The Principle of Isoelectric Focusing A p H






























- Slides: 54


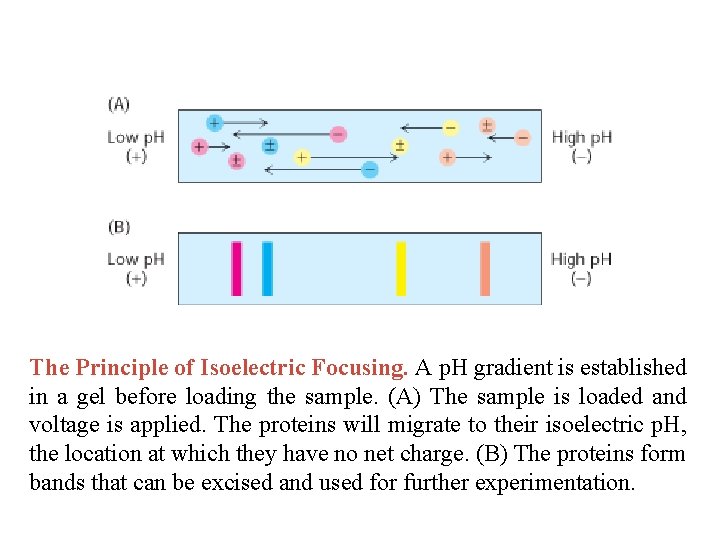
The Principle of Isoelectric Focusing. A p. H gradient is established in a gel before loading the sample. (A) The sample is loaded and voltage is applied. The proteins will migrate to their isoelectric p. H, the location at which they have no net charge. (B) The proteins form bands that can be excised and used for further experimentation.

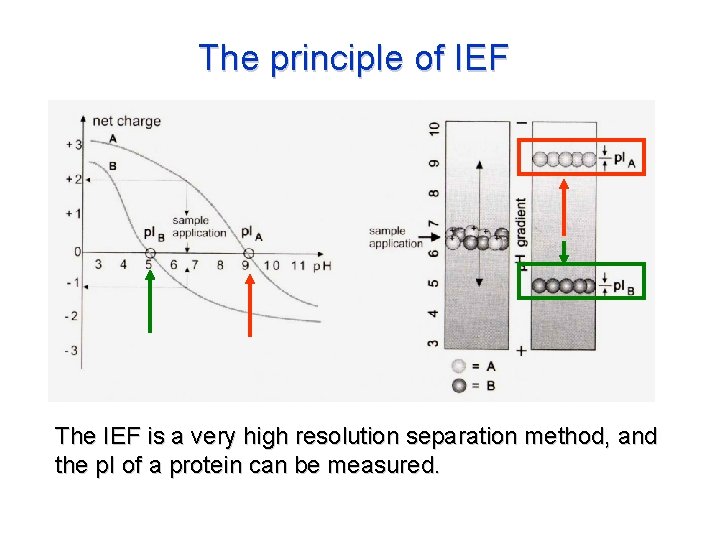
The principle of IEF The IEF is a very high resolution separation method, and the p. I of a protein can be measured.



















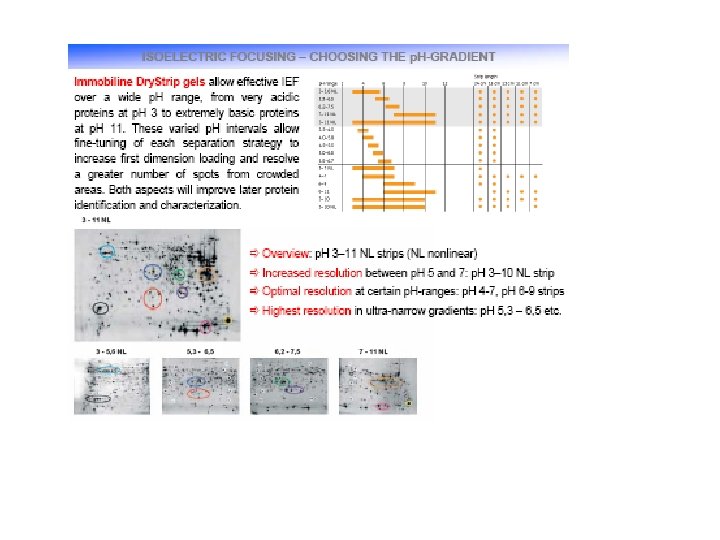
Advantage of IPG strips 1. Industrial standard (GMP) reduce variation. 2. The chemistry of the immobiline is better controllable. 3. The film-supported gel strips are easy to handle. 4. The fixed gradient are consistent during IEF. 5. Stable basic p. H gradient allow reproducible results for basic proteins. 6. High protein loads are achievable. 7. Less protein loss during equilibration in SDS buffer.





General detection methods Coomassie Blue Dyes - commonly used - does not interfere with subsequent protein identification - inexpensive - sensitivity well below silver and fluorescent dyes Silver stain - sensitivity 10 -50 times greater than CB - ability to detect 1 ng of protein - silver diammine/silver nitrate - relatively expensive (reagents/waste disposal) - high background Fluorescent Stains and Dyes - accurately determine changes in protein expression - greater sensitivity than silver stain - DIGE - cost

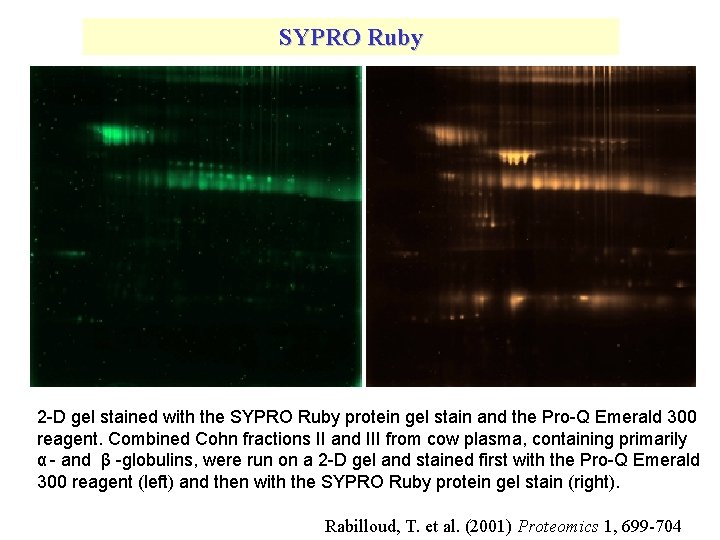
SYPRO Ruby 2 -D gel stained with the SYPRO Ruby protein gel stain and the Pro-Q Emerald 300 reagent. Combined Cohn fractions II and III from cow plasma, containing primarily α - and β -globulins, were run on a 2 -D gel and stained first with the Pro-Q Emerald 300 reagent (left) and then with the SYPRO Ruby protein gel stain (right). Rabilloud, T. et al. (2001) Proteomics 1, 699 -704

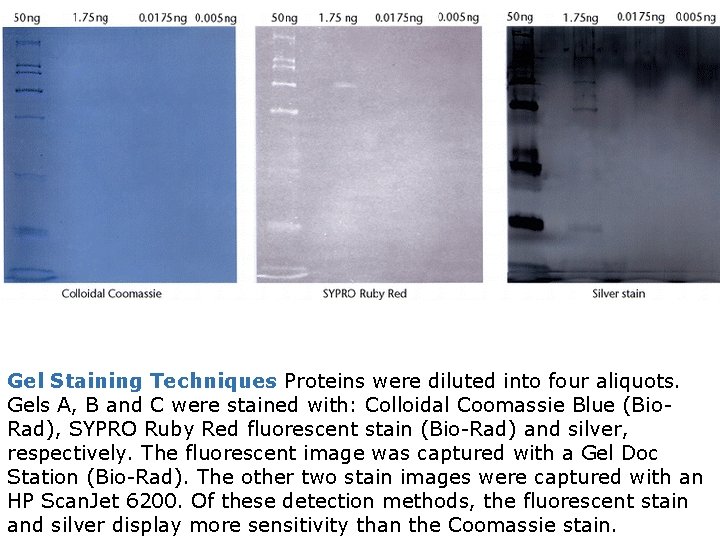
Gel Staining Techniques Proteins were diluted into four aliquots. Gels A, B and C were stained with: Colloidal Coomassie Blue (Bio. Rad), SYPRO Ruby Red fluorescent stain (Bio-Rad) and silver, respectively. The fluorescent image was captured with a Gel Doc Station (Bio-Rad). The other two stain images were captured with an HP Scan. Jet 6200. Of these detection methods, the fluorescent stain and silver display more sensitivity than the Coomassie stain.







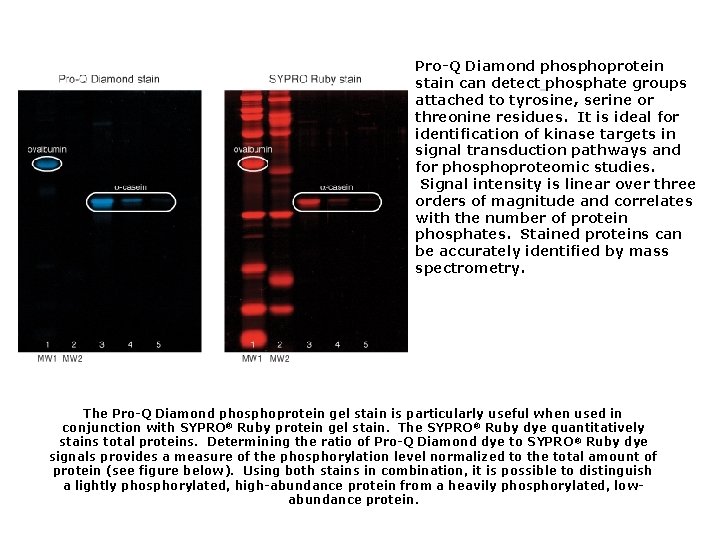
Pro-Q Diamond phosphoprotein stain can detect phosphate groups attached to tyrosine, serine or threonine residues. It is ideal for identification of kinase targets in signal transduction pathways and for phosphoproteomic studies. Signal intensity is linear over three orders of magnitude and correlates with the number of protein phosphates. Stained proteins can be accurately identified by mass spectrometry. The Pro-Q Diamond phosphoprotein gel stain is particularly useful when used in conjunction with SYPRO® Ruby protein gel stain. The SYPRO® Ruby dye quantitatively stains total proteins. Determining the ratio of Pro-Q Diamond dye to SYPRO ® Ruby dye signals provides a measure of the phosphorylation level normalized to the total amount of protein (see figure below). Using both stains in combination, it is possible to distinguish a lightly phosphorylated, high-abundance protein from a heavily phosphorylated, lowabundance protein.

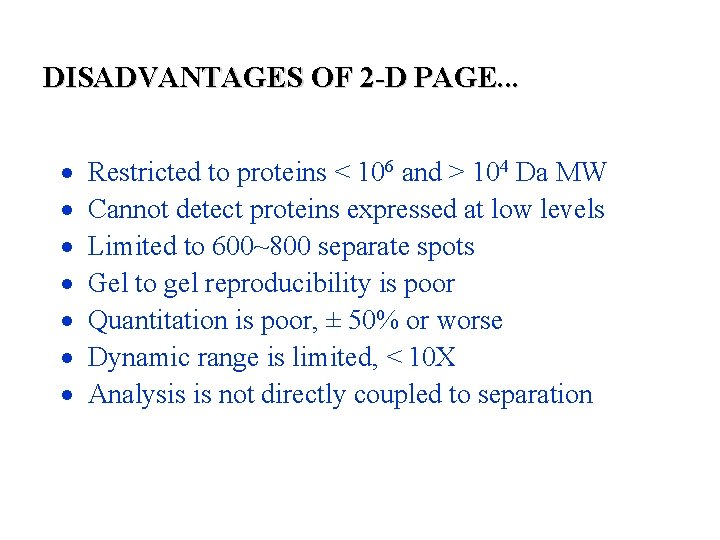
DISADVANTAGES OF 2 -D PAGE. . . Restricted to proteins < 106 and > 104 Da MW Cannot detect proteins expressed at low levels Limited to 600~800 separate spots Gel to gel reproducibility is poor Quantitation is poor, ± 50% or worse Dynamic range is limited, < 10 X Analysis is not directly coupled to separation

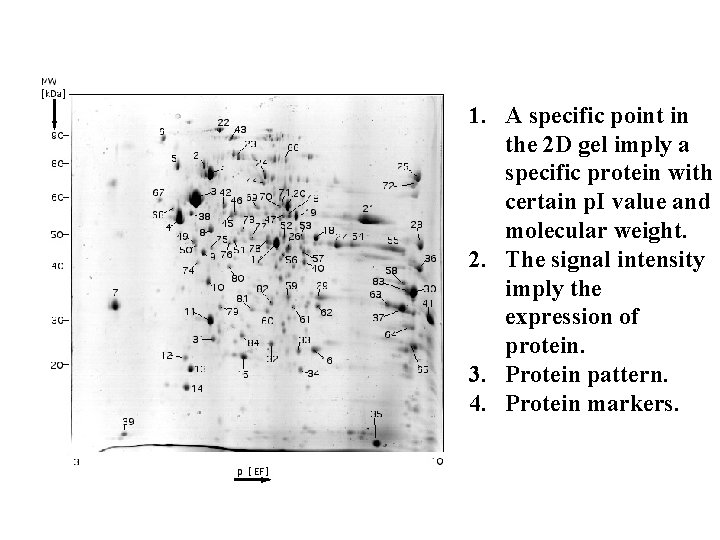
1. A specific point in the 2 D gel imply a specific protein with certain p. I value and molecular weight. 2. The signal intensity imply the expression of protein. 3. Protein pattern. 4. Protein markers.

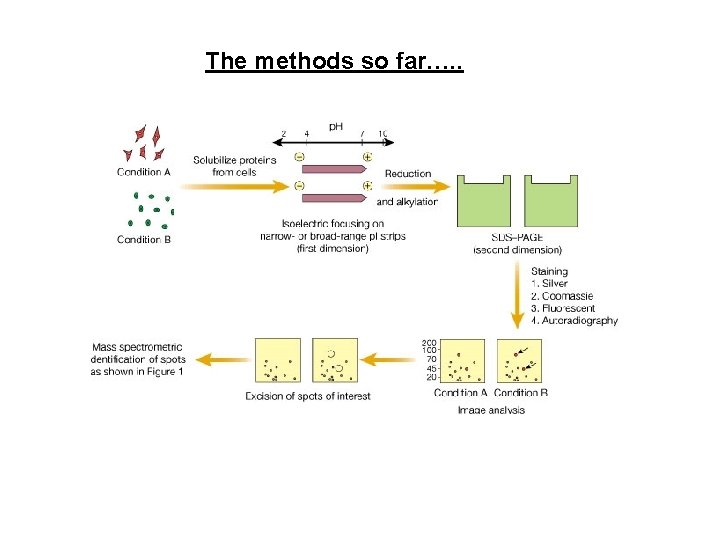
The methods so far…. .


http: //au. expasy. org/melanie/Melanie. htm

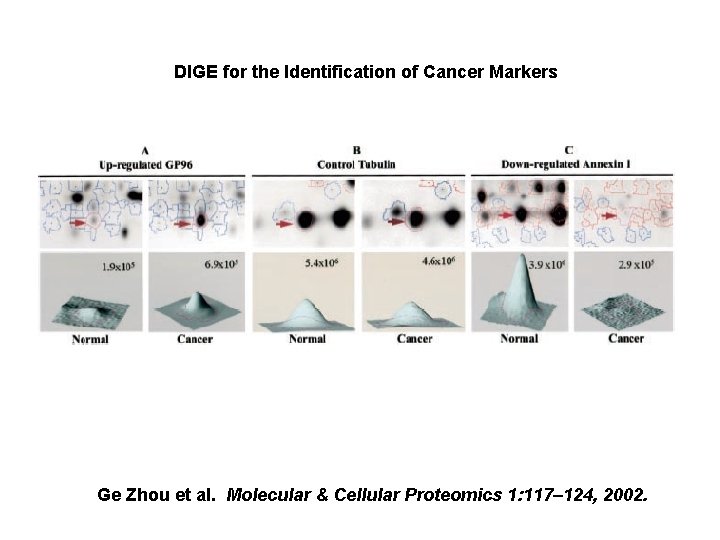
DIGE for the Identification of Cancer Markers Ge Zhou et al. Molecular & Cellular Proteomics 1: 117– 124, 2002.

2 D-PAGE IMAGE ANALYSIS A B Excise spot; elute; digest Extract peptides; MS analyze Protein identification

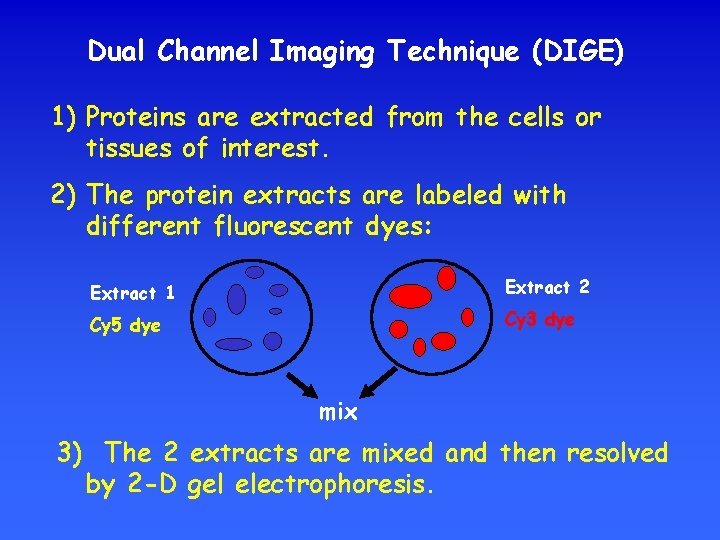
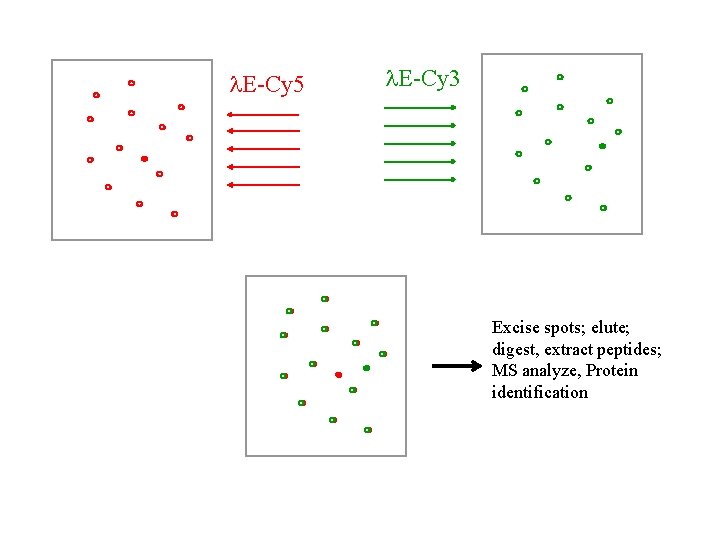
Dual Channel Imaging Technique (DIGE) 1) Proteins are extracted from the cells or tissues of interest. 2) The protein extracts are labeled with different fluorescent dyes: Extract 1 Extract 2 Cy 5 dye Cy 3 dye mix 3) The 2 extracts are mixed and then resolved by 2 -D gel electrophoresis.

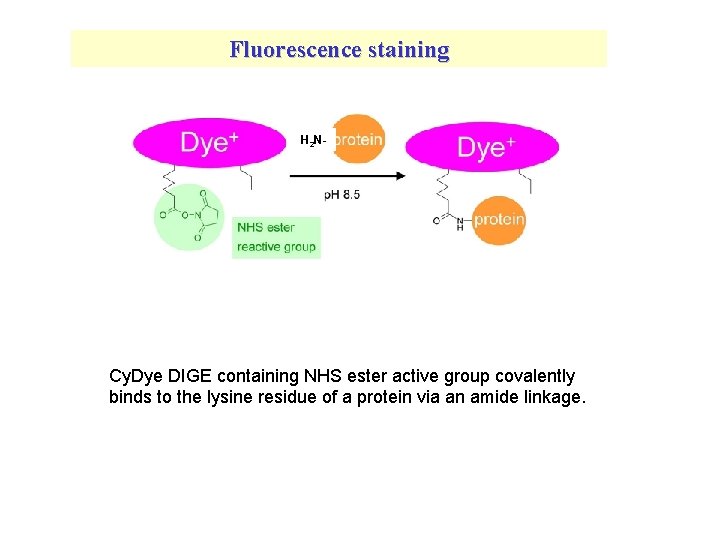
Fluorescence staining H 2 N- Cy. Dye DIGE containing NHS ester active group covalently binds to the lysine residue of a protein via an amide linkage.

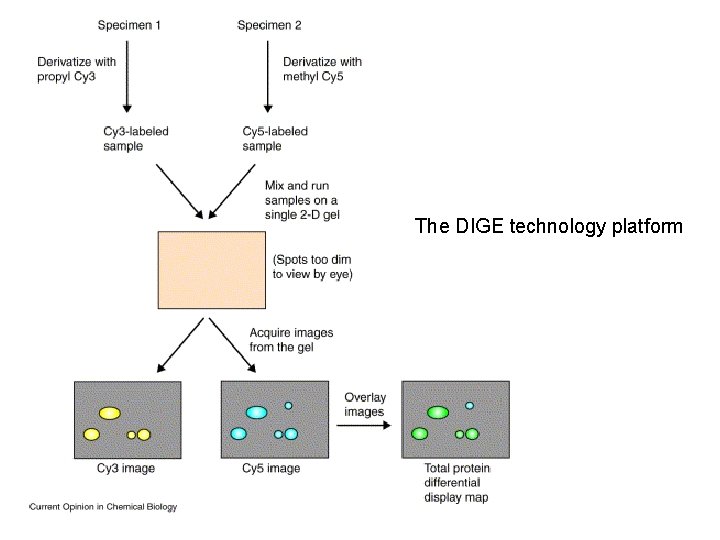
The DIGE technology platform


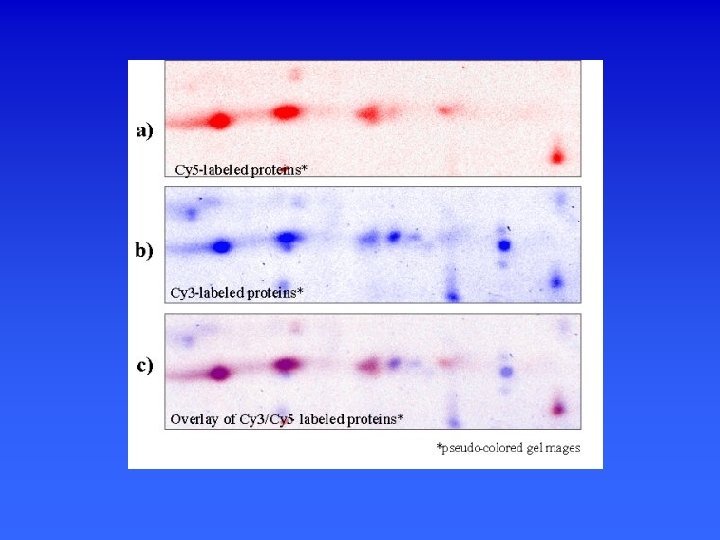
The DIGE technology platform. Two different samples are derivatized with two different fluorophores, combined and then run on a single 2 -D gel. Proteins are detected using a dual laser scanning device or xenon-arc-based instrument equipped with different excitation/emission filters in order to generate two separate images. The images are then matched by a computer-assisted overlay method, signals are normalized, and spots are quantified. Differences in protein expression are identified by evaluation of a pseudo-colored image and data spreadsheet. DIGE technology can maximally evaluate three different samples using Cy 2 -, Cy 3 - and Cy 5 based chemistries

l. E-Cy 5 l. E-Cy 3 Excise spots; elute; digest, extract peptides; MS analyze, Protein identification

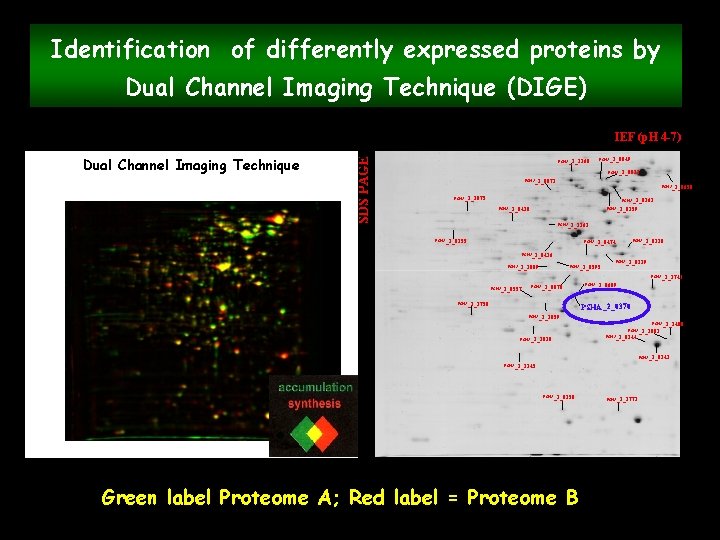
Identification of differently expressed proteins by Dual Channel Imaging Technique (DIGE) Dual Channel Imaging Technique SDS PAGE IEF (p. H 4 -7) PSHA_1_2360 PSHA_1_0849 PSHA_1_0021 PSHA_1_0871 PSHA_1_0658 PSHA_1_1875 PSHA_2_0363 PSHA_2_0420 PSHA_2_0259 PSHA_1_2262 PSHA_1_0255 PSHA_2_0474 PSHA_1_0328 PSHA_1_0416 PSHA_1_3009 PSHA_1_0119 PSHA_1_0595 PSHA_1_1740 PSHA_2_0557 PSHA_1_0870 PSHA_1_2758 PSHA_1_0689 PSHA_2_0370 PSHA_1_1059 PSHA_1_2400 PSHA_1_3038 PSHA_1_3001 PSHA_2_0144 PSHA_1_0341 PSHA_1_2145 PSHA_2_0150 Green label Proteome A; Red label = Proteome B PSHA_1_2771


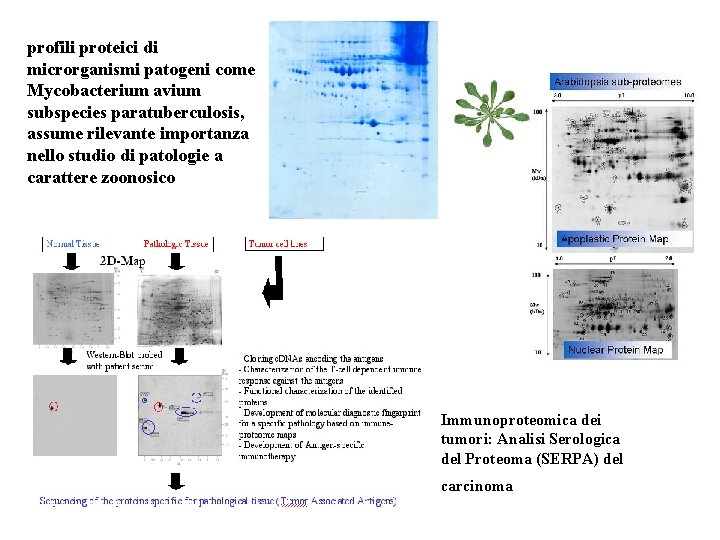
profili proteici di microrganismi patogeni come Mycobacterium avium subspecies paratuberculosis, assume rilevante importanza nello studio di patologie a carattere zoonosico Immunoproteomica dei tumori: Analisi Serologica del Proteoma (SERPA) del carcinoma


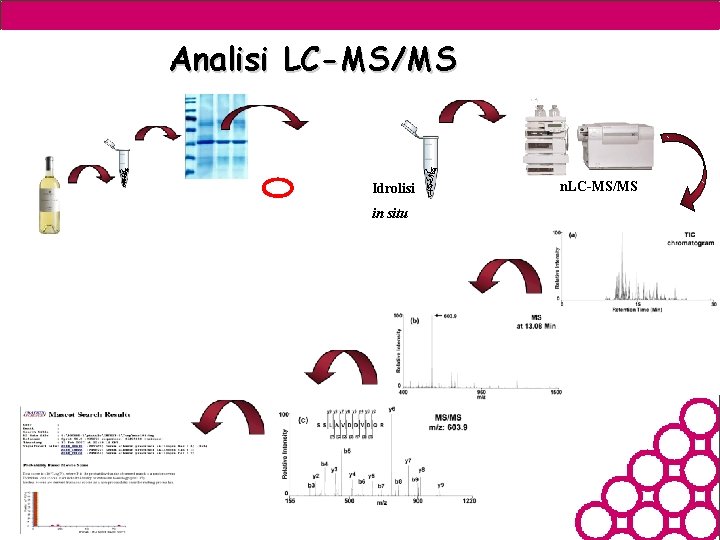
Analisi LC-MS/MS Idrolisi in situ n. LC-MS/MS
 Isoelectric focusing
Isoelectric focusing Isoelectric series
Isoelectric series Amino acid mnemonics
Amino acid mnemonics Isoelectric line
Isoelectric line Isoelectric point of casein
Isoelectric point of casein Jones and bartlett ekg strips 2012
Jones and bartlett ekg strips 2012 When focusing a specimen you should always start with the
When focusing a specimen you should always start with the Focusing on broad organizational needs
Focusing on broad organizational needs Types of microscope
Types of microscope The five focusing steps
The five focusing steps Microscope
Microscope Awareness requires totally focusing attention on the task.
Awareness requires totally focusing attention on the task. Fungsi dari metalic focusing cup adalah
Fungsi dari metalic focusing cup adalah Lwws
Lwws Focusing on broad organizational needs
Focusing on broad organizational needs Graphics display devices
Graphics display devices Explain how irony impacts “the pardoner’s tale.”
Explain how irony impacts “the pardoner’s tale.” Print pushing eye exercise
Print pushing eye exercise Ultrasound
Ultrasound Types of microscope
Types of microscope When focusing a specimen you should always start with the
When focusing a specimen you should always start with the Lời thề hippocrates
Lời thề hippocrates đại từ thay thế
đại từ thay thế Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Môn thể thao bắt đầu bằng từ đua
Môn thể thao bắt đầu bằng từ đua Công của trọng lực
Công của trọng lực Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Sự nuôi và dạy con của hổ
Sự nuôi và dạy con của hổ Thế nào là mạng điện lắp đặt kiểu nổi
Thế nào là mạng điện lắp đặt kiểu nổi Các loại đột biến cấu trúc nhiễm sắc thể
Các loại đột biến cấu trúc nhiễm sắc thể Nguyên nhân của sự mỏi cơ sinh 8
Nguyên nhân của sự mỏi cơ sinh 8 Vẽ hình chiếu đứng bằng cạnh của vật thể
Vẽ hình chiếu đứng bằng cạnh của vật thể Phản ứng thế ankan
Phản ứng thế ankan Gấu đi như thế nào
Gấu đi như thế nào Thiếu nhi thế giới liên hoan
Thiếu nhi thế giới liên hoan điện thế nghỉ
điện thế nghỉ Một số thể thơ truyền thống
Một số thể thơ truyền thống Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Lp html
Lp html Thế nào là số nguyên tố
Thế nào là số nguyên tố đặc điểm cơ thể của người tối cổ
đặc điểm cơ thể của người tối cổ Fecboak
Fecboak Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Glasgow thang điểm
Glasgow thang điểm ưu thế lai là gì
ưu thế lai là gì Hệ hô hấp
Hệ hô hấp Tư thế ngồi viết
Tư thế ngồi viết Cái miệng bé xinh thế chỉ nói điều hay thôi
Cái miệng bé xinh thế chỉ nói điều hay thôi Mật thư anh em như thể tay chân
Mật thư anh em như thể tay chân Bổ thể
Bổ thể Tư thế ngồi viết
Tư thế ngồi viết V cc
V cc Thẻ vin
Thẻ vin Thể thơ truyền thống
Thể thơ truyền thống