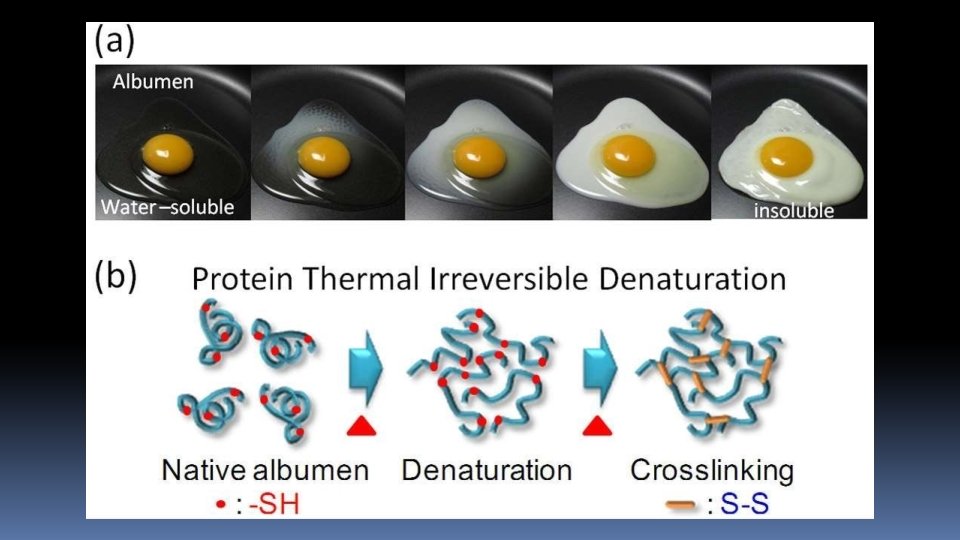
⧗ Denatured protein is more easily digested. This is due to increased exposure of peptide bonds to enzymes Cooking causes protein denaturation and, therefore, cooked food (protein) is more easily digested. ⧗ Denaturation is usually irreversible. For instance, omelet can be prepared from an egg (protein-albumin) but the reversal is not possible. ⧗ Careful denaturation is sometimes reversible (known as renaturation). For example - Hemoglobin undergoes denaturation in the presence of salicylate. By removal of salicylate, hemoglobin is renatured.

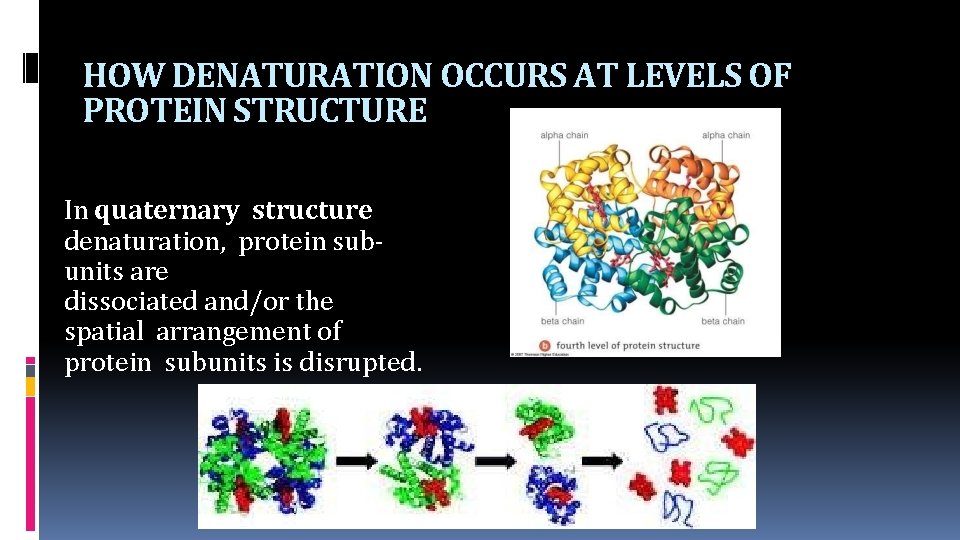
HOW DENATURATION OCCURS AT LEVELS OF PROTEIN STRUCTURE In quaternary structure denaturation, protein subunits are dissociated and/or the spatial arrangement of protein subunits is disrupted.

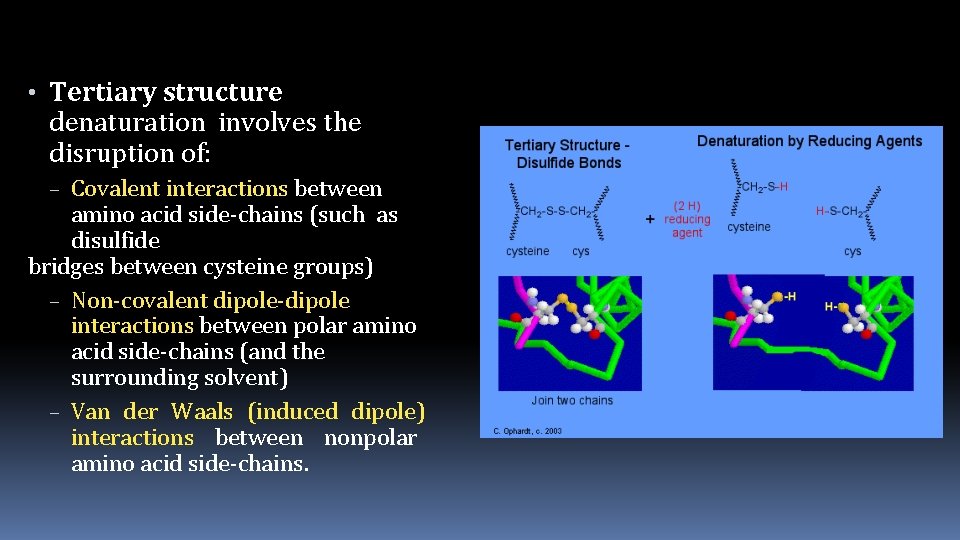
• Tertiary structure denaturation involves the disruption of: – Covalent interactions between amino acid side-chains (such as disulfide bridges between cysteine groups) – Non-covalent dipole-dipole interactions between polar amino acid side-chains (and the surrounding solvent) – Van der Waals (induced dipole) interactions between nonpolar amino acid side-chains.

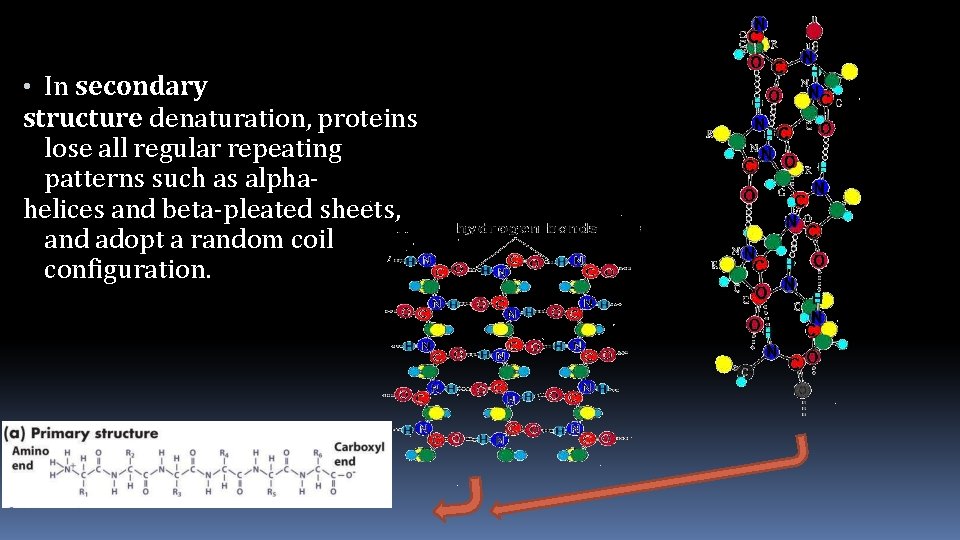
• In secondary structure denaturation, proteins lose all regular repeating patterns such as alphahelices and beta-pleated sheets, and adopt a random coil configuration.

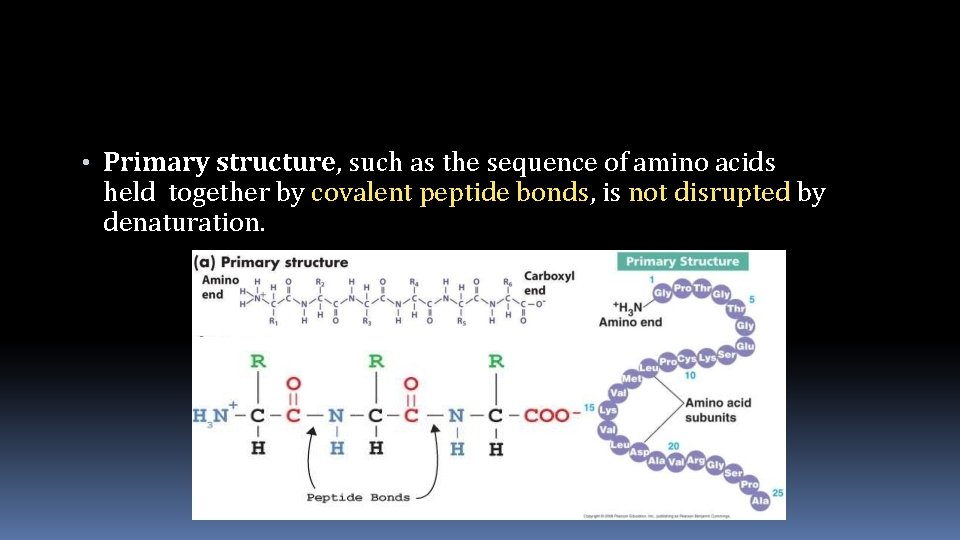
• Primary structure, such as the sequence of amino acids held together by covalent peptide bonds, is not disrupted by denaturation.

LOSS OF FUNCTION • Most biological substrates lose their biological function when denatured. • For example, enzymes lose their activity, because the substrates can no longer bind to the active site, • The denaturing process and the associated loss of activity can be measured using techniques such as • Dual polarization interferometry, • CD [Circular dichroism], and • QCMD [Quality Control for Molecular Diagnostics ].

CONCLUSION / APPLICATIONS OF DENATURATION • The study of Denaturation of protein (transporters, enzymes, hormones) is useful in the field of Proteomics. • To determines the percentage and conc. of alcohol that could lyse the bacterial and viral protein and cell wall but not the skin.

The restoration of native structure of a protein by returning the protein solution to native conditions is known as ‘renaturation’ of the protein. Most of the times, proteins can be renatured simply by reversing the process used to denature them: 1. Gradually decrease temperature of solution: If temperature is reduced too fast, the protein may be trapped in a local minimum rather than reaching native state, with insufficient energy to escape the local minimum. 2. Remove denaturant (organic solvent): Reconstituting the protein in aqueous solution of appropriate p. H and osmolarity is generally sufficient to renature protein. 3. Re-adjust p. H to native conditions: For most aqueous proteins, this is in the range of 6. 5 – 7. 5. However, if the protein in question is from a compartment with a different p. H (such as the thylakoid lumen or lysosome), then renaturation would occur at that p. H. It should be noted that the principles of protein folding and renaturation apply only to aqueous proteins, and not membrane proteins.

REFERENCES: • http: //elmhurst. edu/~chm/vchembook/568 denaturation. html • http: //en. wikipedia. org/wiki/Denaturation_(biochemistry)#Los s_of_function • http: //elmhurst. edu/~chm/vchembook/567 tertprotein. html

THANK YOU
 Coagulation eggs definition
Coagulation eggs definition Alkaloidal reagents denature proteins
Alkaloidal reagents denature proteins Protein denaturation egg white
Protein denaturation egg white Protein hydrolysis
Protein hydrolysis Protein pump vs protein channel
Protein pump vs protein channel Protein-protein docking
Protein-protein docking Thalassemia
Thalassemia Egg white denaturation
Egg white denaturation Which best summarizes the process of protein synthesis?
Which best summarizes the process of protein synthesis? Fictional character μετάφραση
Fictional character μετάφραση Coronoid process and coracoid process
Coronoid process and coracoid process Procedural due process vs substantive due process
Procedural due process vs substantive due process Business process levels
Business process levels Ergodic process in random process
Ergodic process in random process What is process to process delivery
What is process to process delivery Condylar process and coronoid process
Condylar process and coronoid process Stable quality
Stable quality Process-to-process delivery
Process-to-process delivery Sweet process review
Sweet process review Summative and subjective assessment
Summative and subjective assessment Adinda olivia
Adinda olivia Brandi needs her protein
Brandi needs her protein Alpha helix and beta sheet
Alpha helix and beta sheet Eaas benefits
Eaas benefits Institute for protein innovation
Institute for protein innovation Chemistry of life summary
Chemistry of life summary Sel haiwan dan fungsi
Sel haiwan dan fungsi Fibroin secondary structure
Fibroin secondary structure Rna protein synthesis
Rna protein synthesis Protein hydrolysis
Protein hydrolysis Warburg-christian method
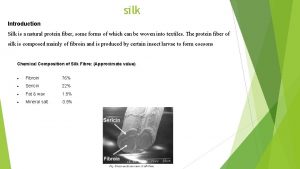
Warburg-christian method Silk is a natural protein fiber some forms of which can be
Silk is a natural protein fiber some forms of which can be Protein fractionation by charge
Protein fractionation by charge