What does the word Promoter mean It is















- Slides: 84

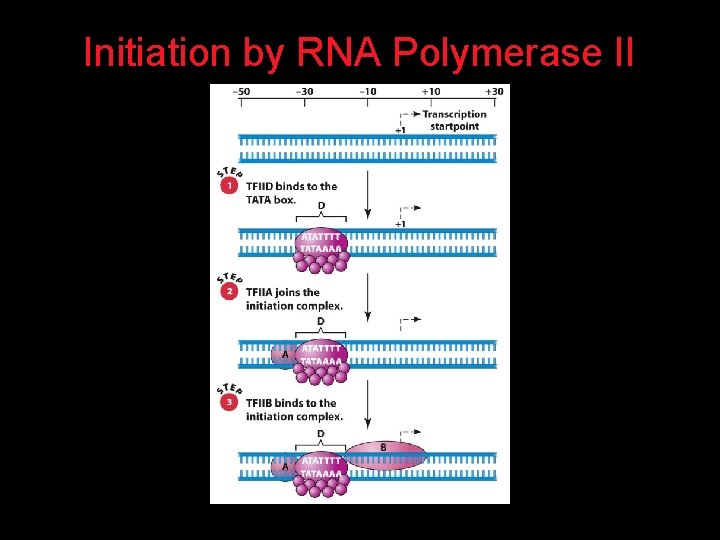
What does the word Promoter mean? It is the place at which RNA Pol II binds. But the word is incorrectly used to describe Enhancers plus Promoter.

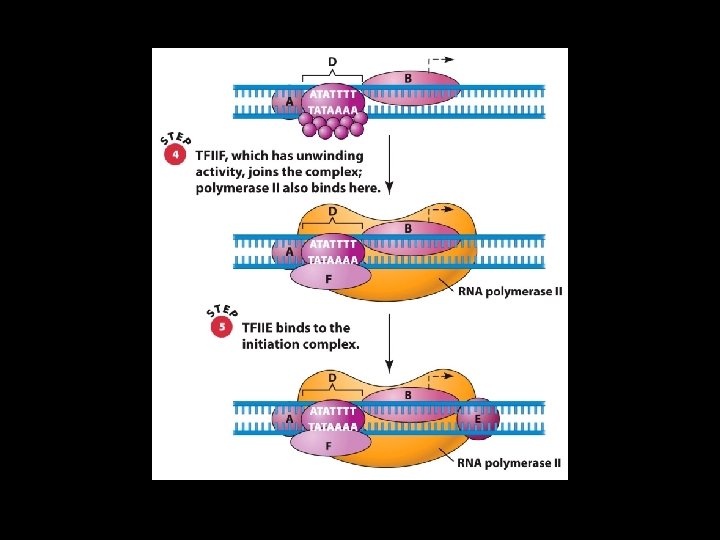
Initiation by RNA Polymerase II



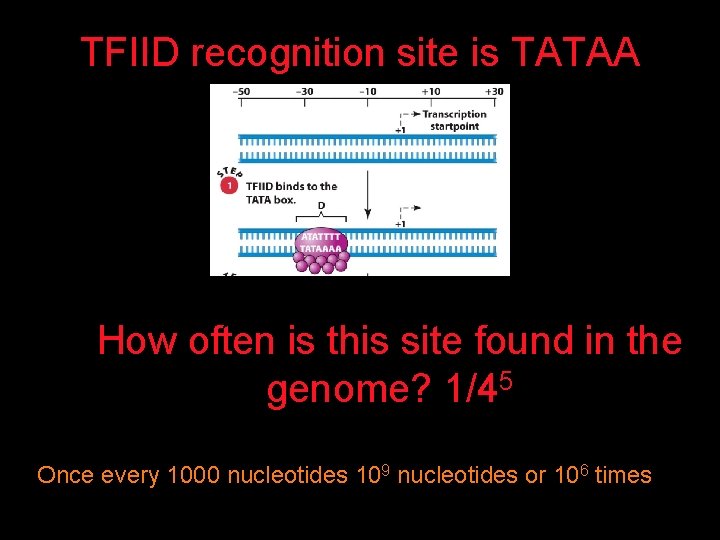
TFIID recognition site is TATAA How often is this site found in the genome? 1/45 Once every 1000 nucleotides 109 nucleotides or 106 times



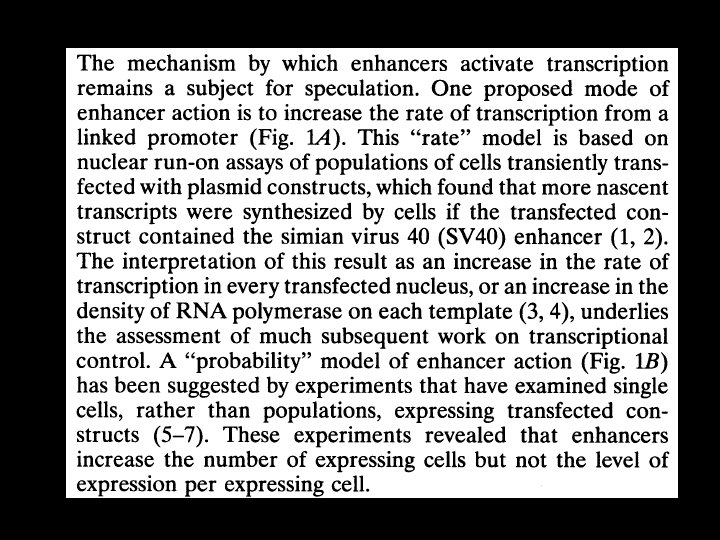
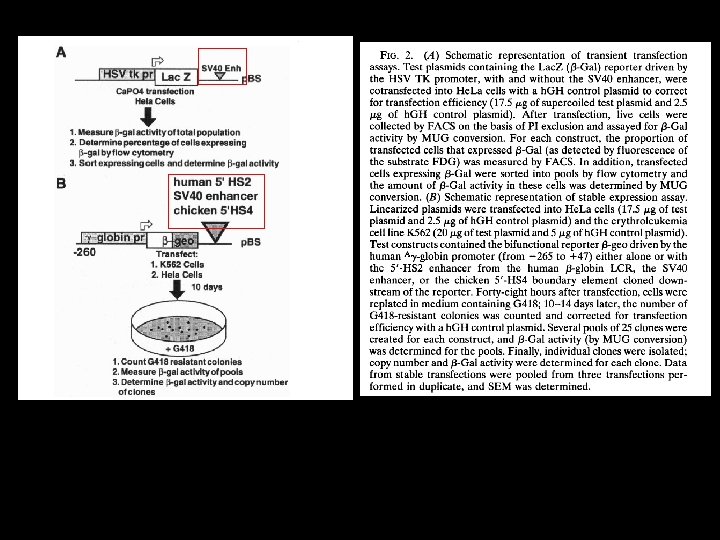
Transient transfection More Cells But on a per cell Basis expression levels of -gal is about the same

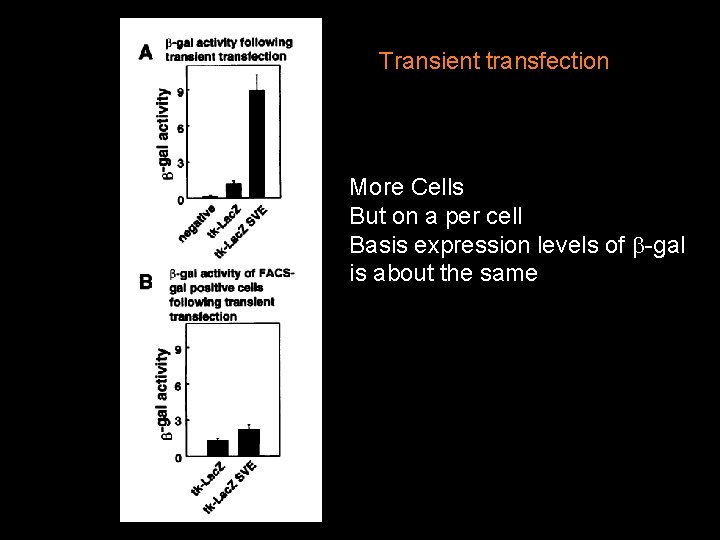
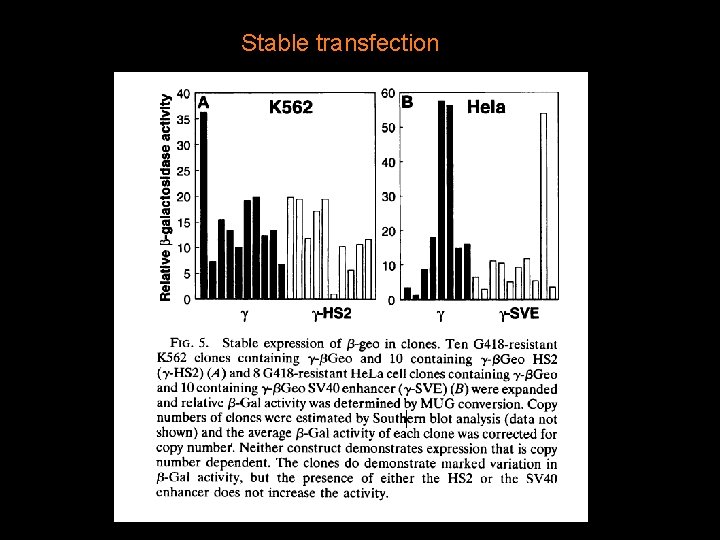
Stable transfection

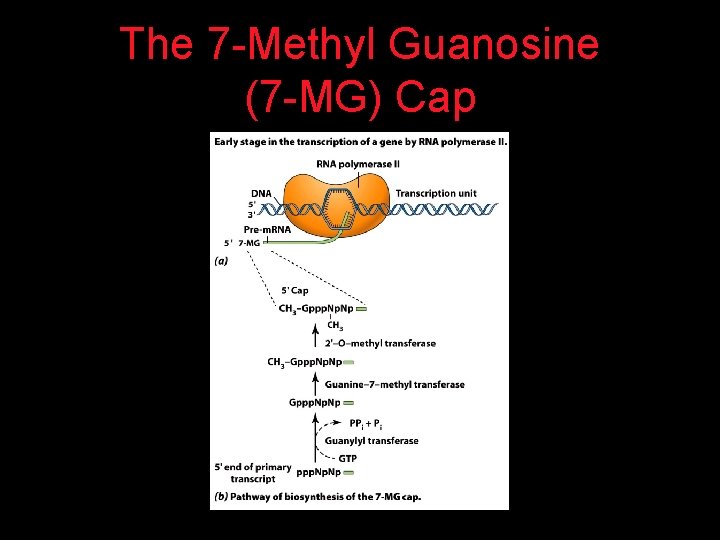
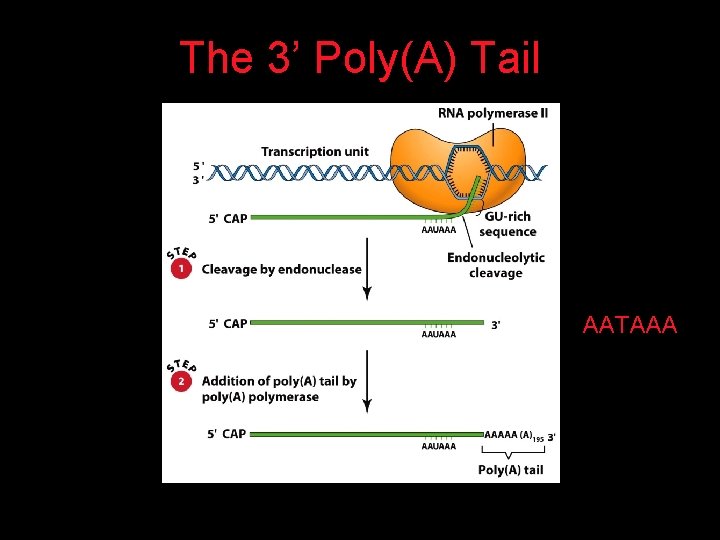
The 7 -Methyl Guanosine (7 -MG) Cap

The 3’ Poly(A) Tail AATAAA

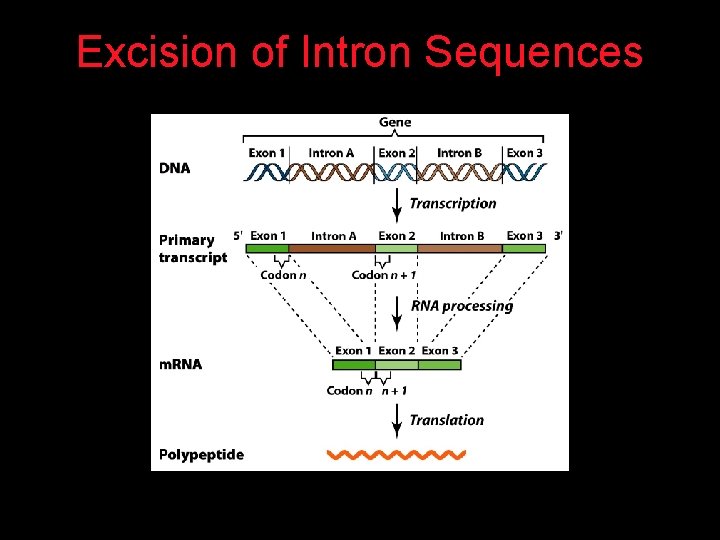
Interrupted Genes in Eukaryotes: Exons and Introns Most eukaryotic genes contain noncoding sequences called introns that interrupt the coding sequences, or exons. The introns are excised from the RNA transcripts prior to their transport to the cytoplasm.

Removal of Intron Sequences by RNA Splicing The noncoding introns are excised from gene transcripts by several different mechanisms.

Excision of Intron Sequences

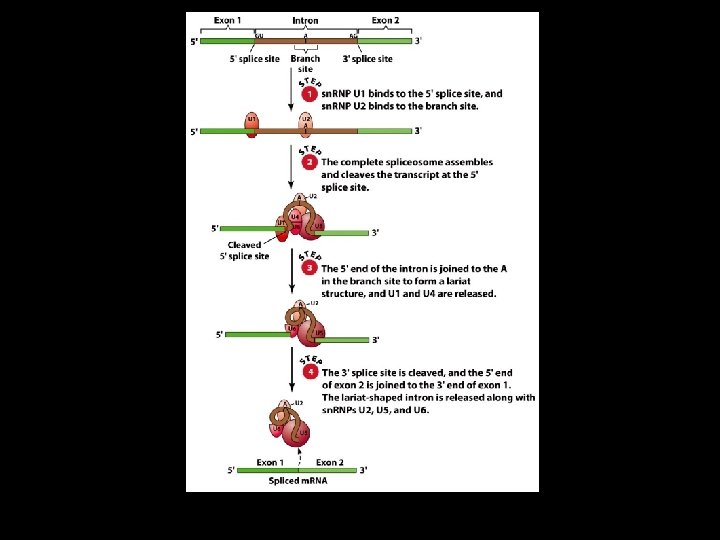
Splicing 4 Removal of introns must be very precise. 4 Conserved sequences for removal of the introns of nuclear m. RNA genes are minimal. – Dinucleotide sequences at the 5’ and 3’ ends of introns. – An A residue about 30 nucleotides upstream from the 3’ splice site is needed for lariat formation.

Types of Intron Excision 4 The introns of t. RNA precursors are excised by precise endonucleolytic cleavage and ligation reactions catalyzed by special splicing endonuclease and ligase activities. 4 The introns of nuclear pre-m. RNA (hn. RNA) transcripts are spliced out in two-step reactions carried out by spliceosomes.

The Spliceosome 4 Five sn. RNAs: U 1, U 2, U 4, U 5, and U 6 4 Some sn. RNAs associate with proteins to form sn. RNAs (small nuclear ribonucleoproteins)


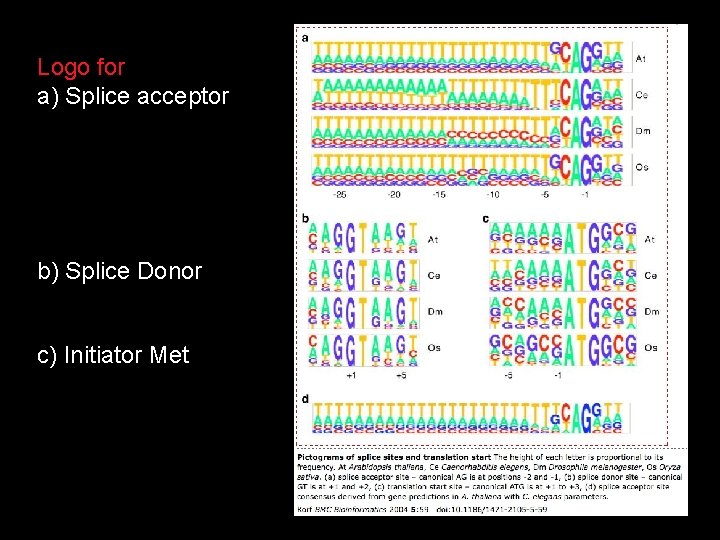
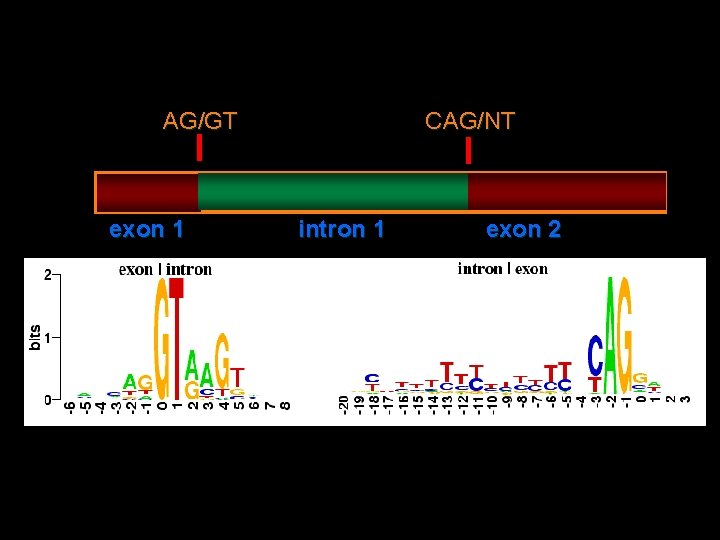
What are Logo plots?

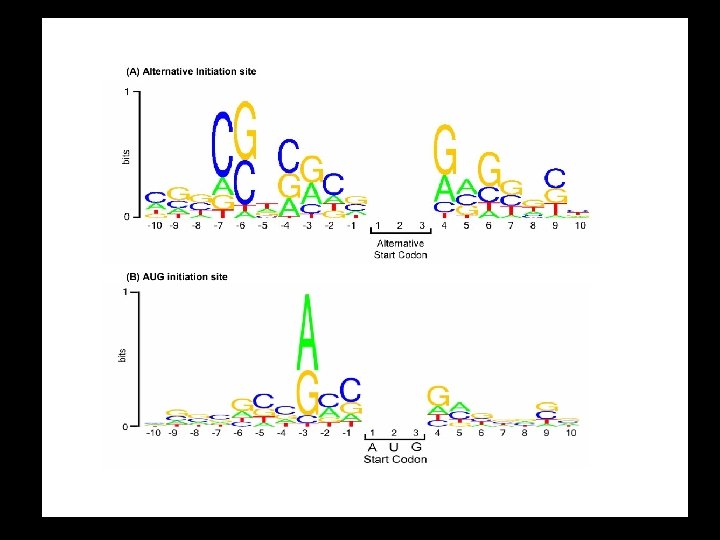
Logo for a) Splice acceptor b) Splice Donor c) Initiator Met

CAG/NT AG/GT exon 1 intron 1 exon 2

Chapter 12 Translation and the Genetic Code

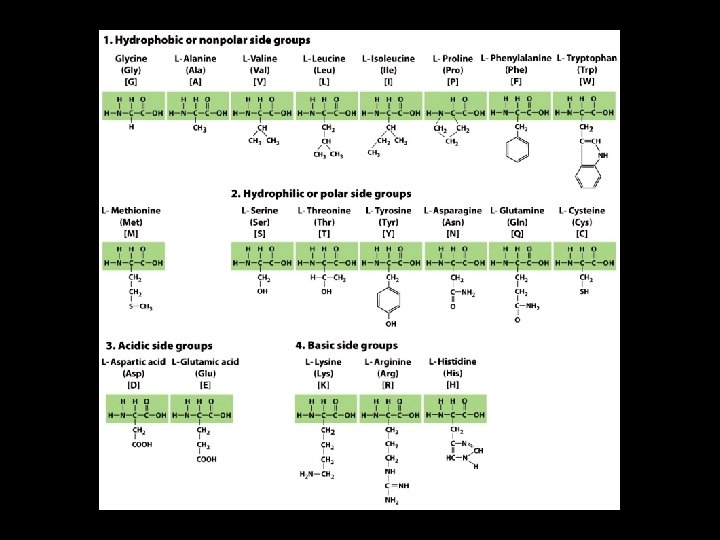
Protein Structure Proteins are complex macromolecules composed of 20 (? ) different amino acids.

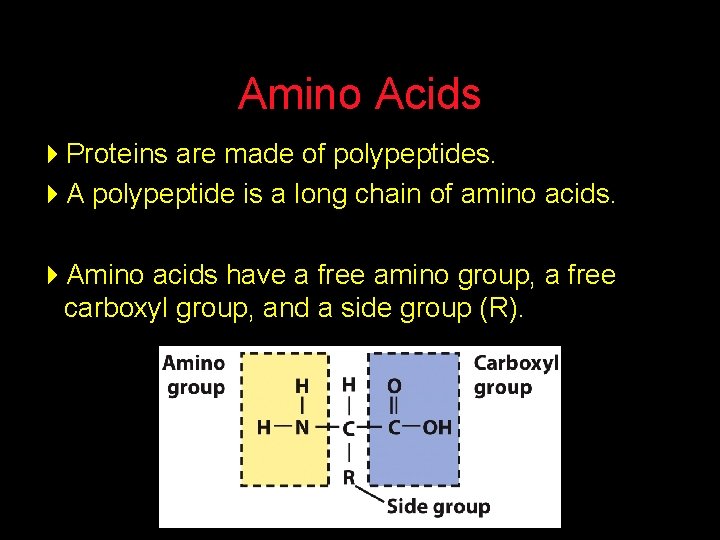
Amino Acids 4 Proteins are made of polypeptides. 4 A polypeptide is a long chain of amino acids. 4 Amino acids have a free amino group, a free carboxyl group, and a side group (R).


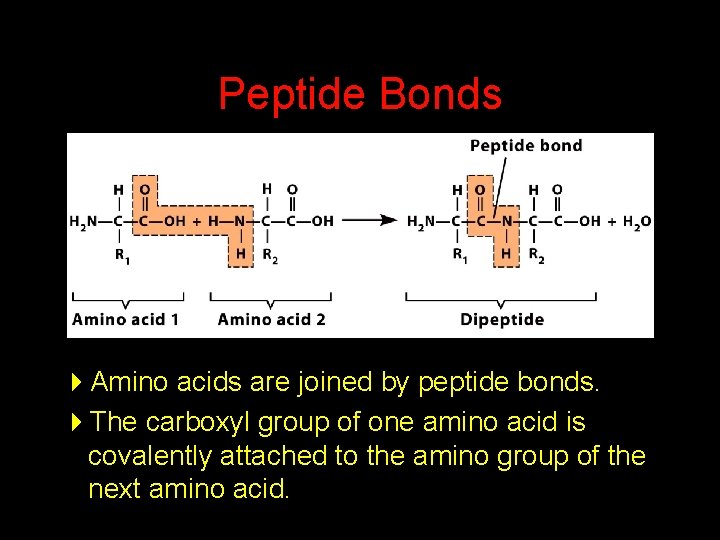
Peptide Bonds 4 Amino acids are joined by peptide bonds. 4 The carboxyl group of one amino acid is covalently attached to the amino group of the next amino acid.

Protein Synthesis: Translation The genetic information in m. RNA molecules is translated into the amino acid sequences of polypeptides according to the specifications of the genetic code.

The Macromolecules of Translation 4 Polypeptides and r. RNA molecules Euk: 28 S, 18 S, 5 S 4 Amino-acid Activating Enzymes 4 t. RNA Molecules 4 Soluble proteins involved in polypeptide chain initiation, elongation, and termination

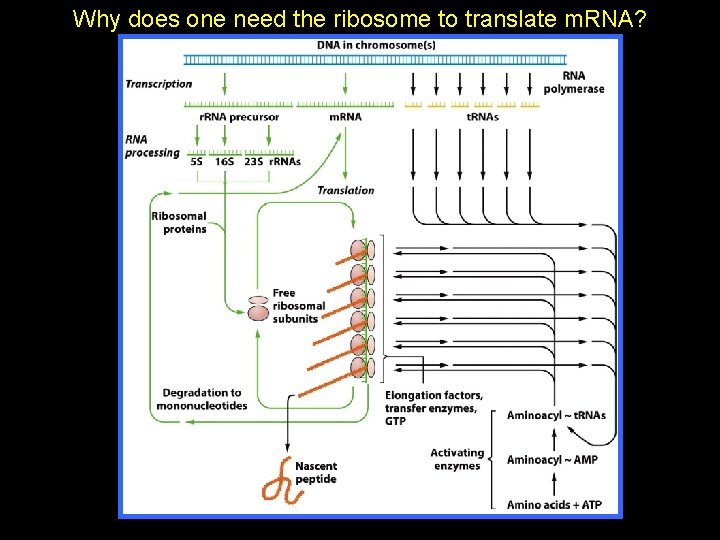
Why does one need the ribosome to translate m. RNA?

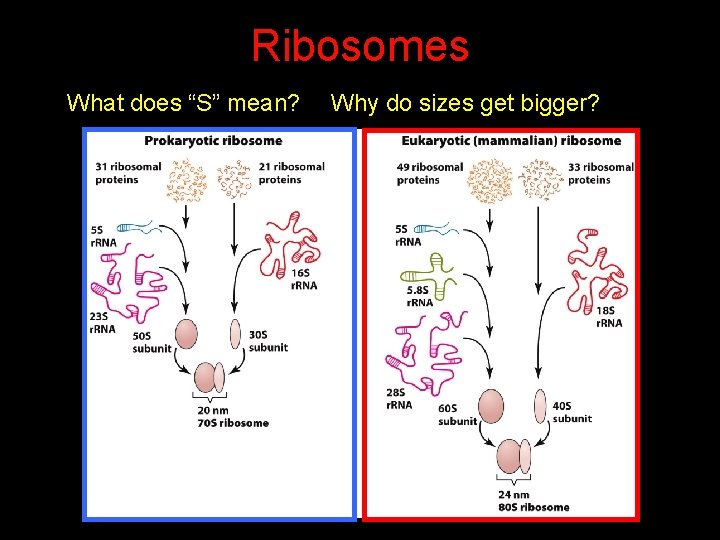
Ribosomes What does “S” mean? Why do sizes get bigger?

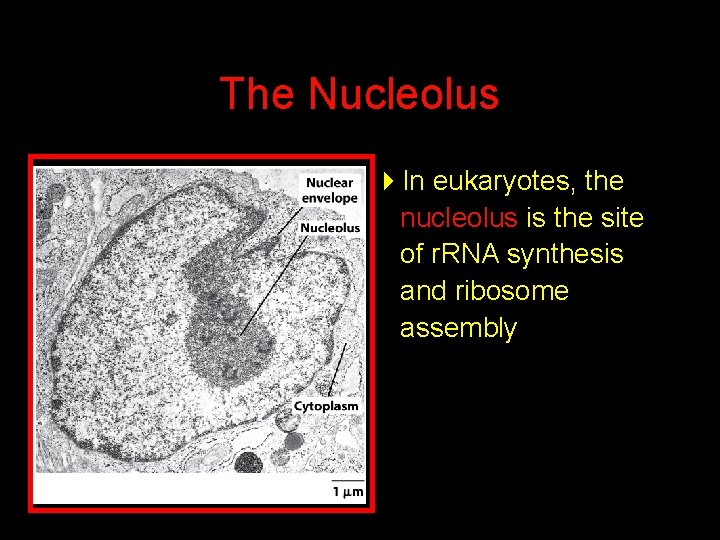
The Nucleolus 4 In eukaryotes, the nucleolus is the site of r. RNA synthesis and ribosome assembly

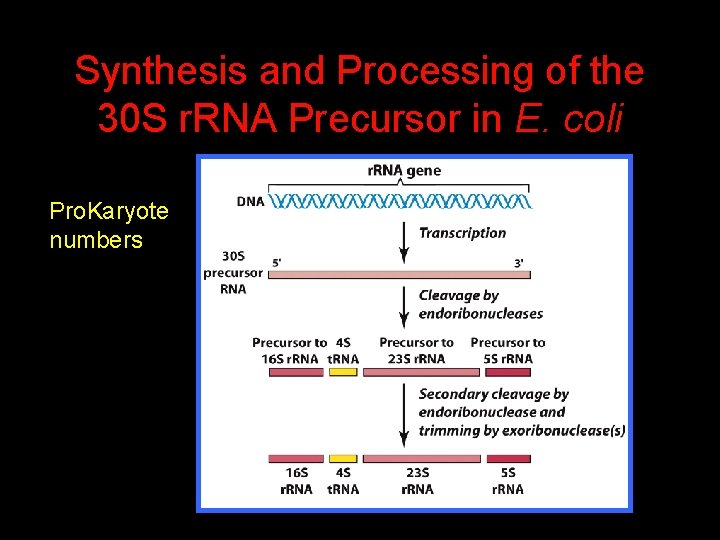
Synthesis and Processing of the 30 S r. RNA Precursor in E. coli Pro. Karyote numbers

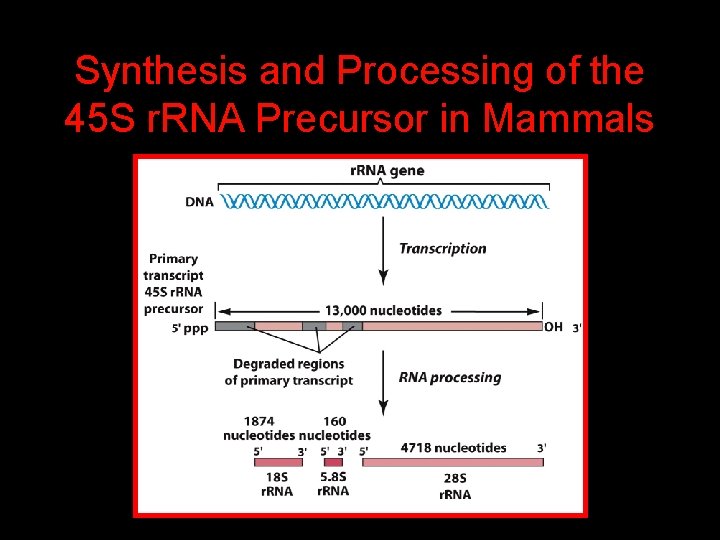
Synthesis and Processing of the 45 S r. RNA Precursor in Mammals

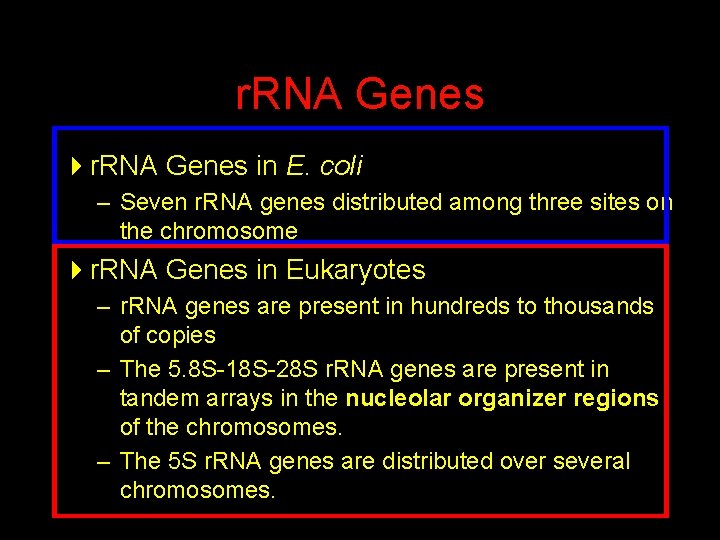
r. RNA Genes 4 r. RNA Genes in E. coli – Seven r. RNA genes distributed among three sites on the chromosome 4 r. RNA Genes in Eukaryotes – r. RNA genes are present in hundreds to thousands of copies – The 5. 8 S-18 S-28 S r. RNA genes are present in tandem arrays in the nucleolar organizer regions of the chromosomes. – The 5 S r. RNA genes are distributed over several chromosomes.

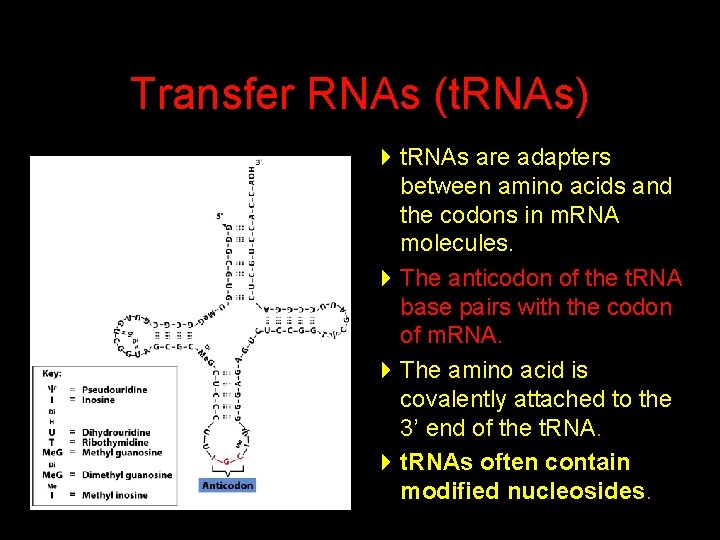
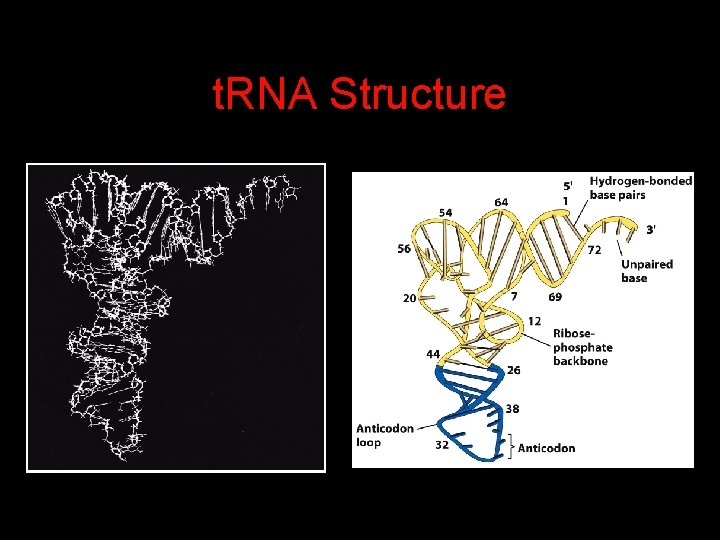
Transfer RNAs (t. RNAs) 4 t. RNAs are adapters between amino acids and the codons in m. RNA molecules. 4 The anticodon of the t. RNA base pairs with the codon of m. RNA. 4 The amino acid is covalently attached to the 3’ end of the t. RNA. 4 t. RNAs often contain modified nucleosides.

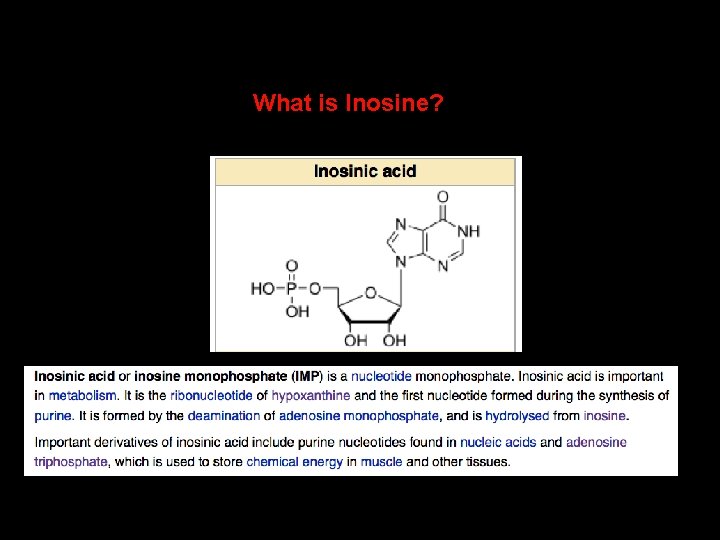
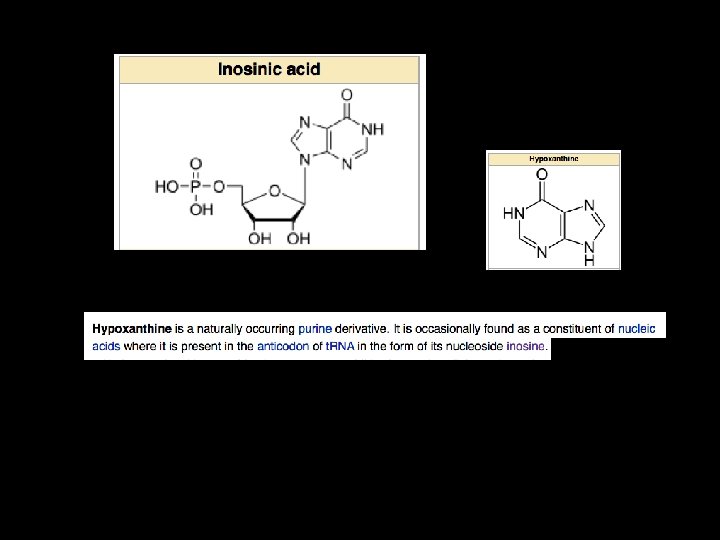
What is Inosine?


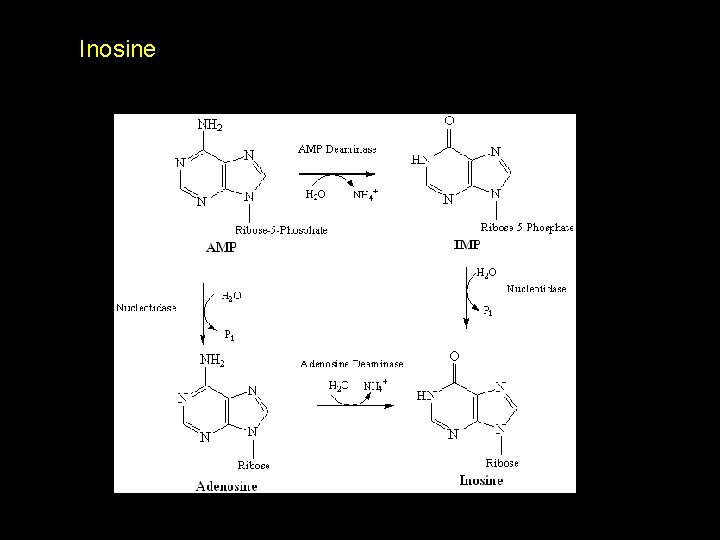
Inosine

t. RNA Structure

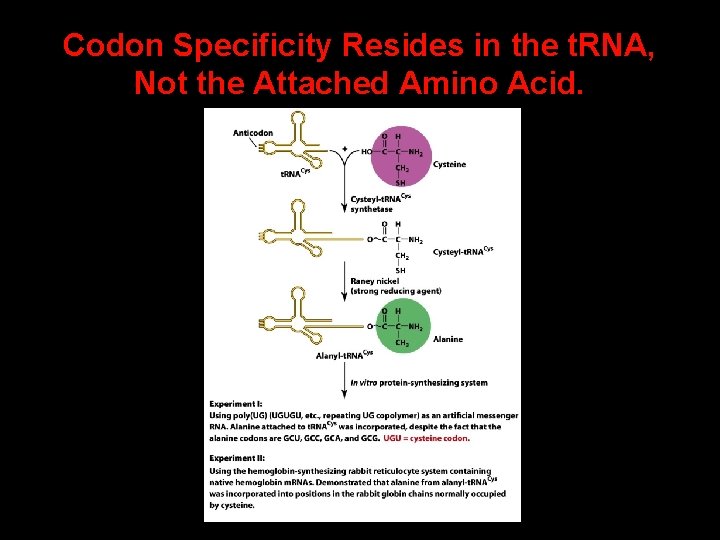
Specificity of t. RNAs 4 t. RNA molecules must have the correct anticodon sequence. 4 t. RNA molecules must be recognized by the correct aminoacyl-t. RNA synthetase. 4 t. RNA molecules must bind to the appropriate sites on the ribosomes.

Codon Specificity Resides in the t. RNA, Not the Attached Amino Acid.

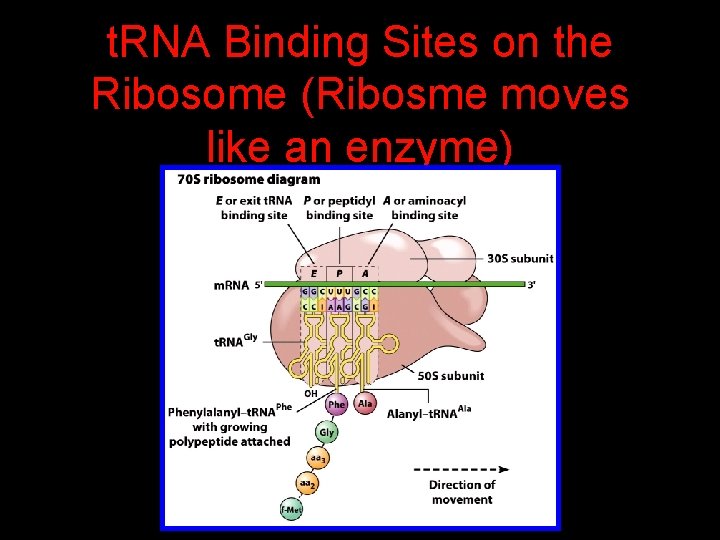
t. RNA Binding Sites on the Ribosome (Ribosme moves like an enzyme)

Stages of Translation 4 Polypeptide Chain Initiation 4 Chain Elongation 4 Chain Termination

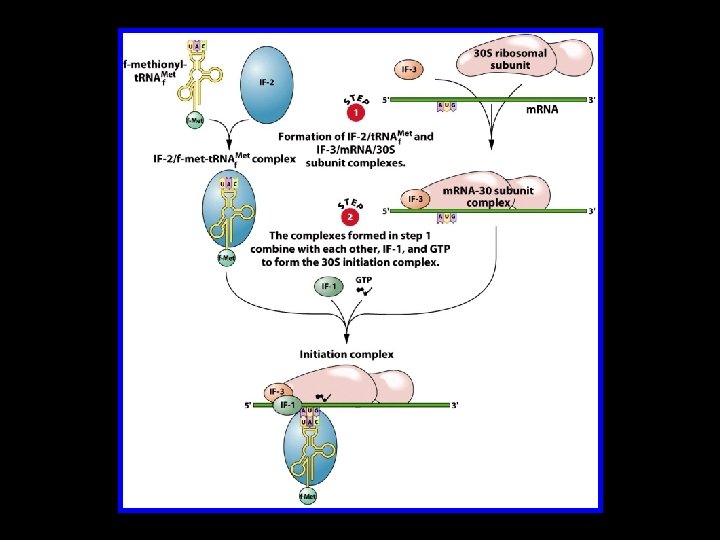
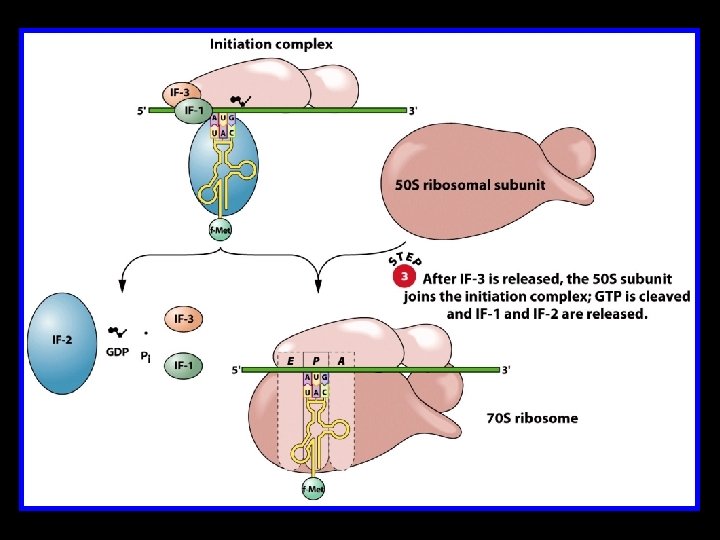
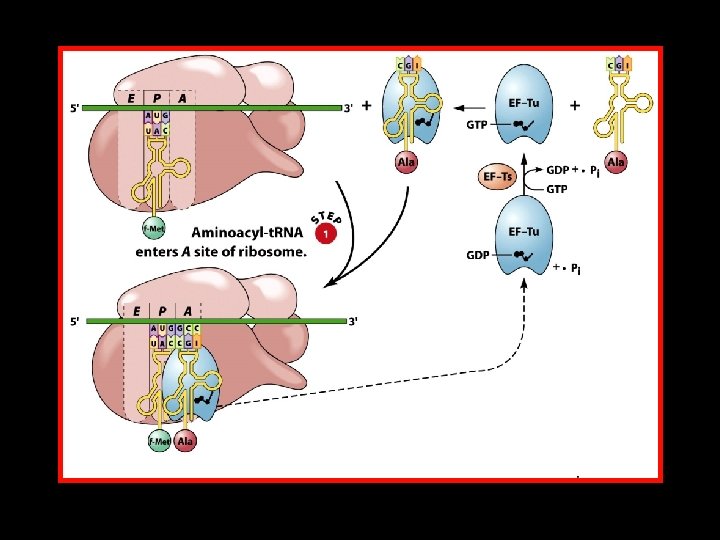
Translation Initiation in E. coli 430 S subunit of the ribosome 4 Initiator t. RNA (t. RNAMet) 4 m. RNA 4 Initiation Factors IF-1, IF-2, and IF-3 4 One molecule of GTP 450 S subunit of the ribosome



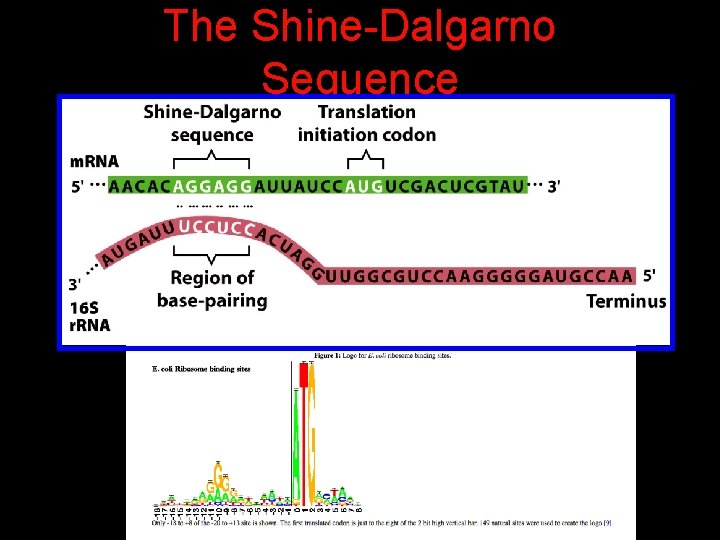
The Shine-Dalgarno Sequence

Translation Initiation in Eukaryotes 4 The amino group of the methionine on the initiator t. RNA is not formylated. 4 The initiation complex forms at the 5’ terminus of the m. RNA, not at the Shine-Dalgarno/AUG translation start site. 4 The initiation complex scans the m. RNA for an AUG initiation codon. Translation usually begins at the first AUG. 4 Kozak’s Rules describe the optimal sequence for efficient translation initiation in eukaryotes.


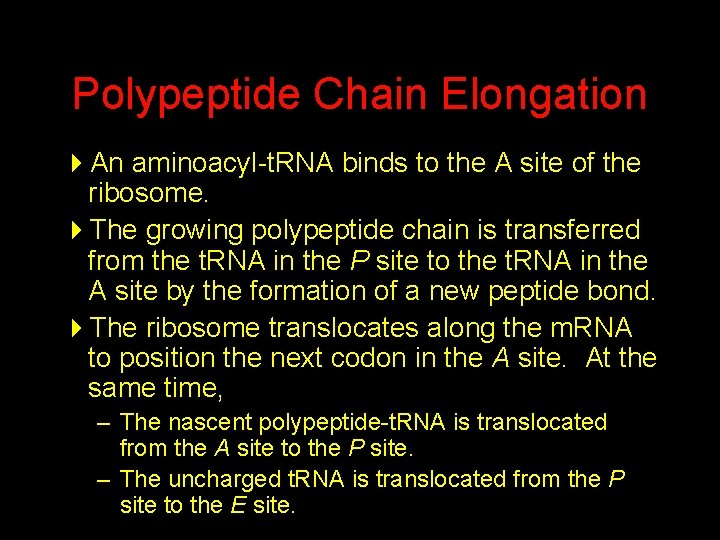
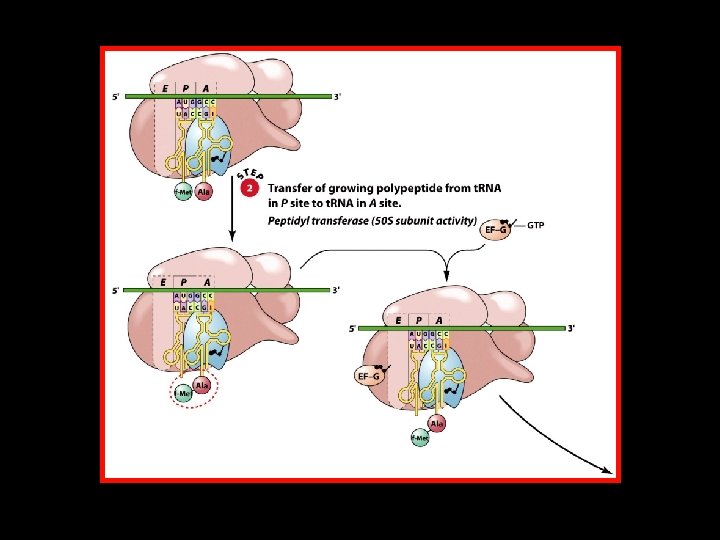
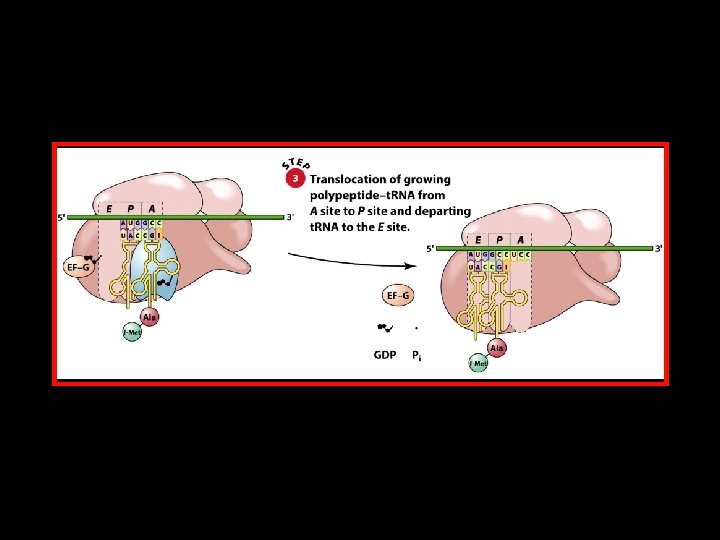
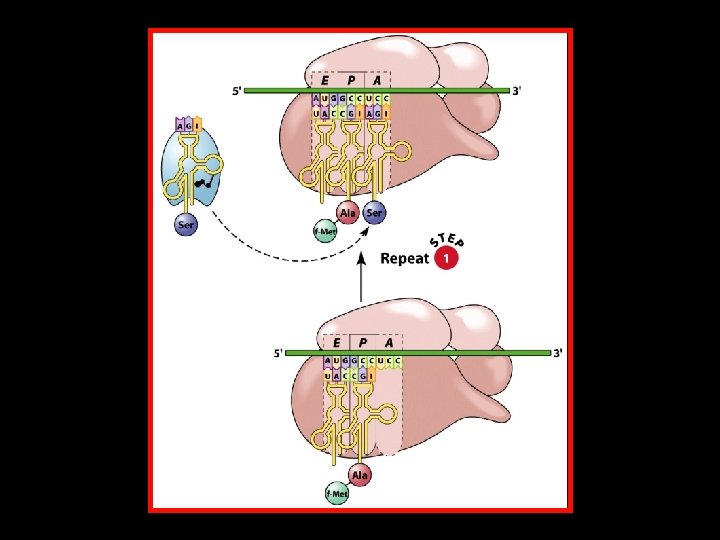
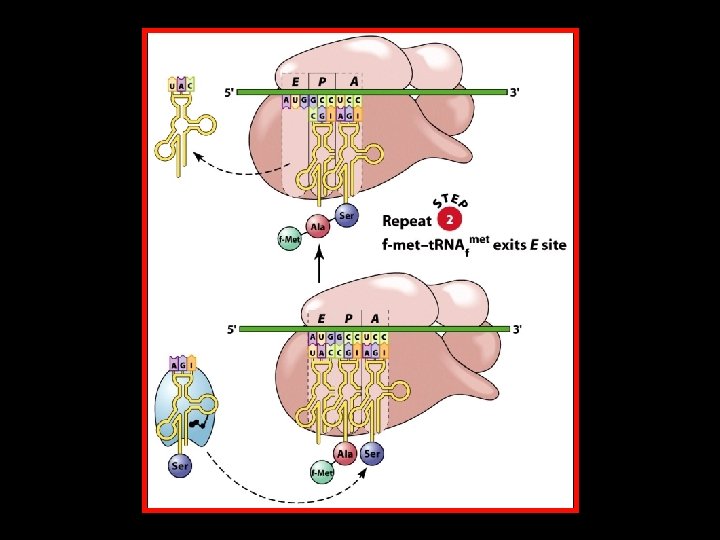
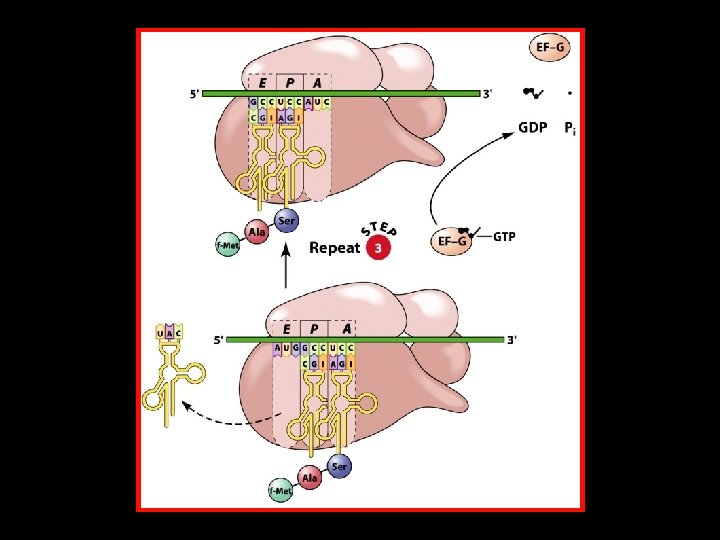
Polypeptide Chain Elongation 4 An aminoacyl-t. RNA binds to the A site of the ribosome. 4 The growing polypeptide chain is transferred from the t. RNA in the P site to the t. RNA in the A site by the formation of a new peptide bond. 4 The ribosome translocates along the m. RNA to position the next codon in the A site. At the same time, – The nascent polypeptide-t. RNA is translocated from the A site to the P site. – The uncharged t. RNA is translocated from the P site to the E site.







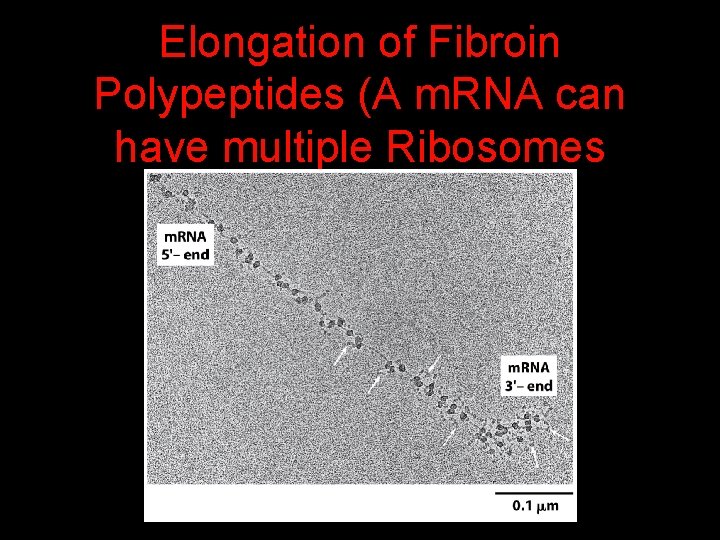
Elongation of Fibroin Polypeptides (A m. RNA can have multiple Ribosomes

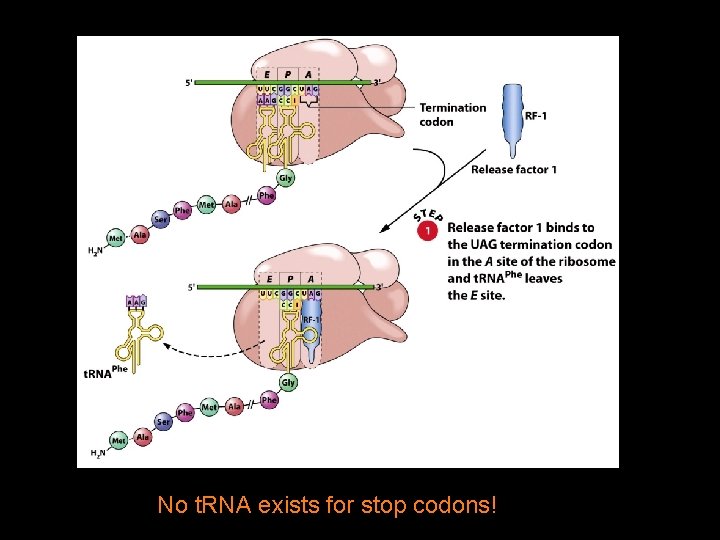
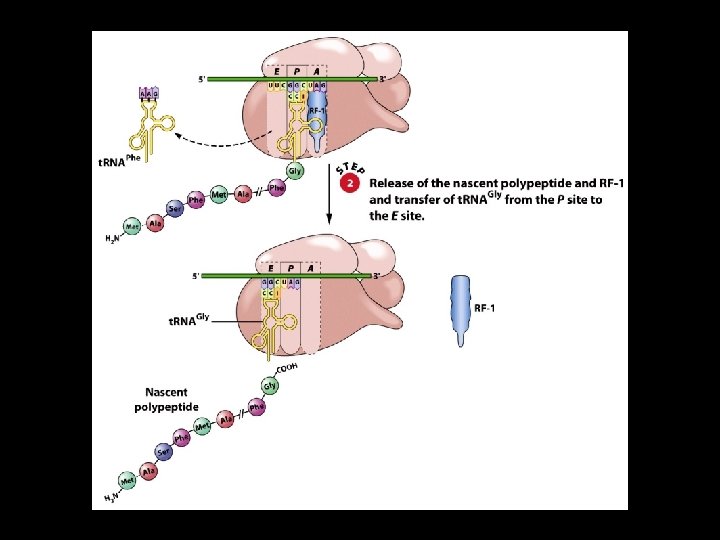
Polypeptide Chain Termination 4 Polypeptide chain termination occurs when a chain-termination codon (stop codon) enters the A site of the ribosome. 4 The stop codons are UAA, UAG, and UGA. 4 When a stop codon is encountered, a release factor binds to the A site. 4 A water molecule is added to the carboxyl terminus of the nascent polypeptide, causing termination.

No t. RNA exists for stop codons!


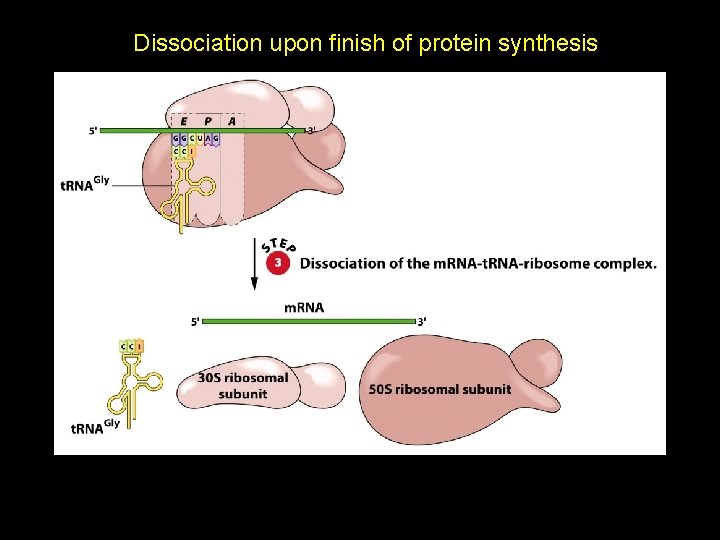
Dissociation upon finish of protein synthesis

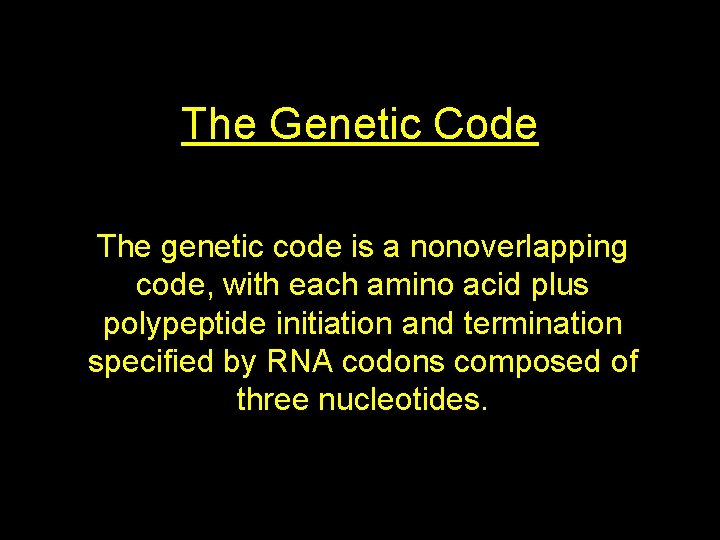
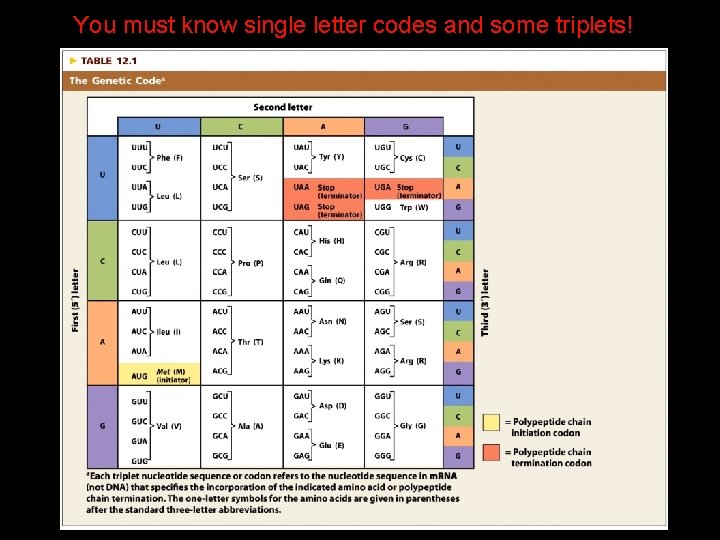
The Genetic Code The genetic code is a nonoverlapping code, with each amino acid plus polypeptide initiation and termination specified by RNA codons composed of three nucleotides.

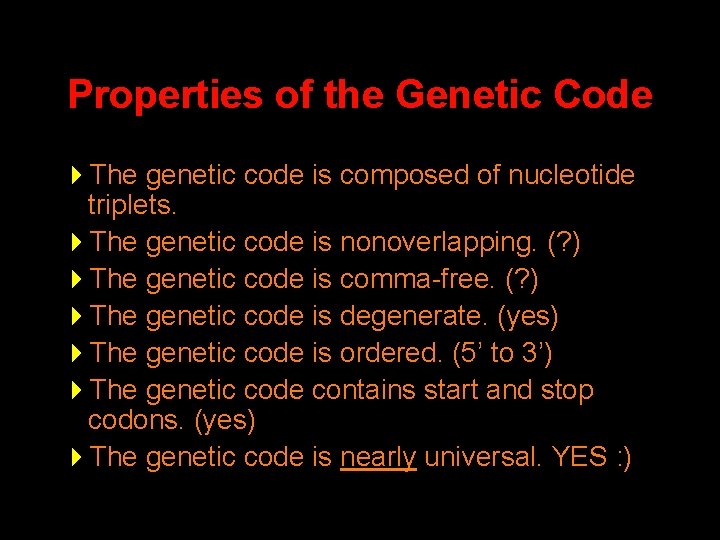
Properties of the Genetic Code 4 The genetic code is composed of nucleotide triplets. 4 The genetic code is nonoverlapping. (? ) 4 The genetic code is comma-free. (? ) 4 The genetic code is degenerate. (yes) 4 The genetic code is ordered. (5’ to 3’) 4 The genetic code contains start and stop codons. (yes) 4 The genetic code is nearly universal. YES : )

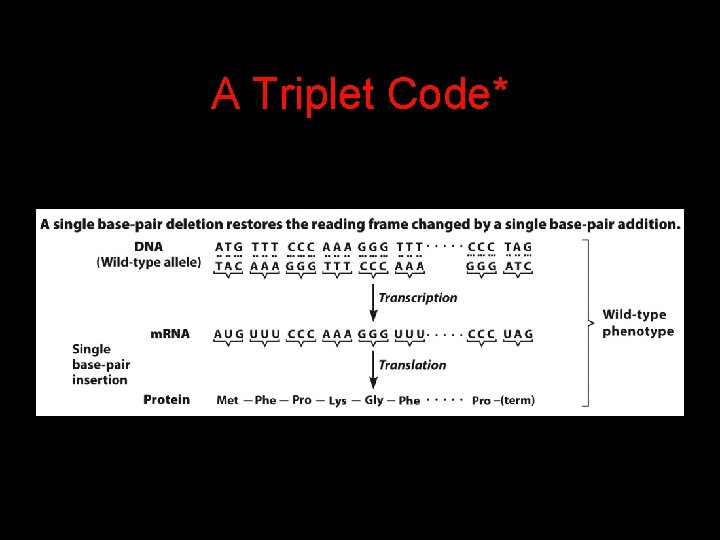
A Triplet Code*

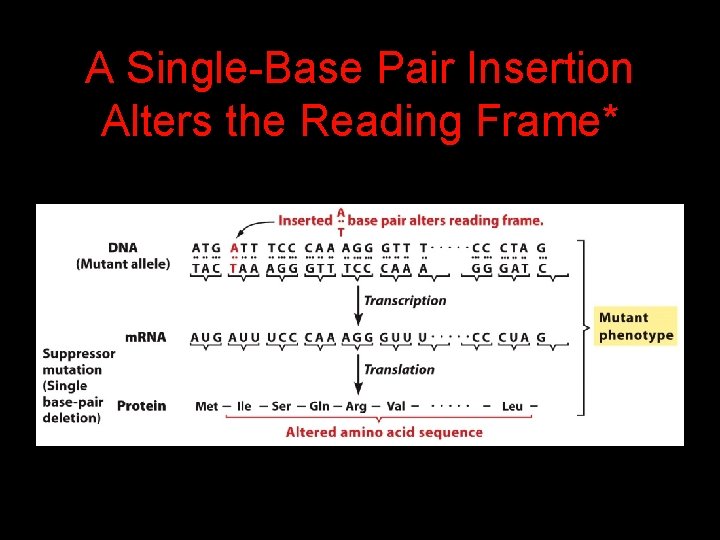
A Single-Base Pair Insertion Alters the Reading Frame*

A suppressor mutation restores the original reading frame. *

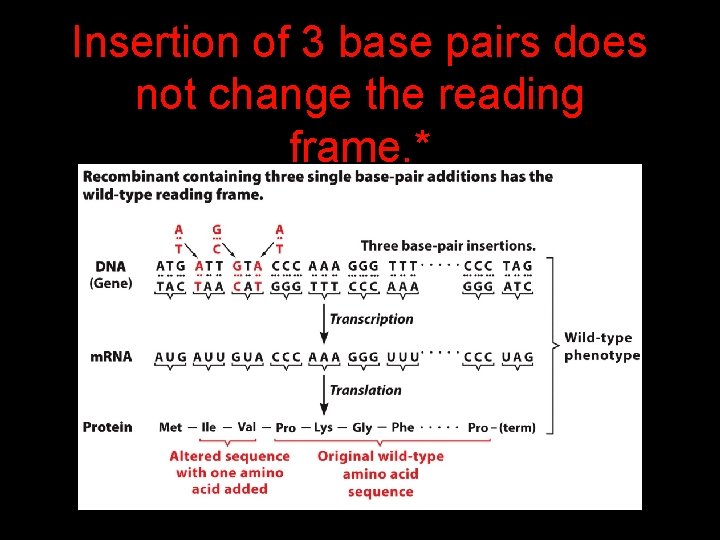
Insertion of 3 base pairs does not change the reading frame. *

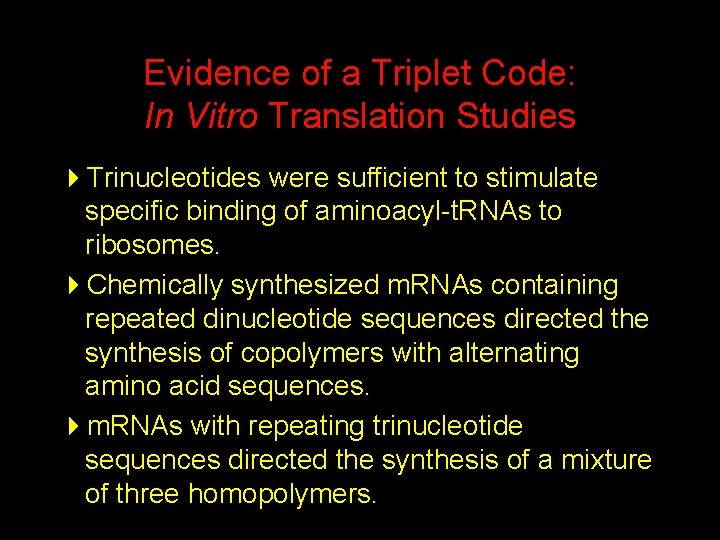
Evidence of a Triplet Code: In Vitro Translation Studies 4 Trinucleotides were sufficient to stimulate specific binding of aminoacyl-t. RNAs to ribosomes. 4 Chemically synthesized m. RNAs containing repeated dinucleotide sequences directed the synthesis of copolymers with alternating amino acid sequences. 4 m. RNAs with repeating trinucleotide sequences directed the synthesis of a mixture of three homopolymers.

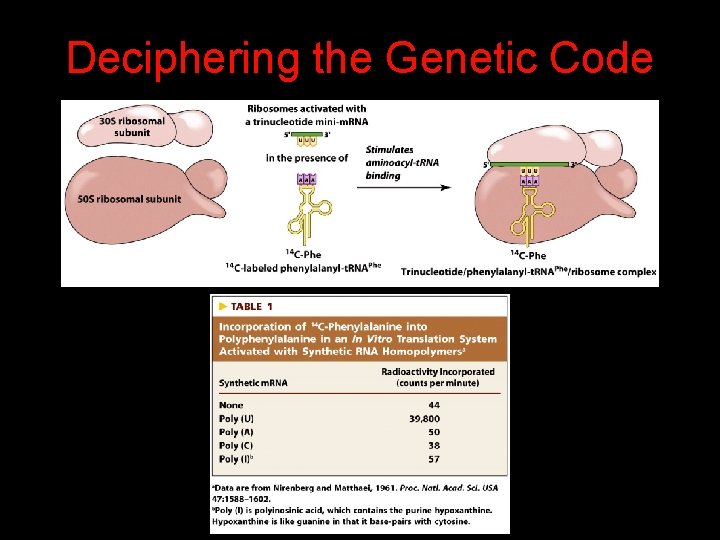
Deciphering the Genetic Code

You must know single letter codes and some triplets!

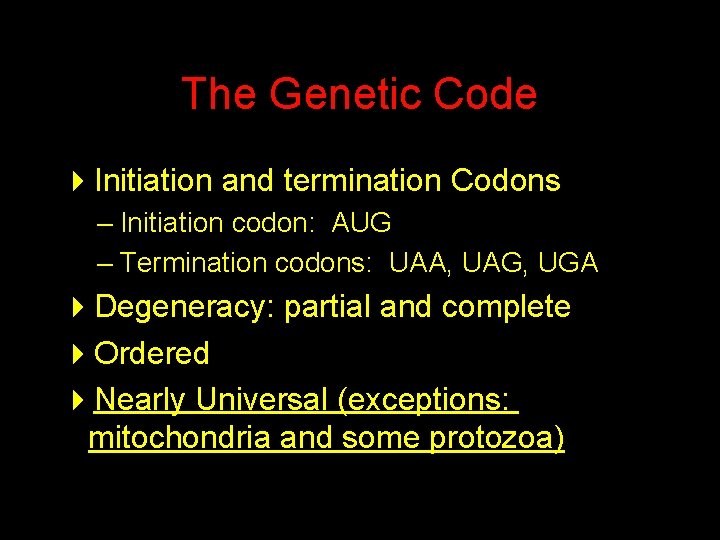
The Genetic Code 4 Initiation and termination Codons – Initiation codon: AUG – Termination codons: UAA, UAG, UGA 4 Degeneracy: partial and complete 4 Ordered 4 Nearly Universal (exceptions: mitochondria and some protozoa)

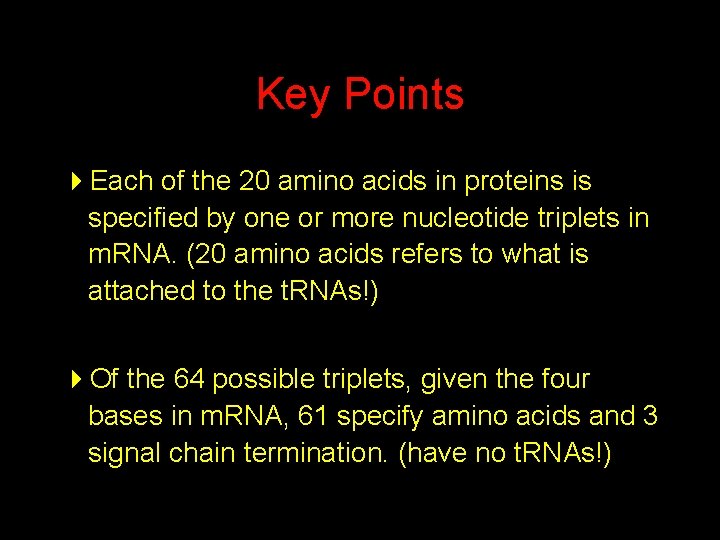
Key Points 4 Each of the 20 amino acids in proteins is specified by one or more nucleotide triplets in m. RNA. (20 amino acids refers to what is attached to the t. RNAs!) 4 Of the 64 possible triplets, given the four bases in m. RNA, 61 specify amino acids and 3 signal chain termination. (have no t. RNAs!)

Key Points 4 The code is nonoverlapping, with each nucleotide part of a single codon, degenerate, with most amino acids specified by two to four codons, and ordered, with similar amino acids specified by related codons. 4 The genetic code is nearly universal; with minor exceptions, the 64 triplets have the same meaning in all organisms. (this is funny)

Do all cells/animals make the same Repertoire of t. RNAs?

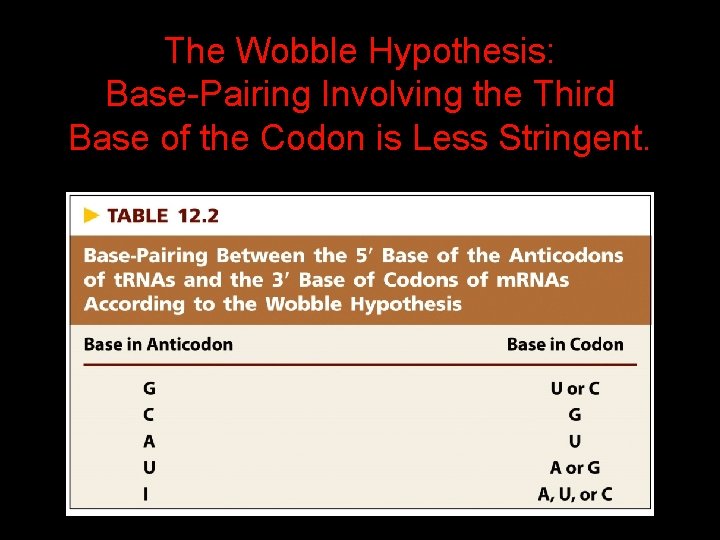
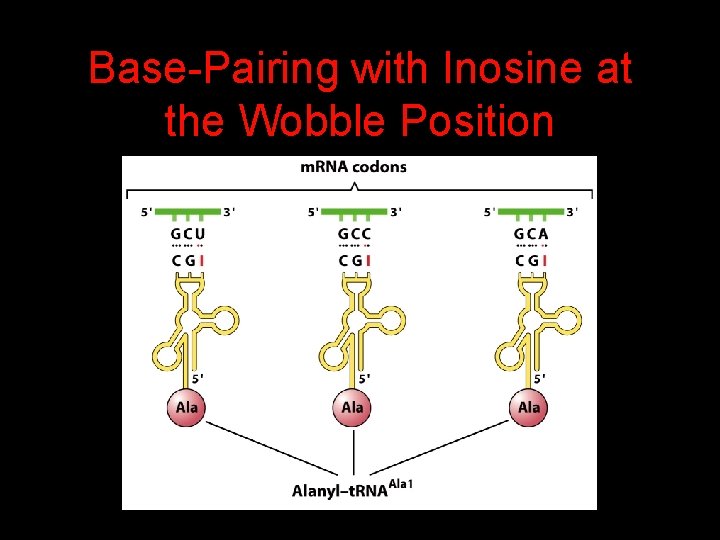
The Wobble Hypothesis: Base-Pairing Involving the Third Base of the Codon is Less Stringent.

Base-Pairing with Inosine at the Wobble Position

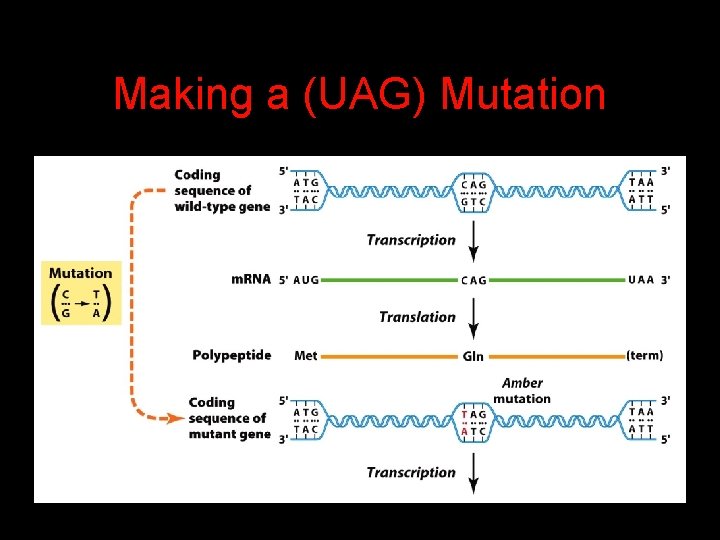
Suppressor Mutations 4 Some mutations in t. RNA genes alter the anticodons and therefore the codons recognized by the mutant t. RNAs. 4 These mutations were initially detected as suppressor mutations that suppressed the effects of other mutations. 4 Example: t. RNA mutations that suppress amber mutations (UAG chain-termination mutations) in the coding sequence of genes.

Making a (UAG) Mutation

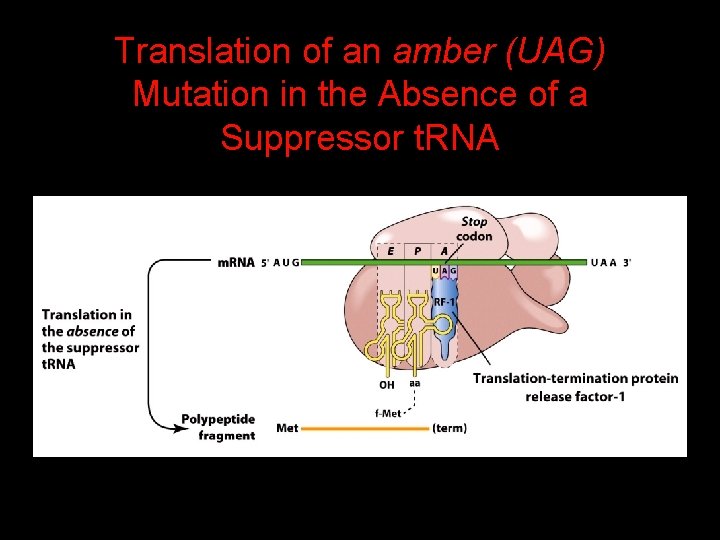
Translation of an amber (UAG) Mutation in the Absence of a Suppressor t. RNA

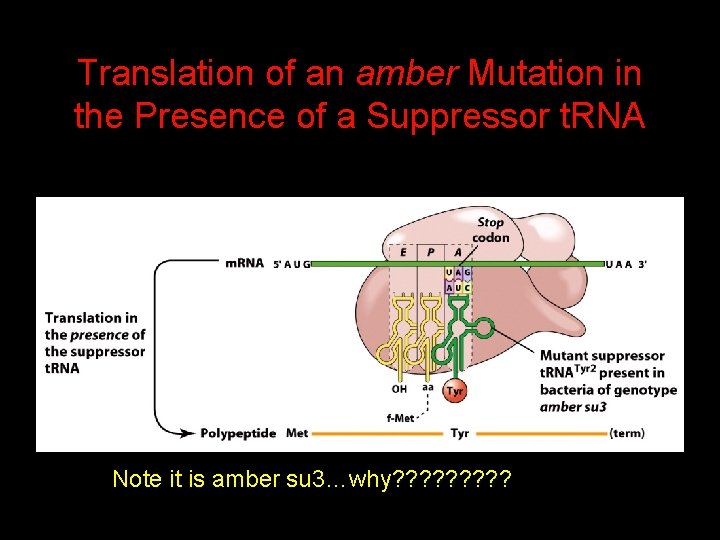
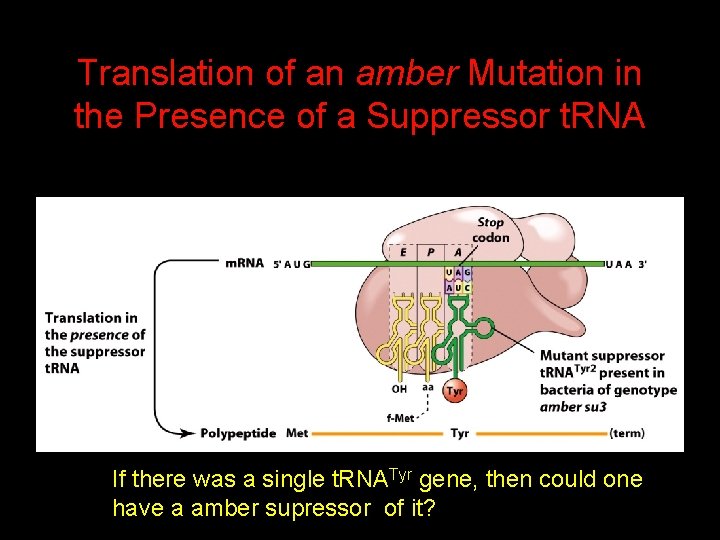
Translation of an amber Mutation in the Presence of a Suppressor t. RNA Note it is amber su 3…why? ? ? ? ?

Translation of an amber Mutation in the Presence of a Suppressor t. RNA If there was a single t. RNATyr gene, then could one have a amber supressor of it?

Historical Comparisons 4 Comparison of the amino acid sequence of bacteriophage MS 2 coat protein and the nucleotide sequence of the gene encoding the protein (Walter Fiers, 1972). Was this first? ? 4 Sickle-cell anemia: comparison of the sequence of the normal and sickle-cell alleles at the amino acid level and at the nucleotide level.

Are the proteins produced a pure reflection of the m. RNA sequence? ? t. RNA environment, protein modifications post-translationally

To Know for Exam RNApol II TATAA CCATGG (Nco I site and Kozak Rule) ATG AGGT…. splice GT……………A………polypyrimidine AG Poly. A recog sequence AATAAA The Reasons why ATG is a single codon and TGG is a single codon.