Chapter 3 Cells The Living Units Part D





- Slides: 61

Chapter 3 Cells: The Living Units: Part D Cell Cycle • Defines changes from formation of the cell until it reproduces • Includes: • Interphase • Cell division (mitotic phase) Copyright © 2010 Pearson Education, Inc.

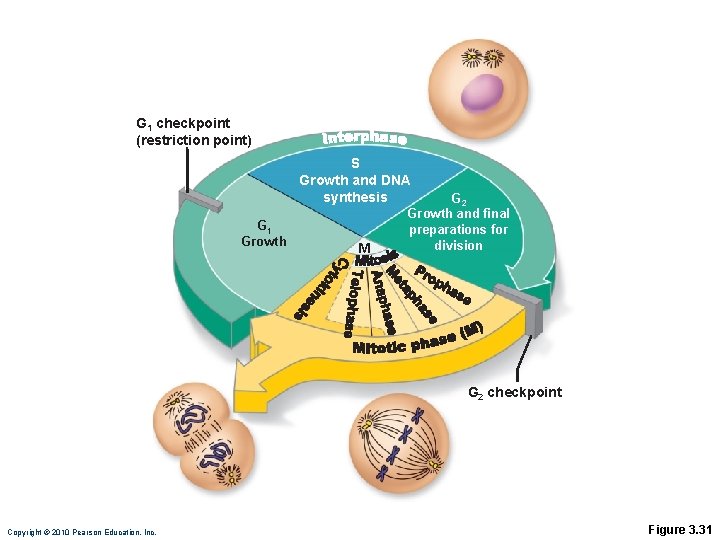
Interphase • Period from cell formation to cell division • Nuclear material called chromatin • Four subphases: • G 1 (gap 1)—vigorous growth and metabolism • G 0—gap phase in cells that permanently cease dividing • S (synthetic)—DNA replication • G 2 (gap 2)—preparation for division Copyright © 2010 Pearson Education, Inc.

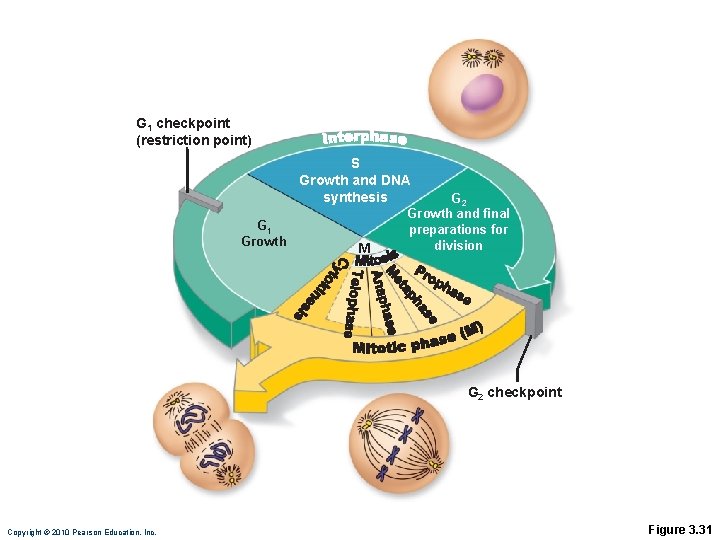
G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 2 checkpoint Copyright © 2010 Pearson Education, Inc. Figure 3. 31

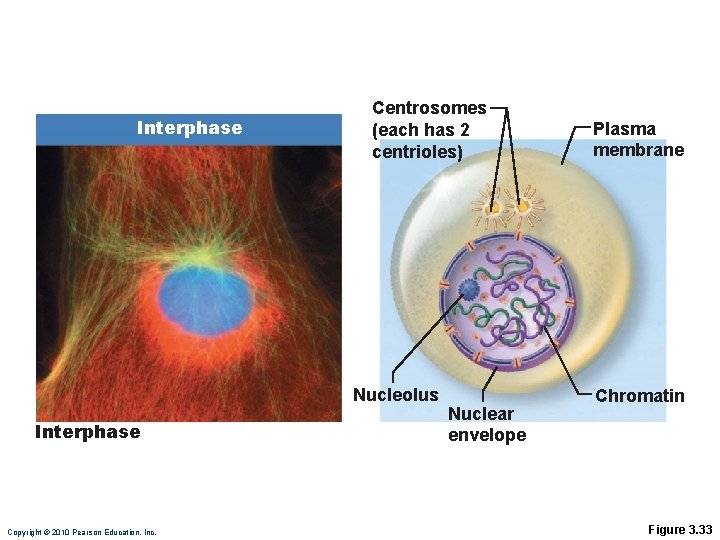
Interphase Centrosomes (each has 2 centrioles) Nucleolus Interphase Copyright © 2010 Pearson Education, Inc. Nuclear envelope Plasma membrane Chromatin Figure 3. 33

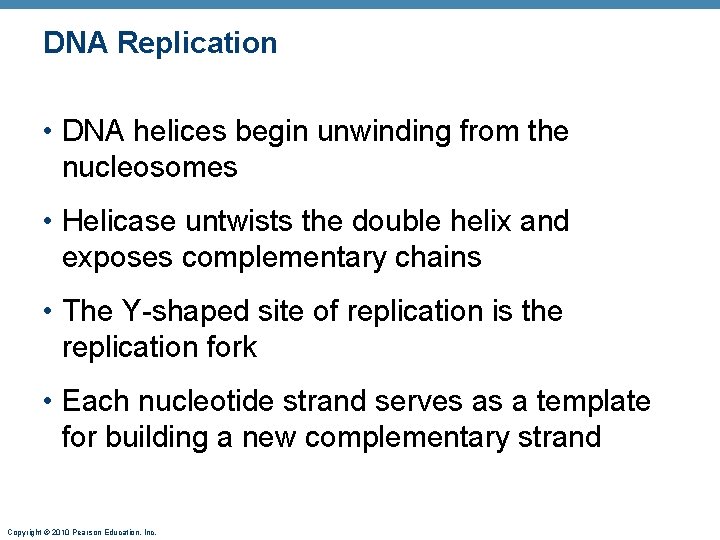
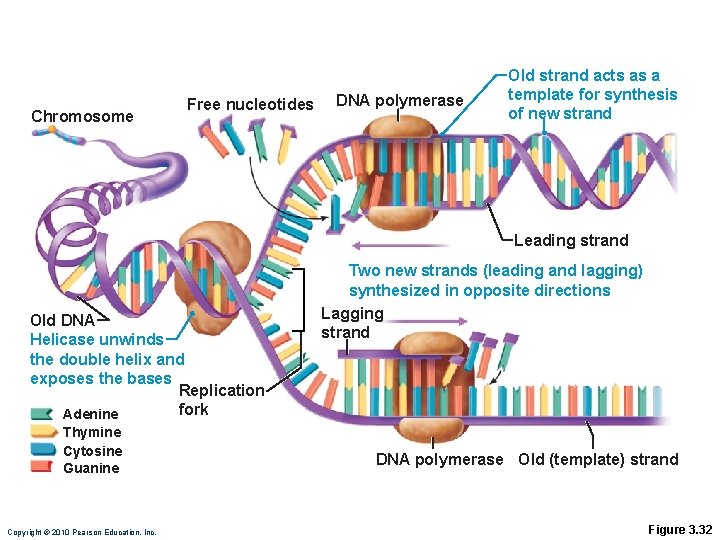
DNA Replication • DNA helices begin unwinding from the nucleosomes • Helicase untwists the double helix and exposes complementary chains • The Y-shaped site of replication is the replication fork • Each nucleotide strand serves as a template for building a new complementary strand Copyright © 2010 Pearson Education, Inc.

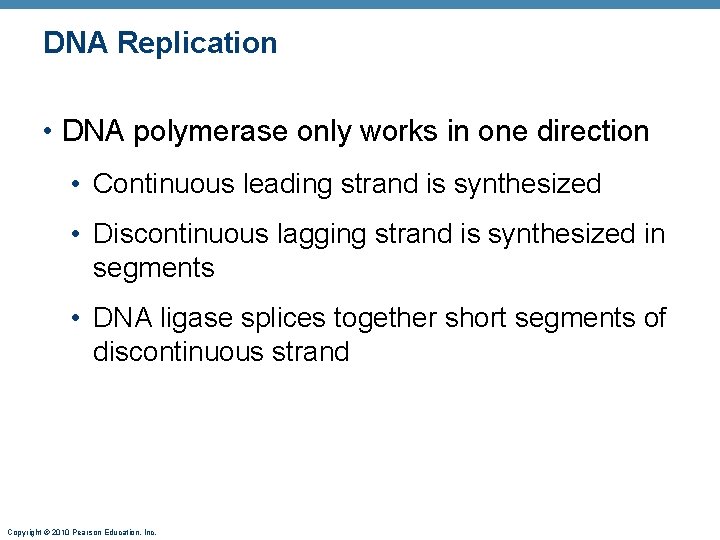
DNA Replication • DNA polymerase only works in one direction • Continuous leading strand is synthesized • Discontinuous lagging strand is synthesized in segments • DNA ligase splices together short segments of discontinuous strand Copyright © 2010 Pearson Education, Inc.

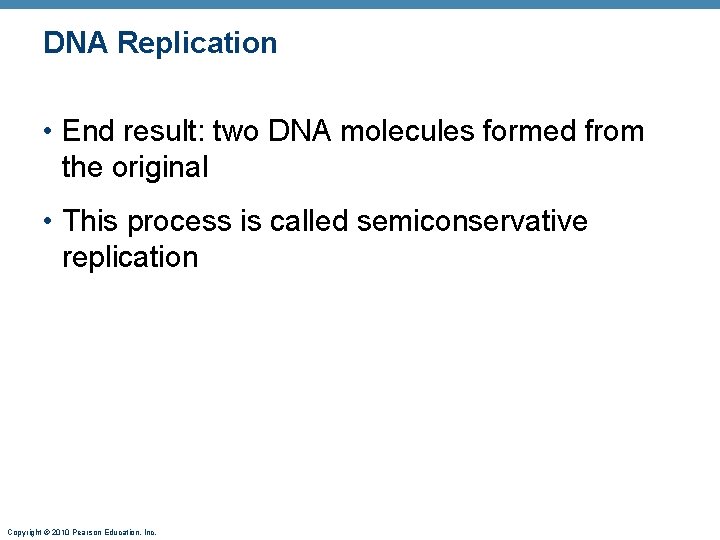
DNA Replication • End result: two DNA molecules formed from the original • This process is called semiconservative replication Copyright © 2010 Pearson Education, Inc.

Chromosome Free nucleotides DNA polymerase Old strand acts as a template for synthesis of new strand Leading strand Old DNA Helicase unwinds the double helix and exposes the bases Replication fork Adenine Thymine Cytosine Guanine Copyright © 2010 Pearson Education, Inc. Two new strands (leading and lagging) synthesized in opposite directions Lagging strand DNA polymerase Old (template) strand Figure 3. 32

DNA Replication PLAY Copyright © 2010 Pearson Education, Inc. Animation: DNA Replication

Cell Division • Mitotic (M) phase of the cell cycle • Essential for body growth and tissue repair • Does not occur in most mature cells of nervous tissue, skeletal muscle, and cardiac muscle Copyright © 2010 Pearson Education, Inc.

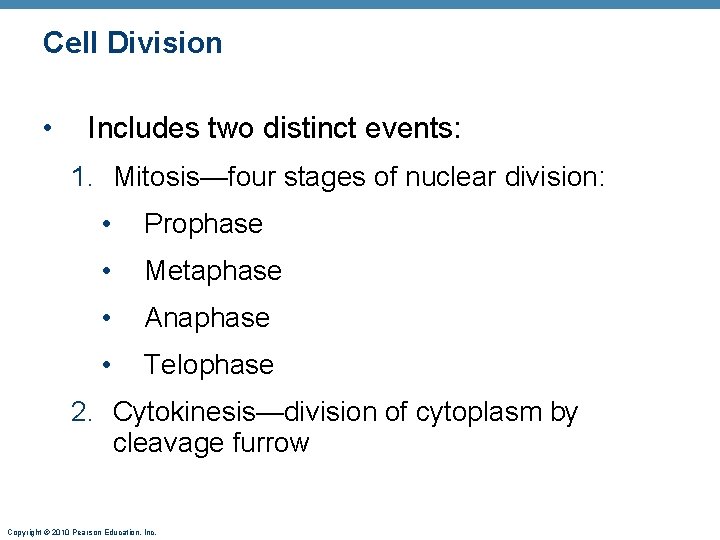
Cell Division • Includes two distinct events: 1. Mitosis—four stages of nuclear division: • Prophase • Metaphase • Anaphase • Telophase 2. Cytokinesis—division of cytoplasm by cleavage furrow Copyright © 2010 Pearson Education, Inc.

G 1 checkpoint (restriction point) S Growth and DNA synthesis G 1 Growth M G 2 Growth and final preparations for division G 2 checkpoint Copyright © 2010 Pearson Education, Inc. Figure 3. 31

Cell Division Copyright © 2010 Pearson Education, Inc. PLAY A&P Flix™: Mitosis PLAY Animation: Mitosis

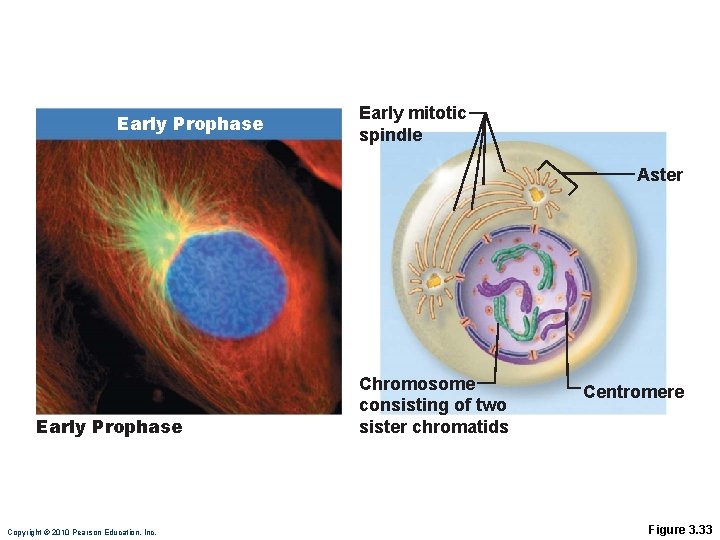
Prophase • Chromosomes become visible, each with two chromatids joined at a centromere • Centrosomes separate and migrate toward opposite poles • Mitotic spindles and asters form Copyright © 2010 Pearson Education, Inc.

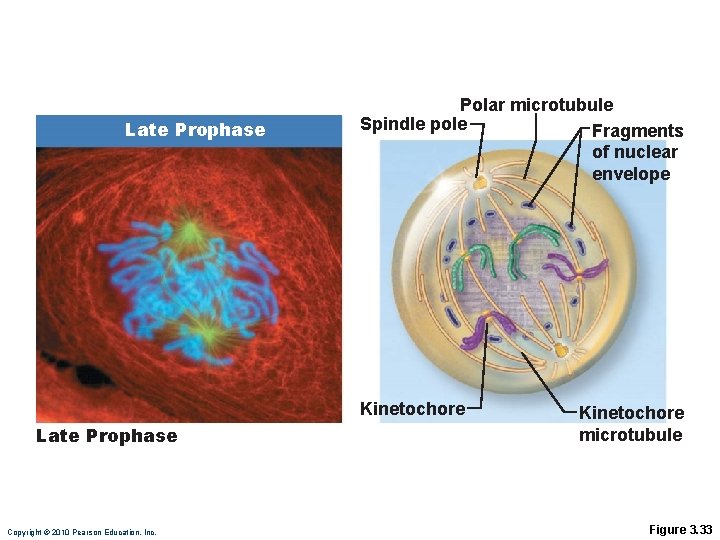
Prophase • Nuclear envelope fragments • Kinetochore microtubules attach to kinetochore of centromeres and draw them toward the equator of the cell • Polar microtubules assist in forcing the poles apart Copyright © 2010 Pearson Education, Inc.

Early Prophase Early mitotic spindle Aster Early Prophase Copyright © 2010 Pearson Education, Inc. Chromosome consisting of two sister chromatids Centromere Figure 3. 33

Late Prophase Polar microtubule Spindle pole Fragments of nuclear envelope Kinetochore Late Prophase Copyright © 2010 Pearson Education, Inc. Kinetochore microtubule Figure 3. 33

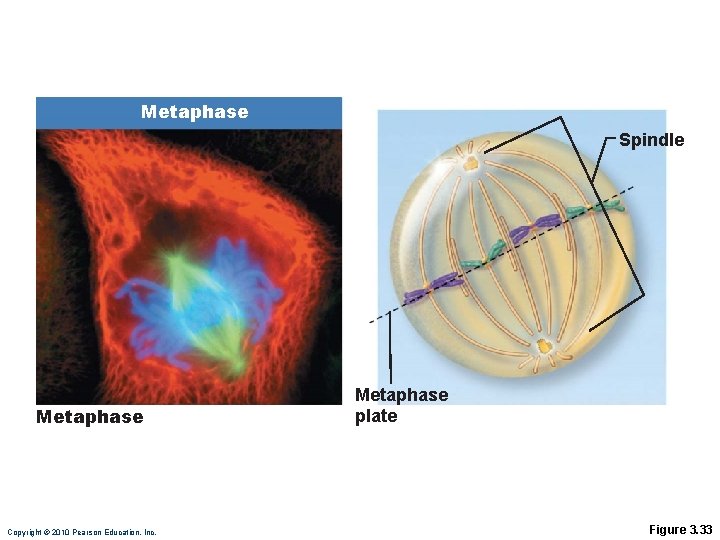
Metaphase • Centromeres of chromosomes are aligned at the equator • This plane midway between the poles is called the metaphase plate Copyright © 2010 Pearson Education, Inc.

Metaphase Spindle Metaphase Copyright © 2010 Pearson Education, Inc. Metaphase plate Figure 3. 33

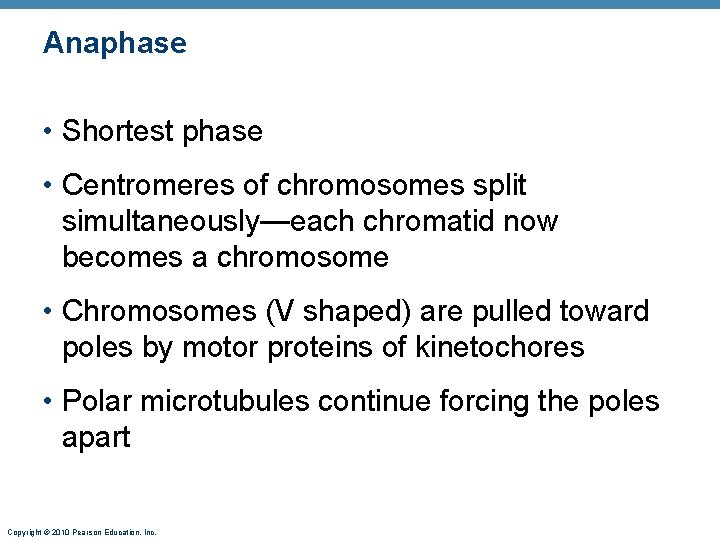
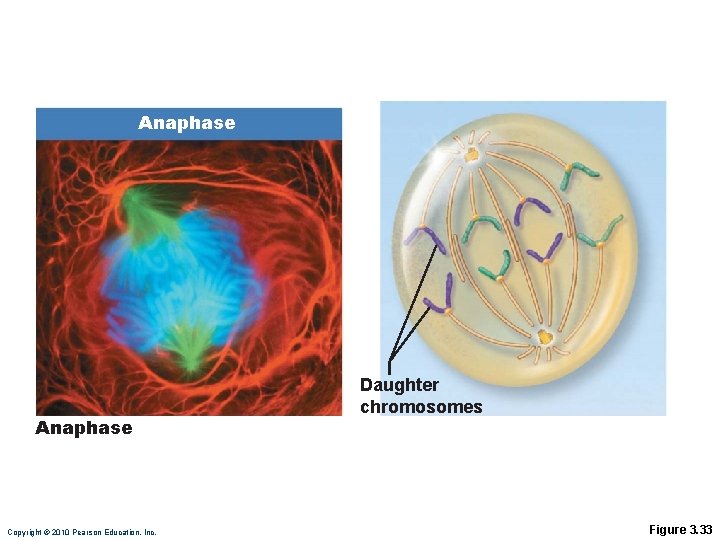
Anaphase • Shortest phase • Centromeres of chromosomes split simultaneously—each chromatid now becomes a chromosome • Chromosomes (V shaped) are pulled toward poles by motor proteins of kinetochores • Polar microtubules continue forcing the poles apart Copyright © 2010 Pearson Education, Inc.

Anaphase Copyright © 2010 Pearson Education, Inc. Daughter chromosomes Figure 3. 33

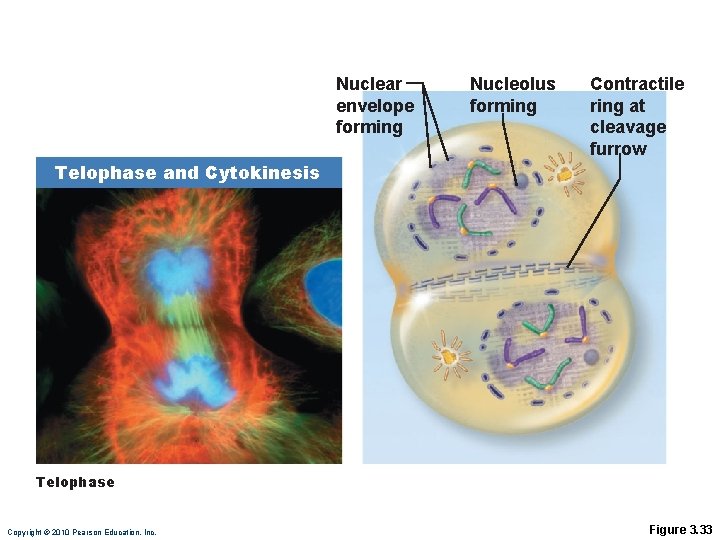
Telophase • Begins when chromosome movement stops • The two sets of chromosomes uncoil to form chromatin • New nuclear membrane forms around each chromatin mass • Nucleoli reappear • Spindle disappears Copyright © 2010 Pearson Education, Inc.

Cytokinesis • Begins during late anaphase • Ring of actin microfilaments contracts to form a cleavage furrow • Two daughter cells are pinched apart, each containing a nucleus identical to the original Copyright © 2010 Pearson Education, Inc.

Nuclear envelope forming Nucleolus forming Contractile ring at cleavage furrow Telophase and Cytokinesis Telophase Copyright © 2010 Pearson Education, Inc. Figure 3. 33

Control of Cell Division • “Go” signals: • Critical volume of cell when area of membrane is inadequate for exchange • Chemicals (e. g. , growth factors, hormones, cyclins, and cyclin-dependent kinases (Cdks)) Copyright © 2010 Pearson Education, Inc.

Control of Cell Division • “Stop” signals: • Contact inhibition • Growth-inhibiting factors produced by repressor genes Copyright © 2010 Pearson Education, Inc.

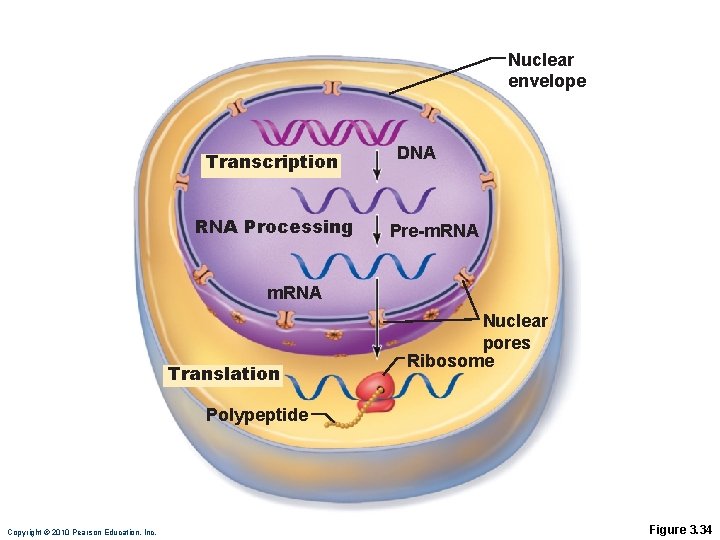
Protein Synthesis • DNA is the master blueprint for protein synthesis • Gene: Segment of DNA with blueprint for one polypeptide • Triplets of nucleotide bases form genetic library • Each triplet specifies coding for an amino acid PLAY Animation: DNA and RNA Copyright © 2010 Pearson Education, Inc.

Nuclear envelope Transcription RNA Processing DNA Pre-m. RNA Translation Nuclear pores Ribosome Polypeptide Copyright © 2010 Pearson Education, Inc. Figure 3. 34

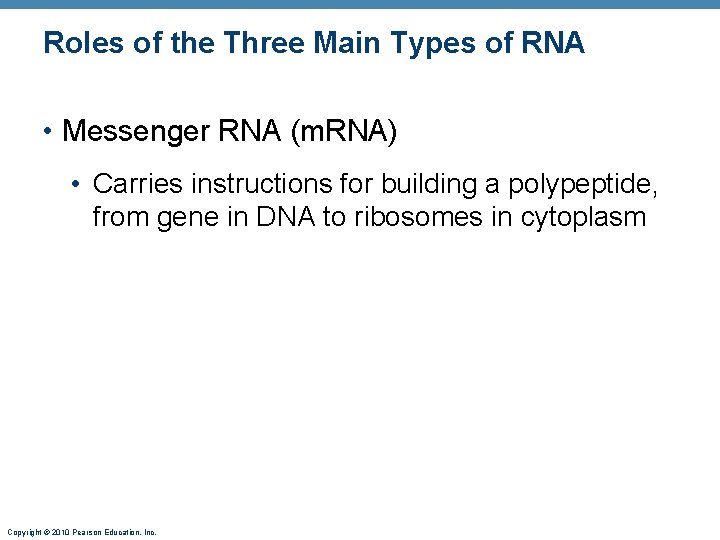
Roles of the Three Main Types of RNA • Messenger RNA (m. RNA) • Carries instructions for building a polypeptide, from gene in DNA to ribosomes in cytoplasm Copyright © 2010 Pearson Education, Inc.

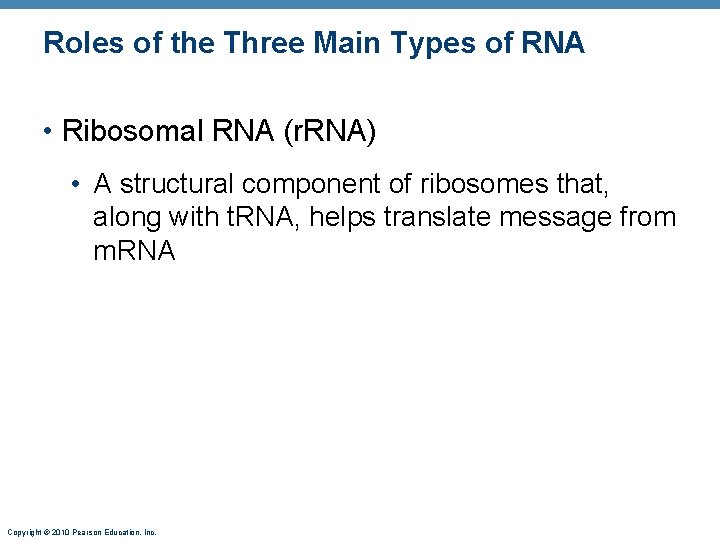
Roles of the Three Main Types of RNA • Ribosomal RNA (r. RNA) • A structural component of ribosomes that, along with t. RNA, helps translate message from m. RNA Copyright © 2010 Pearson Education, Inc.

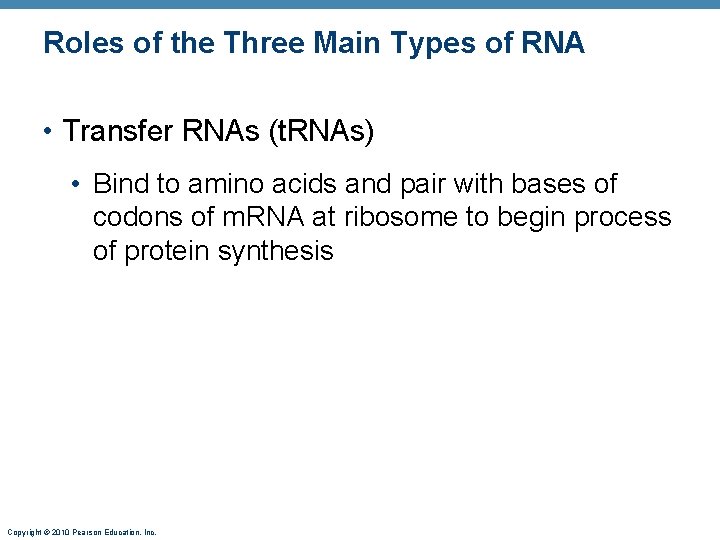
Roles of the Three Main Types of RNA • Transfer RNAs (t. RNAs) • Bind to amino acids and pair with bases of codons of m. RNA at ribosome to begin process of protein synthesis Copyright © 2010 Pearson Education, Inc.

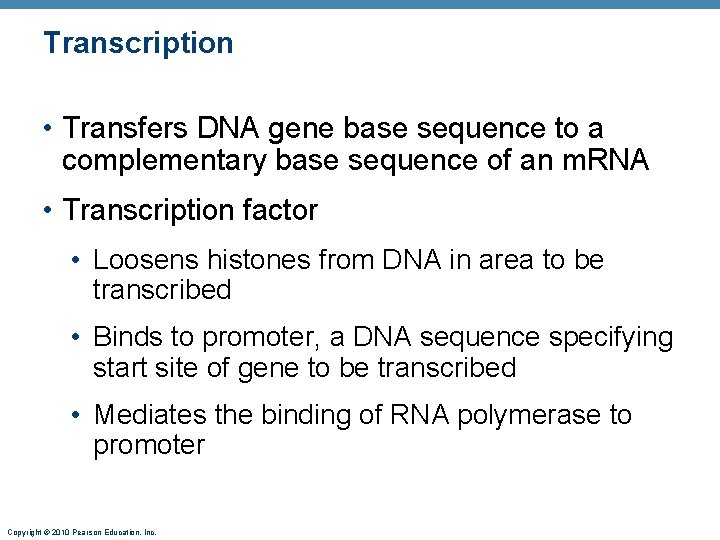
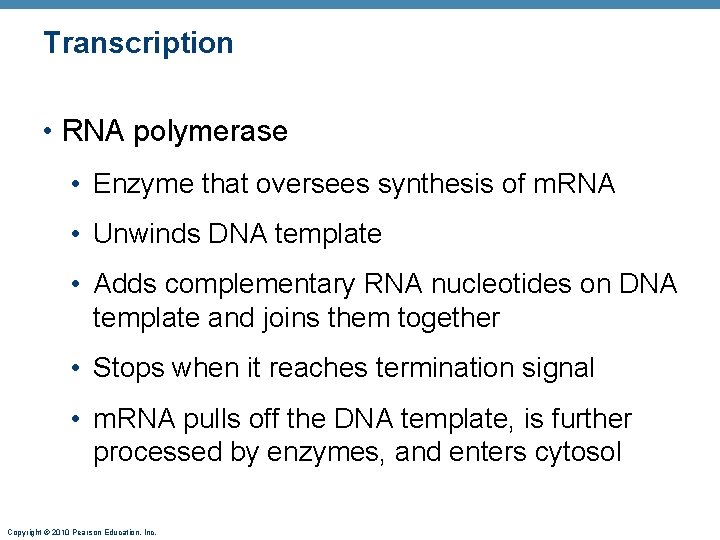
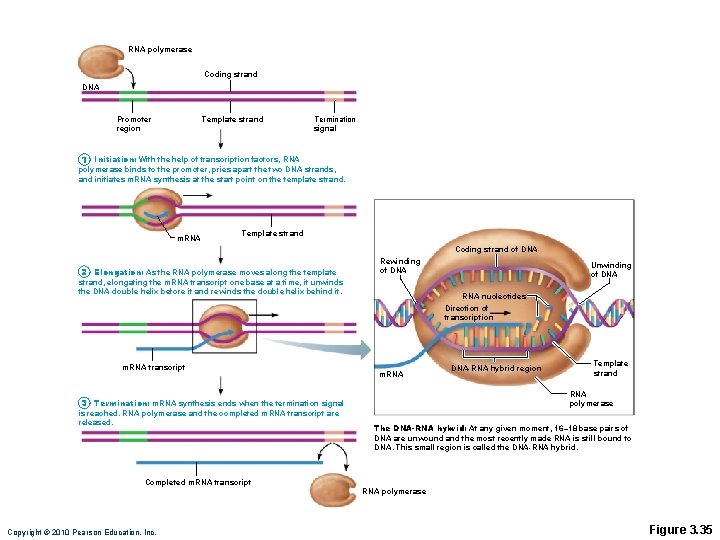
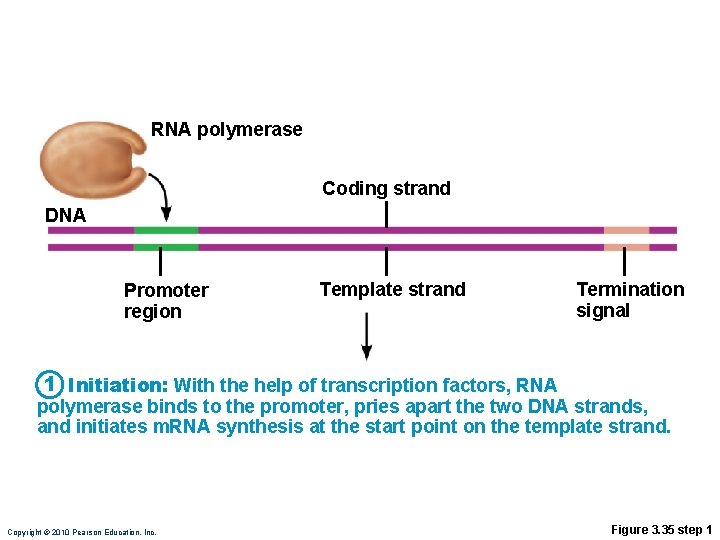
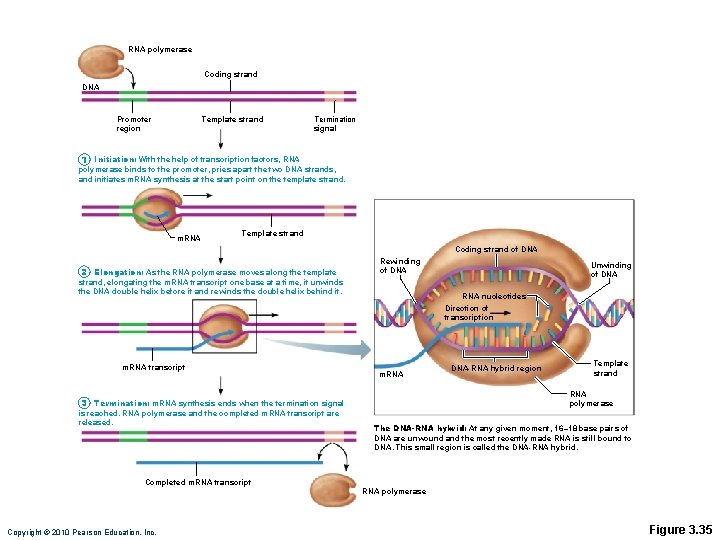
Transcription • Transfers DNA gene base sequence to a complementary base sequence of an m. RNA • Transcription factor • Loosens histones from DNA in area to be transcribed • Binds to promoter, a DNA sequence specifying start site of gene to be transcribed • Mediates the binding of RNA polymerase to promoter Copyright © 2010 Pearson Education, Inc.

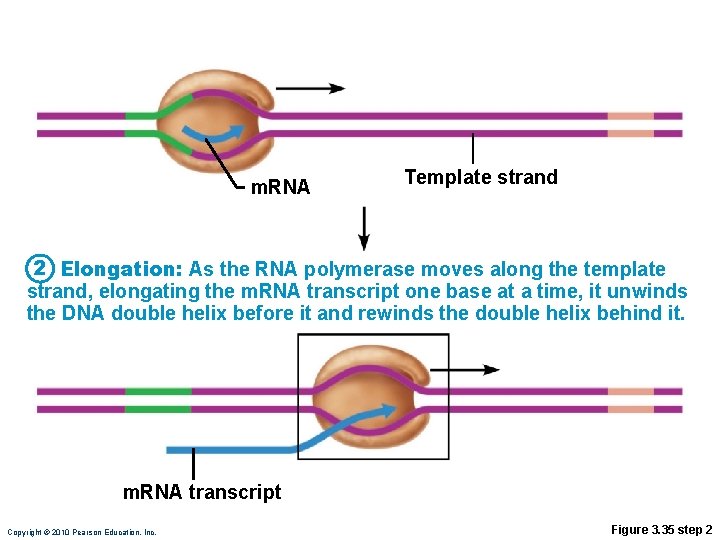
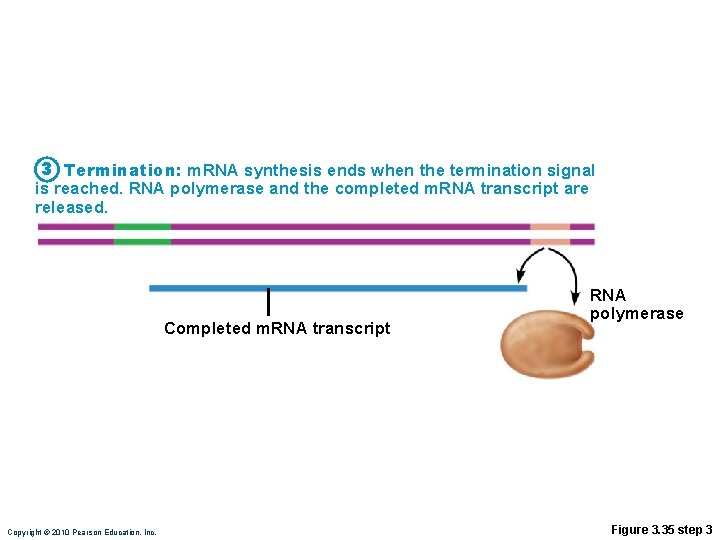
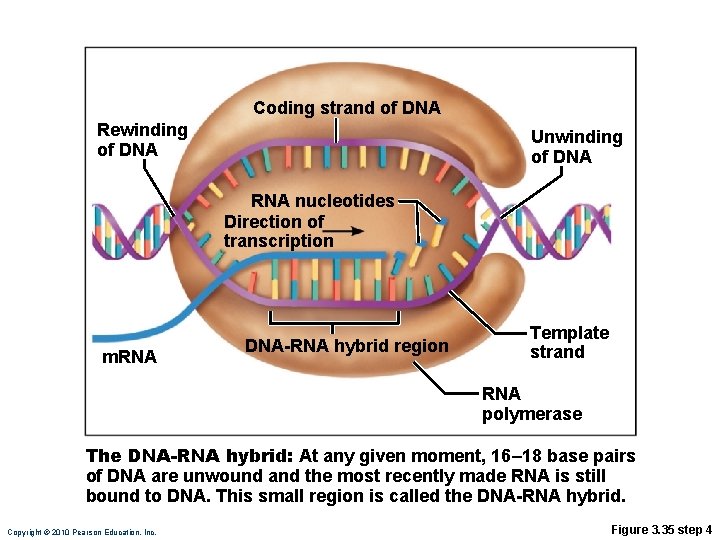
Transcription • RNA polymerase • Enzyme that oversees synthesis of m. RNA • Unwinds DNA template • Adds complementary RNA nucleotides on DNA template and joins them together • Stops when it reaches termination signal • m. RNA pulls off the DNA template, is further processed by enzymes, and enters cytosol Copyright © 2010 Pearson Education, Inc.

RNA polymerase Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates m. RNA synthesis at the start point on the template strand. m. RNA Template strand Coding strand of DNA 2 Elongation: As the RNA polymerase moves along the template Rewinding of DNA strand, elongating the m. RNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. m. RNA transcript RNA nucleotides Direction of transcription m. RNA DNA-RNA hybrid region Template strand RNA polymerase 3 Termination: m. RNA synthesis ends when the termination signal is reached. RNA polymerase and the completed m. RNA transcript are released. Unwinding of DNA The DNA-RNA hybrid: At any given moment, 16– 18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Completed m. RNA transcript RNA polymerase Copyright © 2010 Pearson Education, Inc. Figure 3. 35

RNA polymerase Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates m. RNA synthesis at the start point on the template strand. Copyright © 2010 Pearson Education, Inc. Figure 3. 35 step 1

m. RNA Template strand 2 Elongation: As the RNA polymerase moves along the template strand, elongating the m. RNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. m. RNA transcript Copyright © 2010 Pearson Education, Inc. Figure 3. 35 step 2

3 Termination: m. RNA synthesis ends when the termination signal is reached. RNA polymerase and the completed m. RNA transcript are released. Completed m. RNA transcript Copyright © 2010 Pearson Education, Inc. RNA polymerase Figure 3. 35 step 3

Coding strand of DNA Rewinding of DNA Unwinding of DNA RNA nucleotides Direction of transcription m. RNA DNA-RNA hybrid region Template strand RNA polymerase The DNA-RNA hybrid: At any given moment, 16– 18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Copyright © 2010 Pearson Education, Inc. Figure 3. 35 step 4

RNA polymerase Coding strand DNA Promoter region Template strand Termination signal 1 Initiation: With the help of transcription factors, RNA polymerase binds to the promoter, pries apart the two DNA strands, and initiates m. RNA synthesis at the start point on the template strand. m. RNA Template strand Coding strand of DNA 2 Elongation: As the RNA polymerase moves along the template Rewinding of DNA strand, elongating the m. RNA transcript one base at a time, it unwinds the DNA double helix before it and rewinds the double helix behind it. m. RNA transcript RNA nucleotides Direction of transcription m. RNA DNA-RNA hybrid region Template strand RNA polymerase 3 Termination: m. RNA synthesis ends when the termination signal is reached. RNA polymerase and the completed m. RNA transcript are released. Unwinding of DNA The DNA-RNA hybrid: At any given moment, 16– 18 base pairs of DNA are unwound and the most recently made RNA is still bound to DNA. This small region is called the DNA-RNA hybrid. Completed m. RNA transcript RNA polymerase Copyright © 2010 Pearson Education, Inc. Figure 3. 35

Translation • Converts base sequence of nucleic acids into the amino acid sequence of proteins • Involves m. RNAs, t. RNAs, and r. RNAs Copyright © 2010 Pearson Education, Inc.

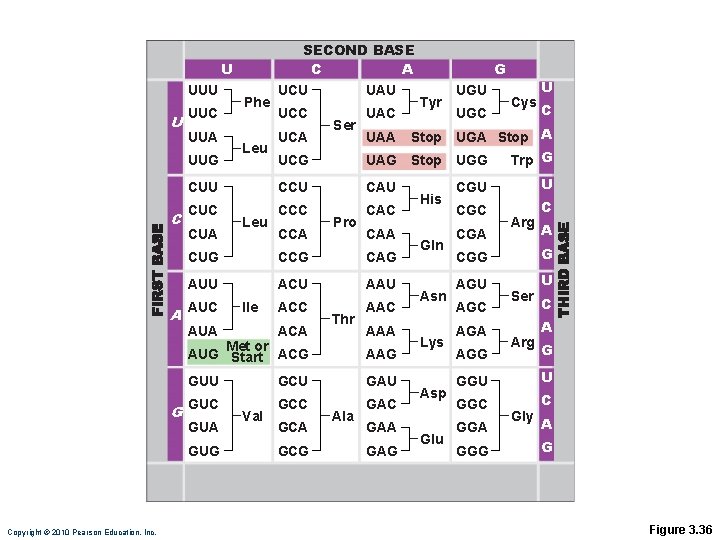
Genetic Code • Each three-base sequence on DNA is represented by a codon • Codon—complementary three-base sequence on m. RNA Copyright © 2010 Pearson Education, Inc.

SECOND BASE C A U UUU U UUC A UAU UCC UAC Ser Cys C CUU CCU CAU CUC CCC CAC CUA Leu CCA Pro UAA CUG CCG CAG AUU ACU AAU ACC AAC AUC Ile AUA ACA Thr AAA AAG GUU GCU GAU GUC GCC GAC GUA GUG Copyright © 2010 Pearson Education, Inc. UGC U UCG Leu UCA Met or AUG Start ACG G Tyr UGU Stop UGA Stop A UAG Stop UGG Trp G UUA UUG C Phe UCU G Val GCA GCG Ala GAA GAG His Gln Asn Lys Asp Glu CGU U CGC C CGA Arg A CGG G AGU U AGC AGA AGG Ser Arg C A G GGU U GGC C GGA GGG Gly A G Figure 3. 36

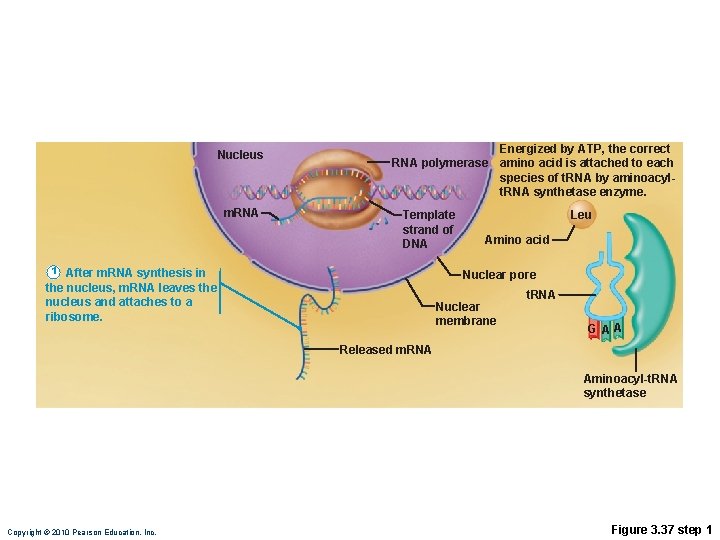
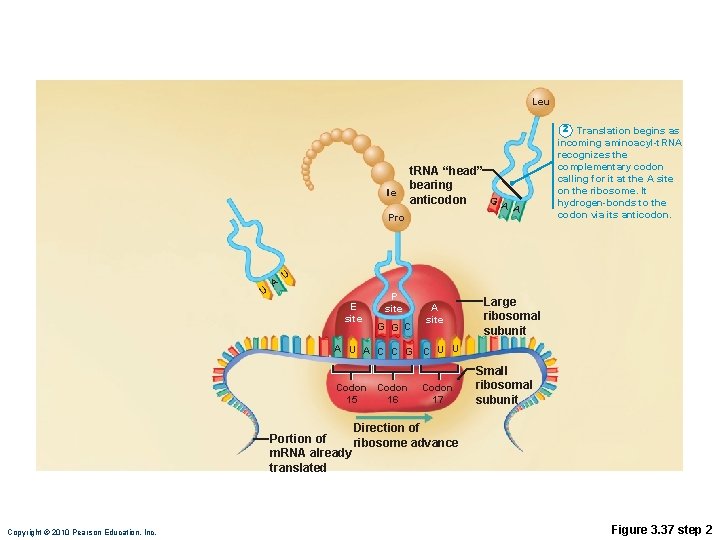
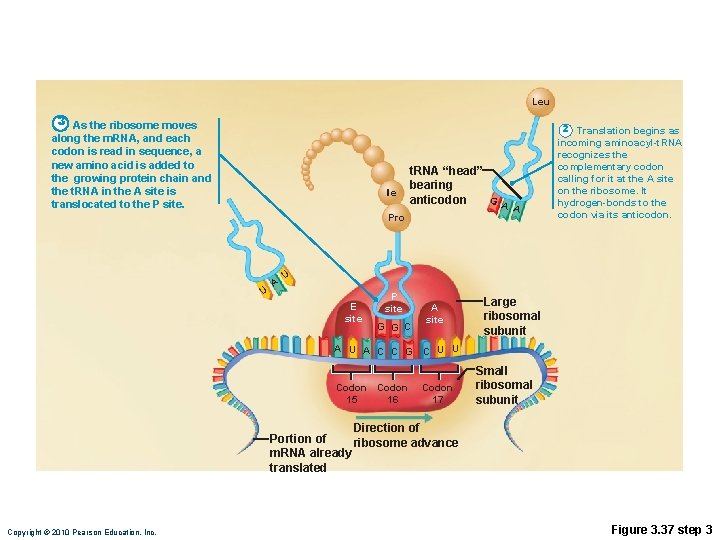
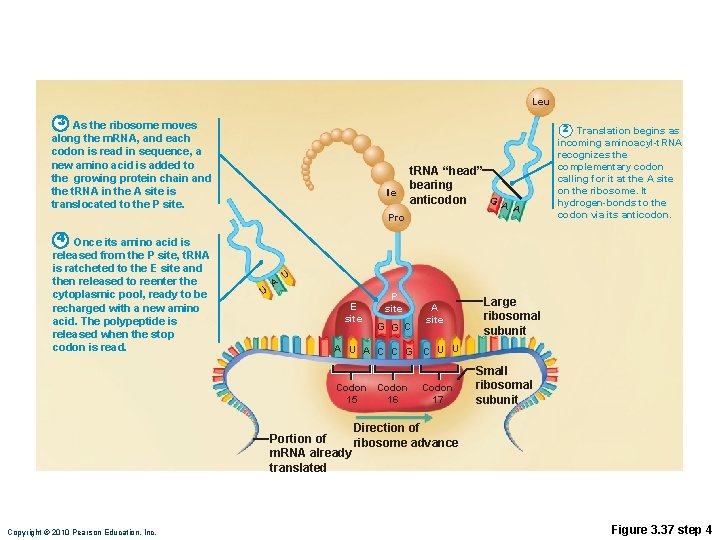
Translation • m. RNA attaches to a small ribosomal subunit that moves along the m. RNA to the start codon • Large ribosomal unit attaches, forming a functional ribosome • Anticodon of a t. RNA binds to its complementary codon and adds its amino acid to the forming protein chain • New amino acids are added by other t. RNAs as ribosome moves along r. RNA, until stop codon is reached Copyright © 2010 Pearson Education, Inc.

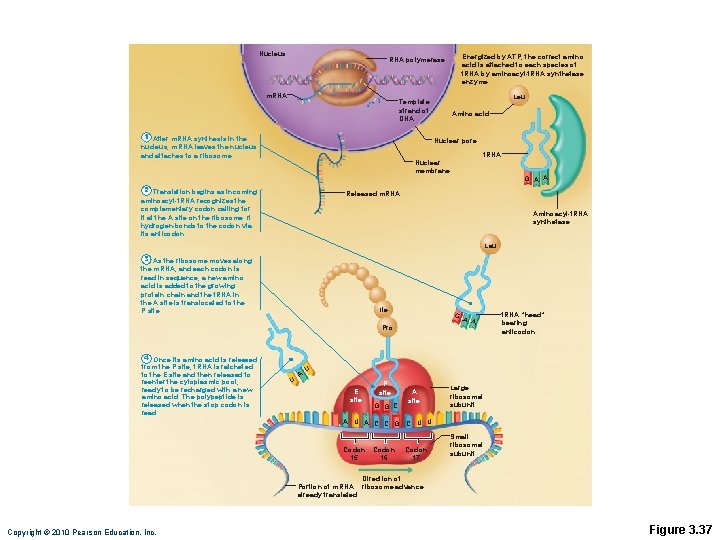
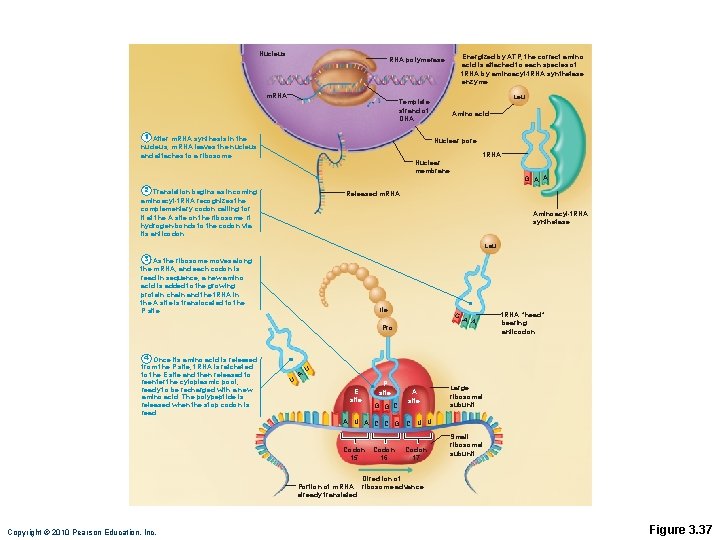
Nucleus Energized by ATP, the correct amino acid is attached to each species of t. RNA by aminoacyl-t. RNA synthetase enzyme. RNA polymerase m. RNA Leu Template strand of DNA 1 After m. RNA synthesis in the nucleus, m. RNA leaves the nucleus and attaches to a ribosome. Amino acid Nuclear pore t. RNA Nuclear membrane 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. G A A Released m. RNA Aminoacyl-t. RNA synthetase Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile G Pro 4 Once its amino acid is released from the P site, t. RNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. The polypeptide is released when the stop codon is read. U A A A t. RNA “head” bearing anticodon U E site P site G G C A site A U A C C G C U U Codon 15 Codon 17 Codon 16 Large ribosomal subunit Small ribosomal subunit Direction of Portion of m. RNA ribosome advance already translated Copyright © 2010 Pearson Education, Inc. Figure 3. 37

Nucleus m. RNA polymerase Template strand of DNA 1 After m. RNA synthesis in the nucleus, m. RNA leaves the nucleus and attaches to a ribosome. Energized by ATP, the correct amino acid is attached to each species of t. RNA by aminoacylt. RNA synthetase enzyme. Leu Amino acid Nuclear pore Nuclear membrane t. RNA GAA Released m. RNA Aminoacyl-t. RNA synthetase Copyright © 2010 Pearson Education, Inc. Figure 3. 37 step 1

Leu Ile Pro U A t. RNA “head” bearing G anticodon A A 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. U E site P site G G C A site Large ribosomal subunit A U A C C G C U U Codon 15 16 Codon 17 Small ribosomal subunit Direction of Portion of ribosome advance m. RNA already translated Copyright © 2010 Pearson Education, Inc. Figure 3. 37 step 2

Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile Pro U A t. RNA “head” bearing G anticodon A A 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. U E site P site G G C A site Large ribosomal subunit A U A C C G C U U Codon 15 16 Codon 17 Small ribosomal subunit Direction of Portion of ribosome advance m. RNA already translated Copyright © 2010 Pearson Education, Inc. Figure 3. 37 step 3

Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile Pro t. RNA “head” bearing G anticodon A A 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. 4 Once its amino acid is released from the P site, t. RNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. The polypeptide is released when the stop codon is read. U A U E site P site G G C A site Large ribosomal subunit A U A C C G C U U Codon 15 16 Codon 17 Small ribosomal subunit Direction of Portion of ribosome advance m. RNA already translated Copyright © 2010 Pearson Education, Inc. Figure 3. 37 step 4

Nucleus Energized by ATP, the correct amino acid is attached to each species of t. RNA by aminoacyl-t. RNA synthetase enzyme. RNA polymerase m. RNA Leu Template strand of DNA 1 After m. RNA synthesis in the nucleus, m. RNA leaves the nucleus and attaches to a ribosome. Amino acid Nuclear pore t. RNA Nuclear membrane 2 Translation begins as incoming aminoacyl-t. RNA recognizes the complementary codon calling for it at the A site on the ribosome. It hydrogen-bonds to the codon via its anticodon. G A A Released m. RNA Aminoacyl-t. RNA synthetase Leu 3 As the ribosome moves along the m. RNA, and each codon is read in sequence, a new amino acid is added to the growing protein chain and the t. RNA in the A site is translocated to the P site. Ile G Pro 4 Once its amino acid is released from the P site, t. RNA is ratcheted to the E site and then released to reenter the cytoplasmic pool, ready to be recharged with a new amino acid. The polypeptide is released when the stop codon is read. U A A A t. RNA “head” bearing anticodon U E site P site G G C A site A U A C C G C U U Codon 15 Codon 17 Codon 16 Large ribosomal subunit Small ribosomal subunit Direction of Portion of m. RNA ribosome advance already translated Copyright © 2010 Pearson Education, Inc. Figure 3. 37

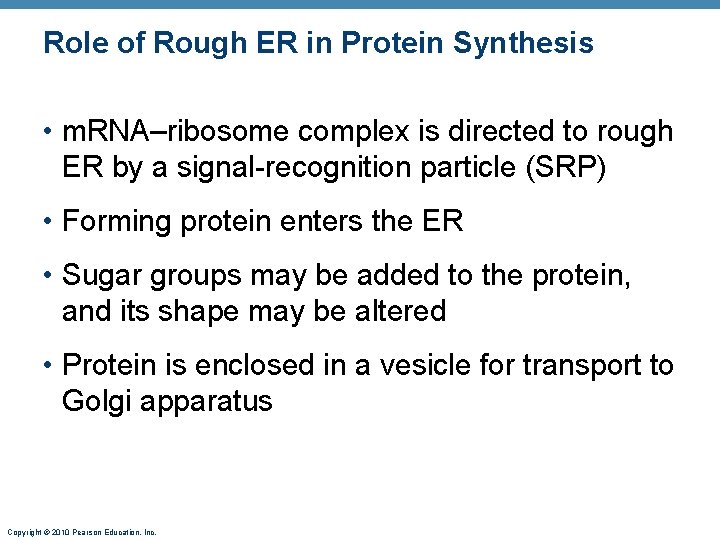
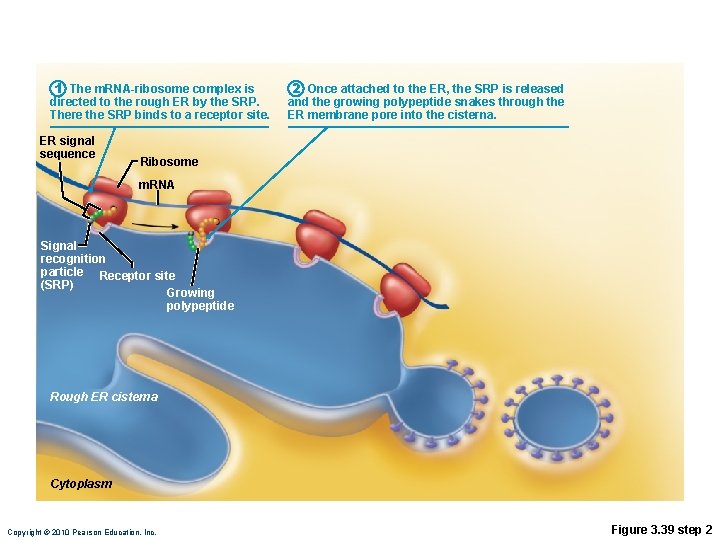
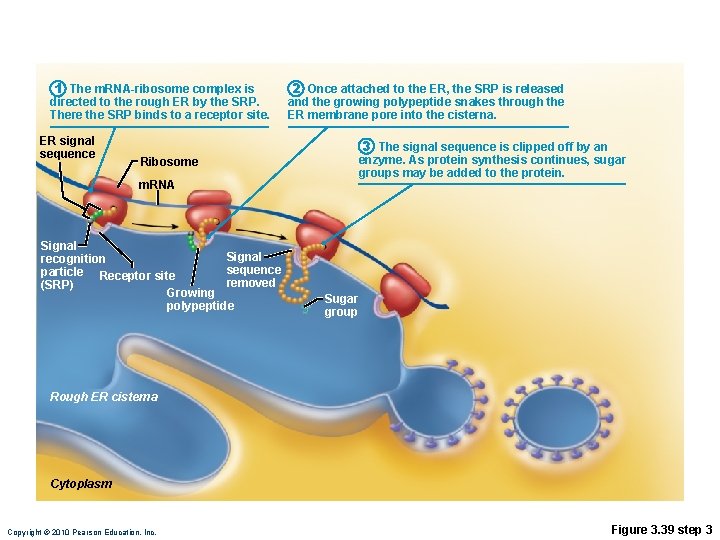
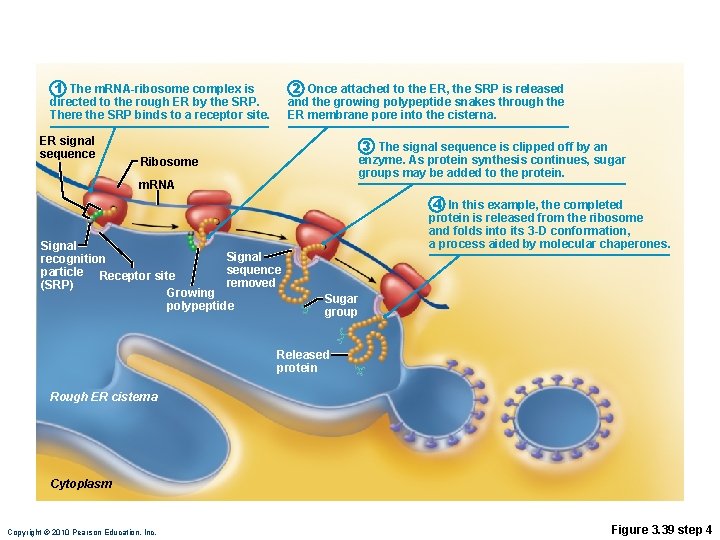
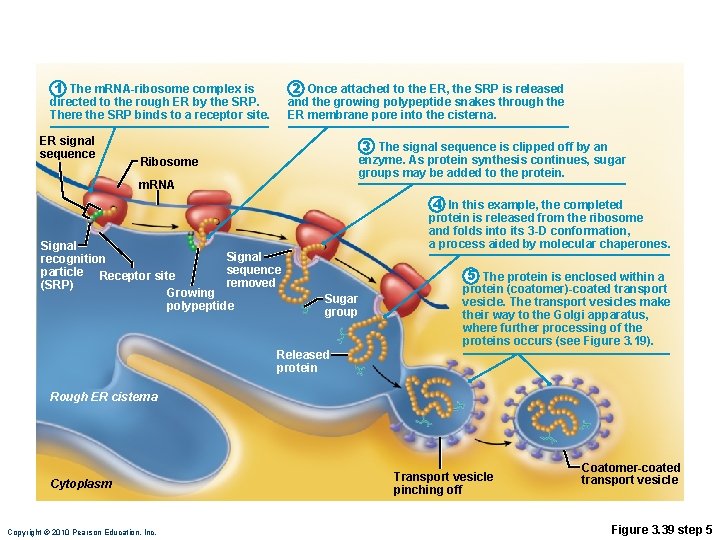
Role of Rough ER in Protein Synthesis • m. RNA–ribosome complex is directed to rough ER by a signal-recognition particle (SRP) • Forming protein enters the ER • Sugar groups may be added to the protein, and its shape may be altered • Protein is enclosed in a vesicle for transport to Golgi apparatus Copyright © 2010 Pearson Education, Inc.

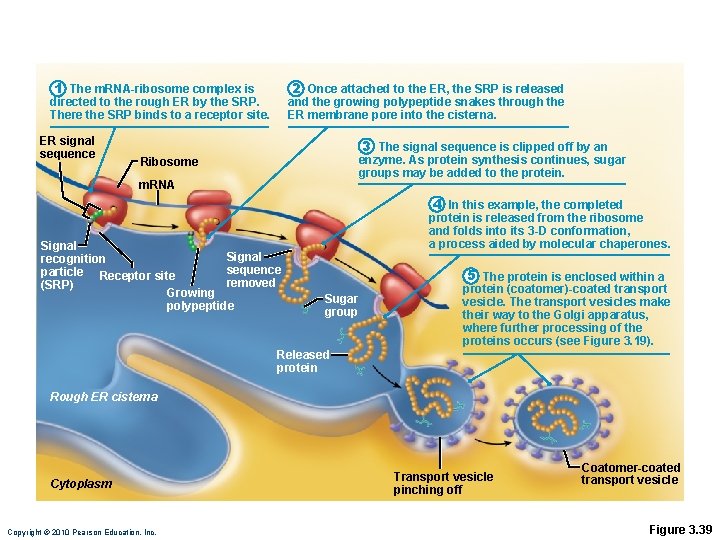
1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3 -D conformation, a process aided by molecular chaperones. Sugar group 5 The protein is enclosed within a protein (coatomer)-coated transport vesicle. The transport vesicles make their way to the Golgi apparatus, where further processing of the proteins occurs (see Figure 3. 19). Released protein Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Transport vesicle pinching off Coatomer-coated transport vesicle Figure 3. 39

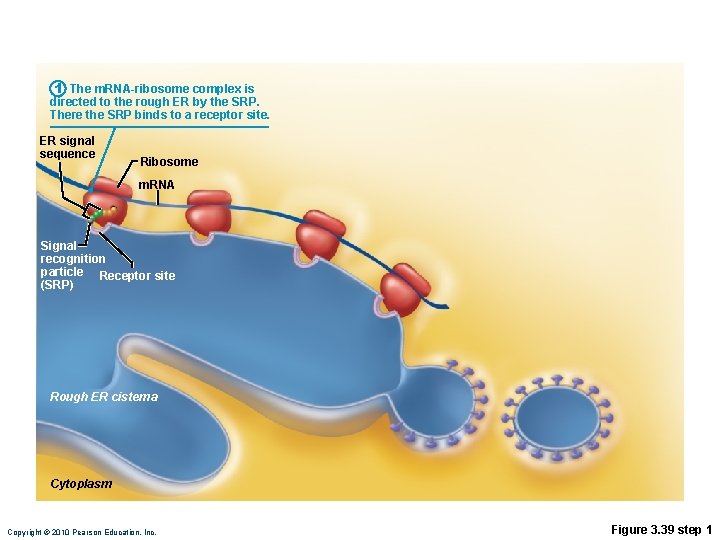
1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence Ribosome m. RNA Signal recognition particle Receptor site (SRP) Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Figure 3. 39 step 1

1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. Ribosome m. RNA Signal recognition particle Receptor site (SRP) Growing polypeptide Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Figure 3. 39 step 2

1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide Sugar group Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Figure 3. 39 step 3

1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3 -D conformation, a process aided by molecular chaperones. Sugar group Released protein Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Figure 3. 39 step 4

1 The m. RNA-ribosome complex is directed to the rough ER by the SRP. There the SRP binds to a receptor site. ER signal sequence 2 Once attached to the ER, the SRP is released and the growing polypeptide snakes through the ER membrane pore into the cisterna. 3 The signal sequence is clipped off by an enzyme. As protein synthesis continues, sugar groups may be added to the protein. Ribosome m. RNA Signal recognition sequence particle Receptor site removed (SRP) Growing polypeptide 4 In this example, the completed protein is released from the ribosome and folds into its 3 -D conformation, a process aided by molecular chaperones. Sugar group 5 The protein is enclosed within a protein (coatomer)-coated transport vesicle. The transport vesicles make their way to the Golgi apparatus, where further processing of the proteins occurs (see Figure 3. 19). Released protein Rough ER cisterna Cytoplasm Copyright © 2010 Pearson Education, Inc. Transport vesicle pinching off Coatomer-coated transport vesicle Figure 3. 39 step 5

Other Roles of DNA • Intron (“junk”) regions of DNA code for other types of RNA: • Antisense RNA • Prevents protein-coding RNA from being translated • Micro. RNA • Small RNAs that interfere with m. RNAs made by certain exons • Riboswitches • Folded RNAs that act as switches regulating protein synthesis in response to environmental conditions Copyright © 2010 Pearson Education, Inc.

Cytosolic Protein Degradation • Nonfunctional organelle proteins are degraded by lysosomes • Ubiquitin tags damaged or unneeded soluble proteins in cytosol; they are digested by enzymes of proteasomes Copyright © 2010 Pearson Education, Inc.

Extracellular Materials • Body fluids (interstitial fluid, blood plasma, and cerebrospinal fluid) • Cellular secretions (intestinal and gastric fluids, saliva, mucus, and serous fluids) • Extracellular matrix (abundant jellylike mesh containing proteins and polysaccharides in contact with cells) Copyright © 2010 Pearson Education, Inc.

Developmental Aspects of Cells • All cells of the body contain the same DNA but are not identical • Chemical signals in the embryo channel cells into specific developmental pathways by turning some genes off • Development of specific and distinctive features in cells is called cell differentiation • Elimination of excess, injured, or aged cells occurs through programmed rapid cell death (apoptosis) followed by phagocytosis Copyright © 2010 Pearson Education, Inc.

Theories of Cell Aging • Wear and tear theory: Little chemical insults and free radicals have cumulative effects • Immune system disorders: Autoimmune responses and progressive weakening of the immune response • Genetic theory: Cessation of mitosis and cell aging are programmed into genes. Telomeres (strings of nucleotides on the ends of chromosomes) may determine the number of times a cell can divide. Copyright © 2010 Pearson Education, Inc.