Introduction to RealTime PCR Dr Chaim Wachtel April























![Quality Control • Spectroscopic methods – Concentration, [NA] = A 260 x e mg/ml Quality Control • Spectroscopic methods – Concentration, [NA] = A 260 x e mg/ml](https://slidetodoc.com/presentation_image_h/ad0a8dd132e8db8fa50cd6e2b69e12ee/image-39.jpg)



- Slides: 69

Introduction to Real-Time PCR Dr. Chaim Wachtel April 11, 2013

Real-Time PCR • What is it? • How does it work • How do you properly perform an experiment • Analysis

The Nobel Prize in Chemistry 1993 was awarded "for contributions to the developments of methods within DNA-based chemistry" jointly with one half to Kary B. Mullis "for his invention of the polymerase chain reaction (PCR) method"and with one half to Michael Smith "for his fundamental contributions to the establishment of oligonucleotide-based, site-directed mutagenesis and its development for protein studies". Michael Smith

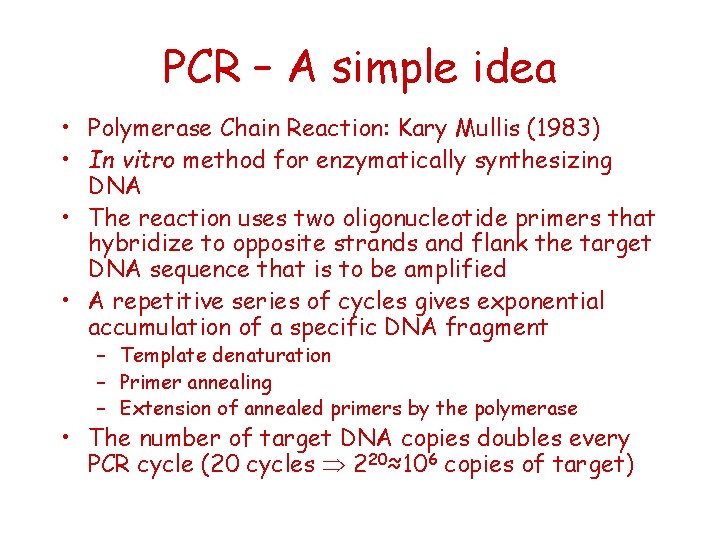
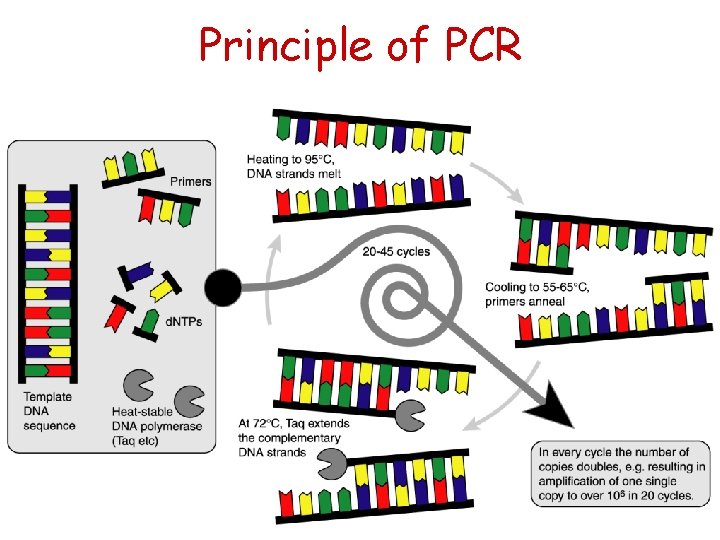
PCR – A simple idea • Polymerase Chain Reaction: Kary Mullis (1983) • In vitro method for enzymatically synthesizing DNA • The reaction uses two oligonucleotide primers that hybridize to opposite strands and flank the target DNA sequence that is to be amplified • A repetitive series of cycles gives exponential accumulation of a specific DNA fragment – Template denaturation – Primer annealing – Extension of annealed primers by the polymerase • The number of target DNA copies doubles every PCR cycle (20 cycles 220≈106 copies of target)

Principle of PCR

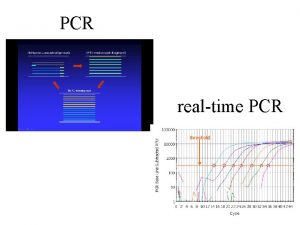
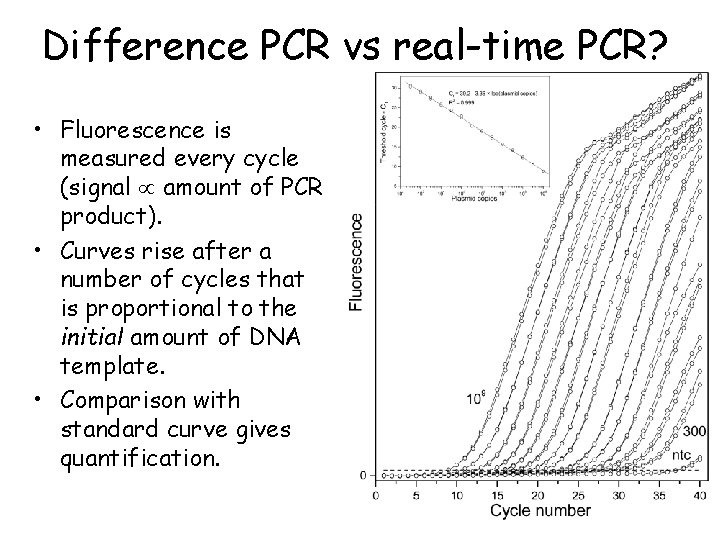
Difference PCR vs real-time PCR? • Fluorescence is measured every cycle (signal amount of PCR product). • Curves rise after a number of cycles that is proportional to the initial amount of DNA template. • Comparison with standard curve gives quantification.

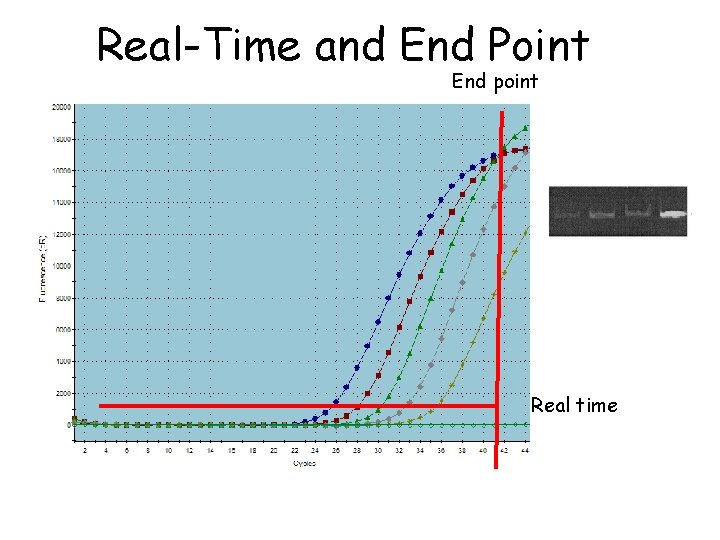
Real-Time and End Point End point Real time

MIQE: the minimum information for the publication of q. PCR experiments. http: //www. rdml. org/miqe. php




The m. RNA of the Arabidopsis Gene FT Moves from Leaf to Shoot Apex and Induces Flowering Tao Huang, Henrik Böhlenius, Sven Eriksson, François Parcy, and Ove Nilsson Science 9 September 2005: 1694 -1696. 2005: Signaling Breakthroughs of the Year

Retraction WE WISH TO RETRACT OUR RESEARCH ARTICLE “THE MRNA OF THE ARABIDOPSIS GENE FT MOVES from leaf to shoot apex and induces flowering” (1). After the first author (T. H. ) left the Umeå Plant Science Centre for another position, analysis of his original data revealed several anomalies. It is apparent from these files that data from the real-time RT-PCR were analyzed incorrectly. Certain data points were removed, while other data points were given increased weight in the statistical analysis. When all the primary real-time RT-PCR data are subjected to correct statistical analysis, most of the reported significant differences between time points disappear. Because of this, we are retracting the paper in its entirety.

Real-Time Machines • How do they work • What can you do with one – Gene expression – SNP detection – DNA detection (quantify) • How do you use them – Experiment design • Everything you need to know and more about RNA and RT-PCR

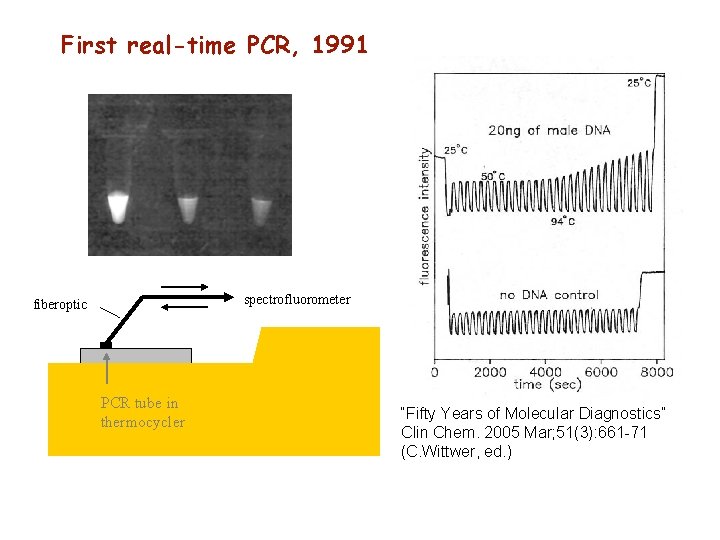
First real-time PCR, 1991 spectrofluorometer fiberoptic PCR tube in thermocycler “Fifty Years of Molecular Diagnostics” Clin Chem. 2005 Mar; 51(3): 661 -71 (C. Wittwer, ed. )

First commercial real-time PCR instruments ABI 7700 – laser/fiberoptic-based ABI 5700 – CCD camera-based Idaho Technology Light. Cycler – capillary tubes

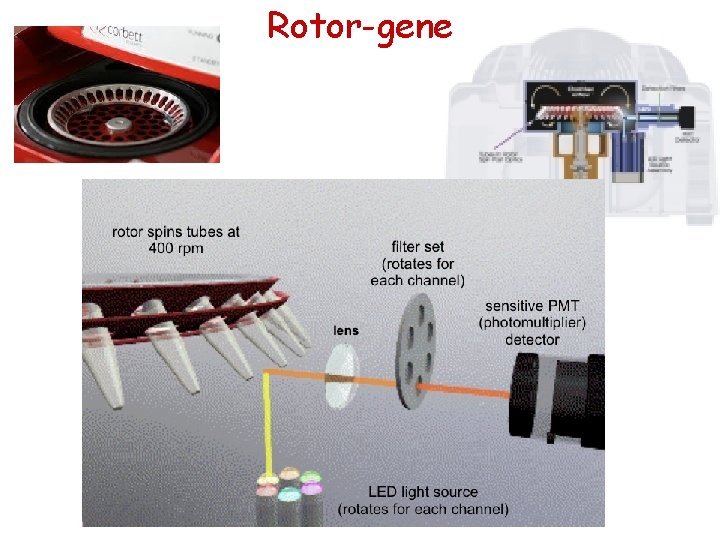
RT-PCR machines at Bar Ilan AB Step. One. Plus Fast Real-Time PCR System 7900 HT Fast Real-Time PCR System (Sol Efroni’s lab) Qiagen’s Rotor-gene (Oren Levy’s lab) Bio-Rad CFX-96 Thermo Piko. Real (Bachelet Lab)

Rotor-gene

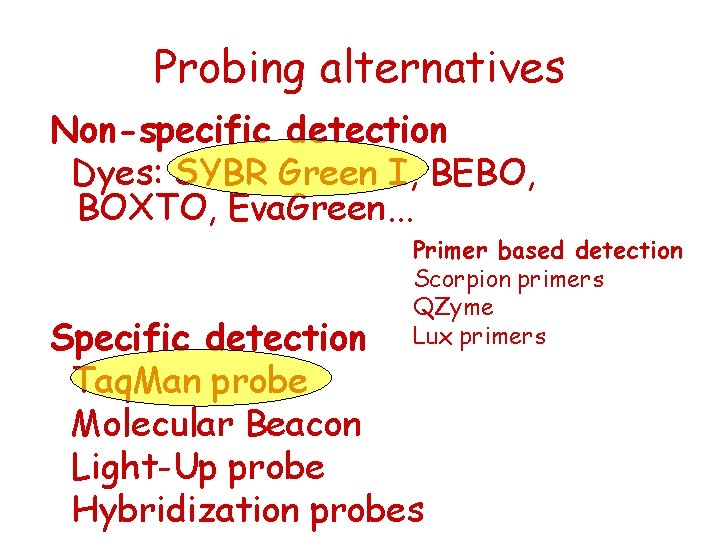
Probing alternatives Non-specific detection Dyes: SYBR Green I, BEBO, BOXTO, Eva. Green. . . Primer based detection Scorpion primers QZyme Lux primers Specific detection Taq. Man probe Molecular Beacon Light-Up probe Hybridization probes

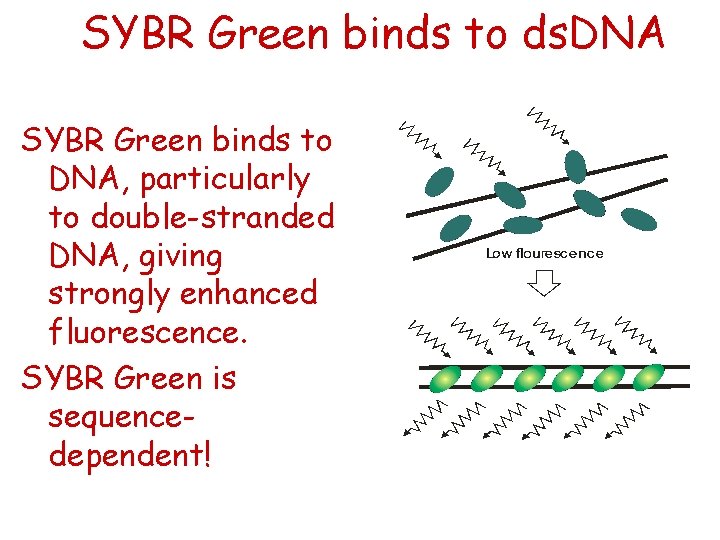
SYBR Green binds to ds. DNA SYBR Green binds to DNA, particularly to double-stranded DNA, giving strongly enhanced fluorescence. SYBR Green is sequencedependent!

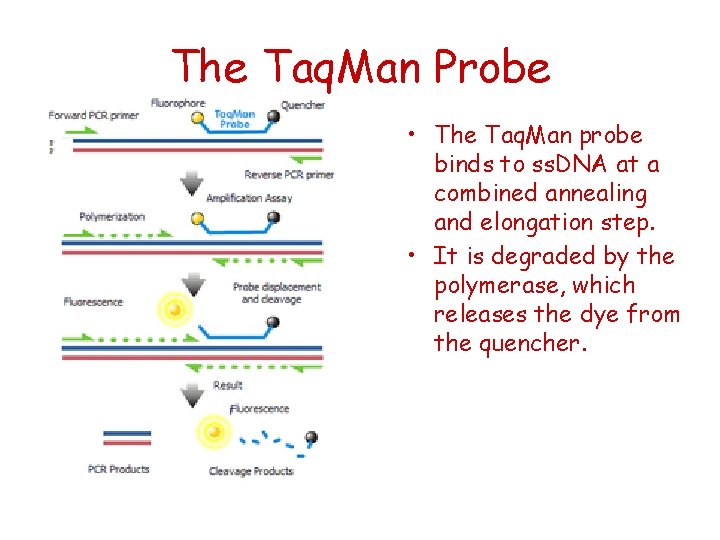
The Taq. Man Probe • The Taq. Man probe binds to ss. DNA at a combined annealing and elongation step. • It is degraded by the polymerase, which releases the dye from the quencher.

Multiplex Q-PCR • Detection of two (or more) different target sequences in the same reaction.

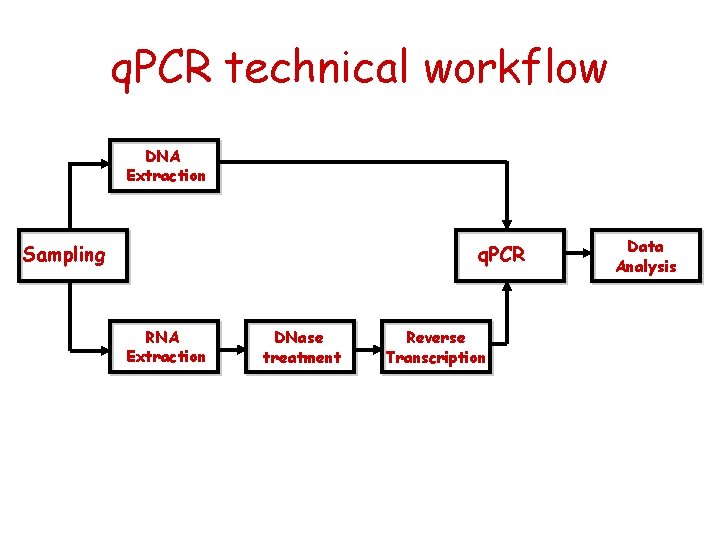
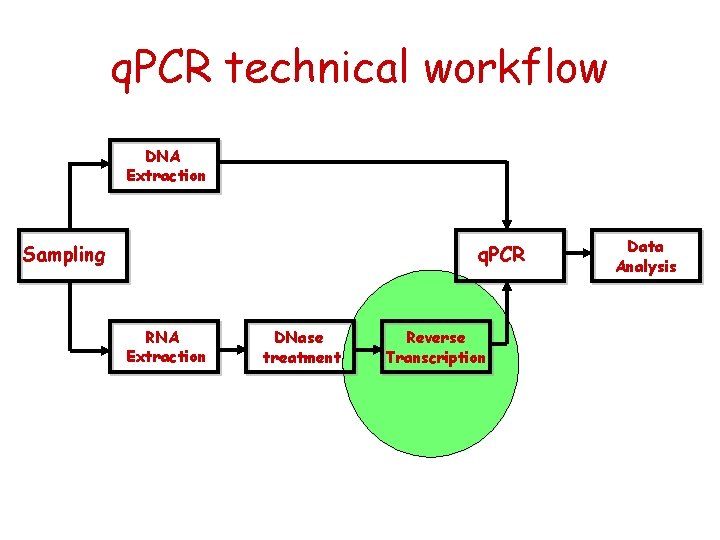
q. PCR technical workflow DNA Extraction Sampling q. PCR RNA Extraction DNase treatment Reverse Transcription Data Analysis

Nucleic acid isolation and purification

Overview • Sampling • Accessibility and lysis • Commonly used techniques • RNA considerations • Quality control

Why sample preparation? • Make target available • Remove inhibitors • Remove fluorescent contaminants • Preserve target integrity • Concentrate target

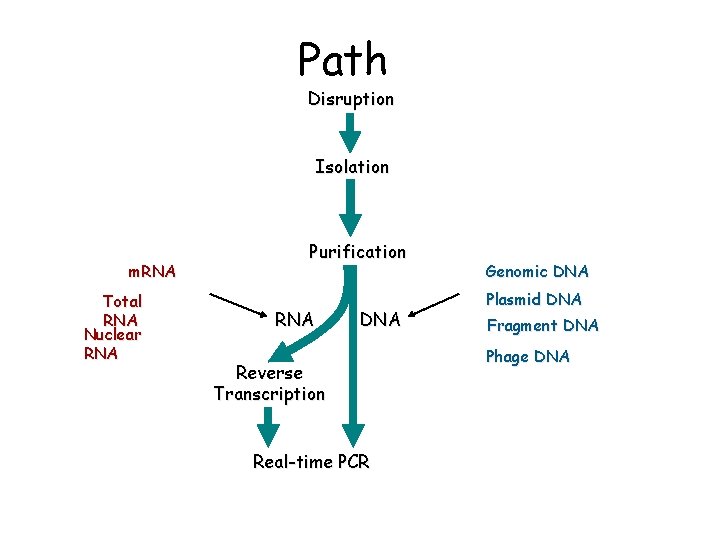
Path Disruption Isolation m. RNA Total RNA Nuclear RNA Purification RNA DNA Reverse Transcription Real-time PCR Genomic DNA Plasmid DNA Fragment DNA Phage DNA

Accessibility Sample disruption and homogenization – Mechanical • Grinding, Sonication, Vortexing, Polytron – Physical • Freezing – Enzymatic • Proteinase K, Lysozyme, Collagenase – Chemical • Guanidinium isothiocyanate (GITC), Alkali treatment, CTAB

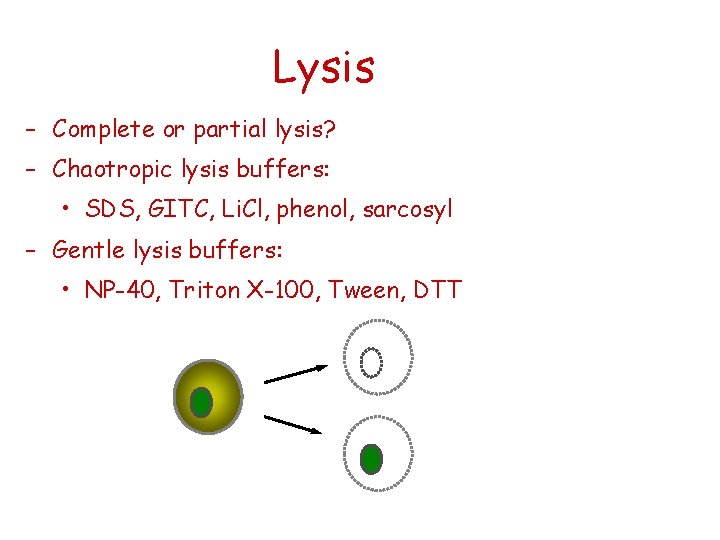
Lysis – Complete or partial lysis? – Chaotropic lysis buffers: • SDS, GITC, Li. Cl, phenol, sarcosyl – Gentle lysis buffers: • NP-40, Triton X-100, Tween, DTT

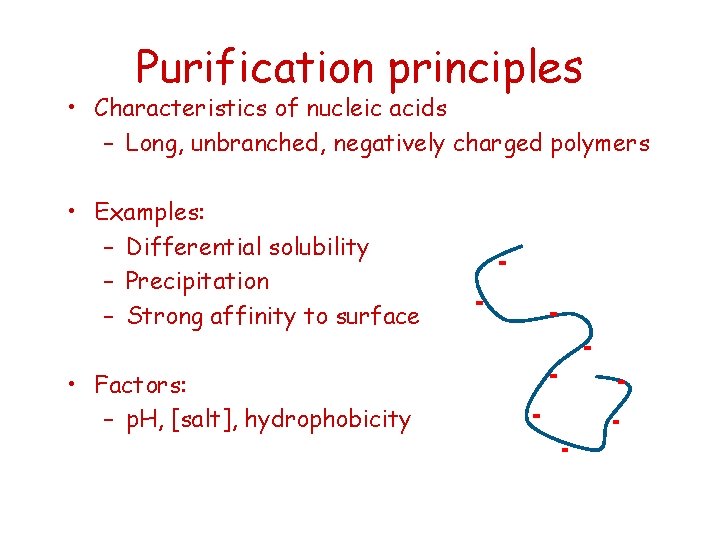
Purification principles • Characteristics of nucleic acids – Long, unbranched, negatively charged polymers • Examples: – Differential solubility – Precipitation – Strong affinity to surface • Factors: – p. H, [salt], hydrophobicity

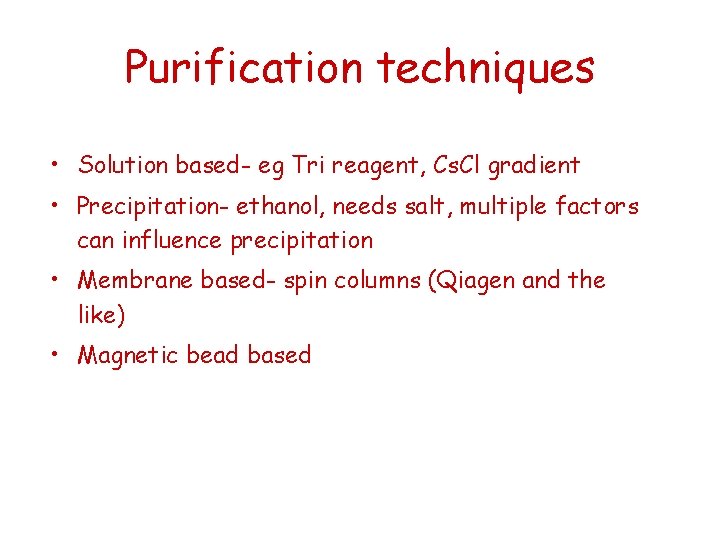
Purification techniques • Solution based- eg Tri reagent, Cs. Cl gradient • Precipitation- ethanol, needs salt, multiple factors can influence precipitation • Membrane based- spin columns (Qiagen and the like) • Magnetic bead based

Solution based isolation • Most methods use hazardous reagents • Phenol/Chloroform extraction – Proteins, lipids, polysaccharides go into the organic phase or in the interphase. – DNA/RNA remains in aqueous phase • Caesium chloride density gradient ultracentrifugation – Time consuming • Acid guanidine phenol chloroform extraction – Commonly called TRIzol

Precipitation purification • Nucleic acids precipitate in alcohols • Salt (Na. Cl, Na. Ac) facilitates the process • Important factors: Temperature, time, p. H, and amount

Membrane based isolation • Anion exchange technology • Spin column / silica gel membrane – Chaotropic salts (e. g. Na. I or guanidine hydrochloride) bind H 2 O molecules – Loss of water from DNA changes shape and charge – DNA binds reversibly to silica membrane

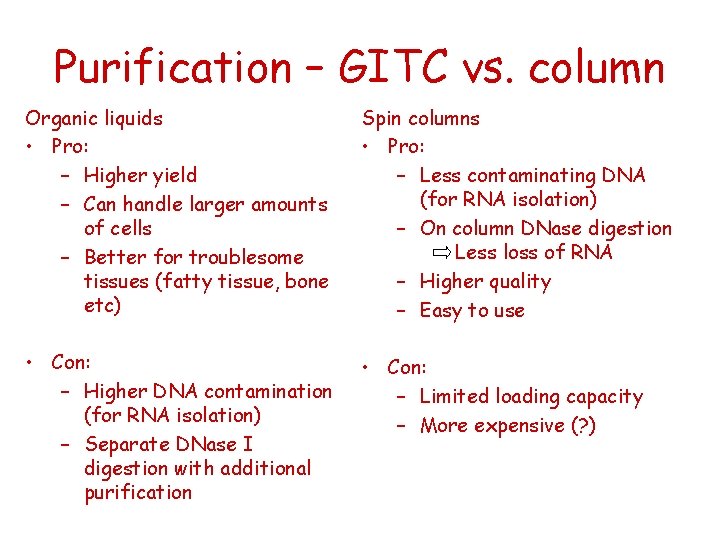
Purification – GITC vs. column Organic liquids • Pro: – Higher yield – Can handle larger amounts of cells – Better for troublesome tissues (fatty tissue, bone etc) Spin columns • Pro: – Less contaminating DNA (for RNA isolation) – On column DNase digestion Less loss of RNA – Higher quality – Easy to use • Con: – Higher DNA contamination (for RNA isolation) – Separate DNase I digestion with additional purification • Con: – Limited loading capacity – More expensive (? )

RNA Considerations • RNA is chemically and biologically less stable than DNA • Extrinsic and intrinsic ribonucleases (RNases) – Specific and Nonspecific inhibitors

Stabilizing conditions • Work on ice • Process immediately • Flash freeze sample in liquid nitrogen and store at -70°C until later use • Store samples in stabilization buffer

Storage of nucleic acids • Nuclease-free plasticware • Eluted in nuclease-free water, TE or sodium citrate solution • RNA: • Neutral p. H to avoid degradation • Aliquot sample to avoid multiple freeze-thaw cycles • Isolated RNA should be stored at -20 deg C or -70 deg C for even better protection in ethanol and not water.
![Quality Control Spectroscopic methods Concentration NA A 260 x e mgml Quality Control • Spectroscopic methods – Concentration, [NA] = A 260 x e mg/ml](https://slidetodoc.com/presentation_image_h/ad0a8dd132e8db8fa50cd6e2b69e12ee/image-39.jpg)
Quality Control • Spectroscopic methods – Concentration, [NA] = A 260 x e mg/ml – Purity: A 260 / A 280 (≈1. 8 for DNA, 2. 0 for RNA) • Dyes – Quantification by fluorescence of DNA/RNAbinding dyes (Qubit) • Electrophoresis (28 S and 18 S bands)

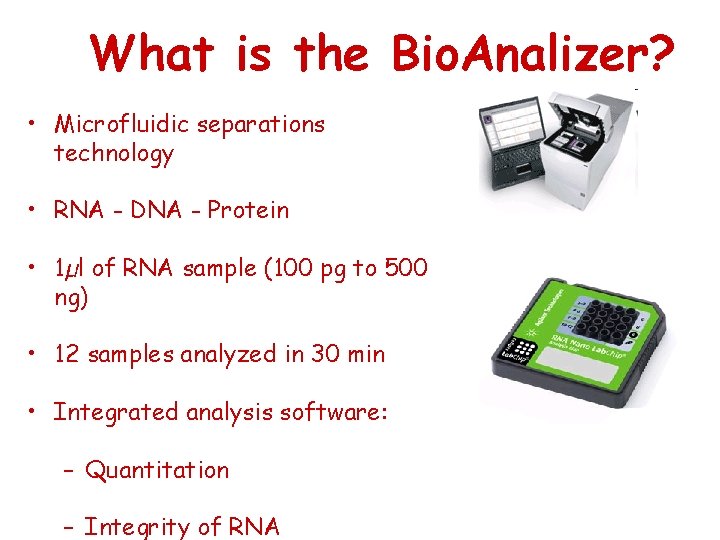
What is the Bio. Analizer? • Microfluidic separations technology • RNA - DNA - Protein • 1µl of RNA sample (100 pg to 500 ng) • 12 samples analyzed in 30 min • Integrated analysis software: – Quantitation – Integrity of RNA

Bioanalyzer




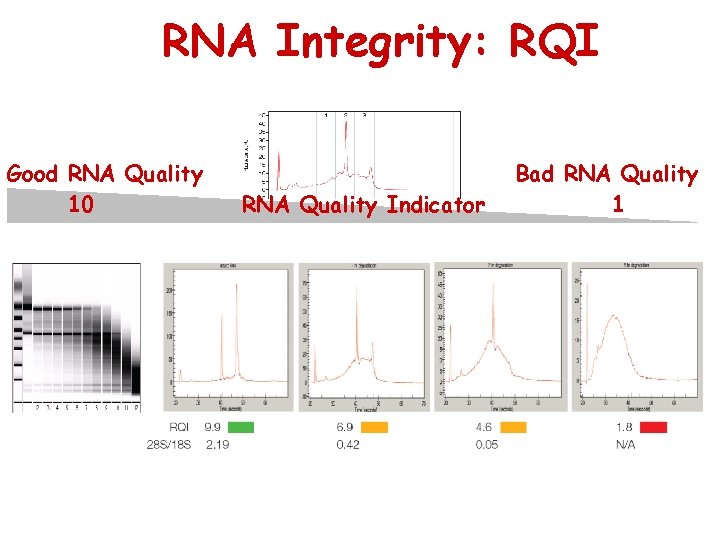
RNA Integrity: RQI Good RNA Quality 10 RNA Quality Indicator Bad RNA Quality 1

Publications on RNA integrity

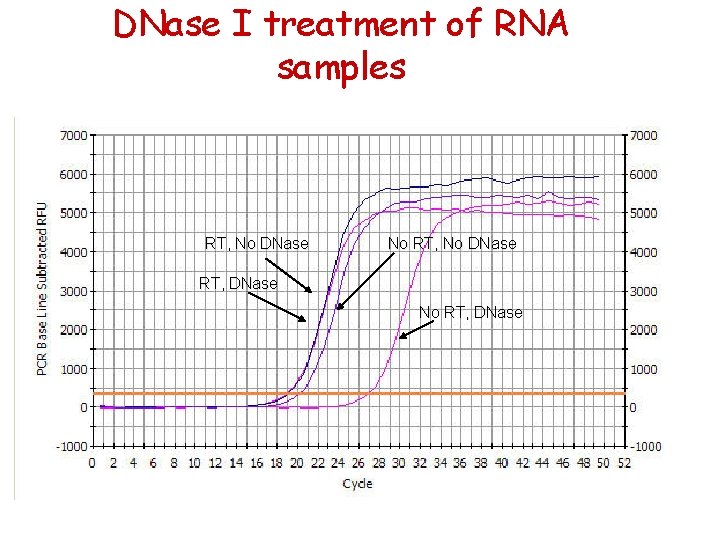
DNase I treatment of RNA samples RT, No DNase No RT, No DNase RT, DNase No RT, DNase

q. PCR technical workflow DNA Extraction Sampling q. PCR RNA Extraction DNase treatment Reverse Transcription Data Analysis

Reverse transcription RT

Outline • Priming efficiency • Reproducibility • Properties of Reverse transcriptase • RNA concentrations

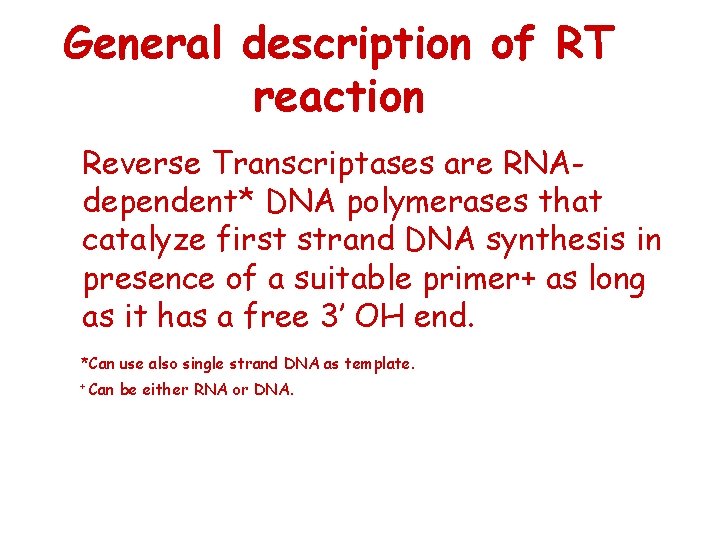
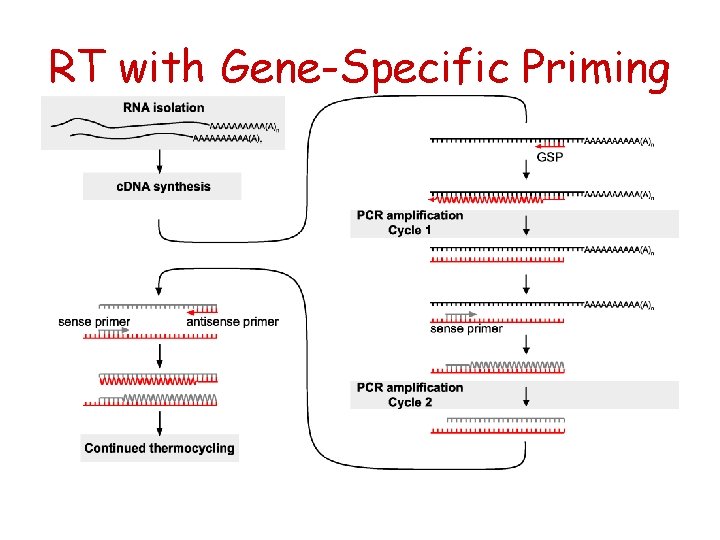
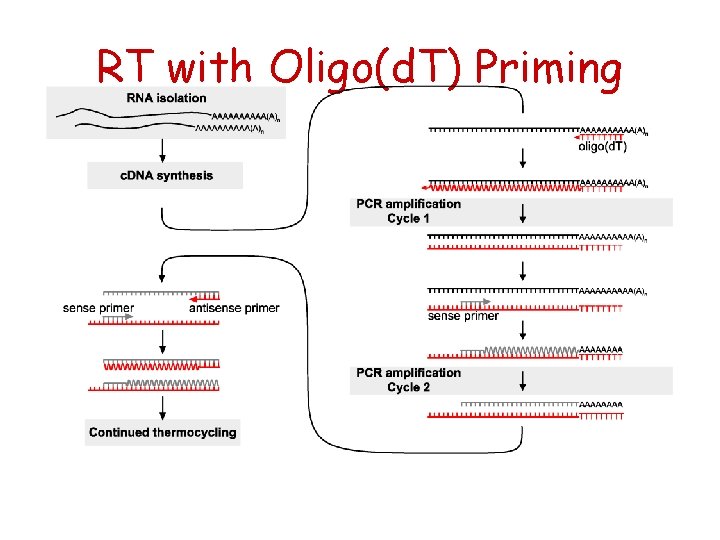
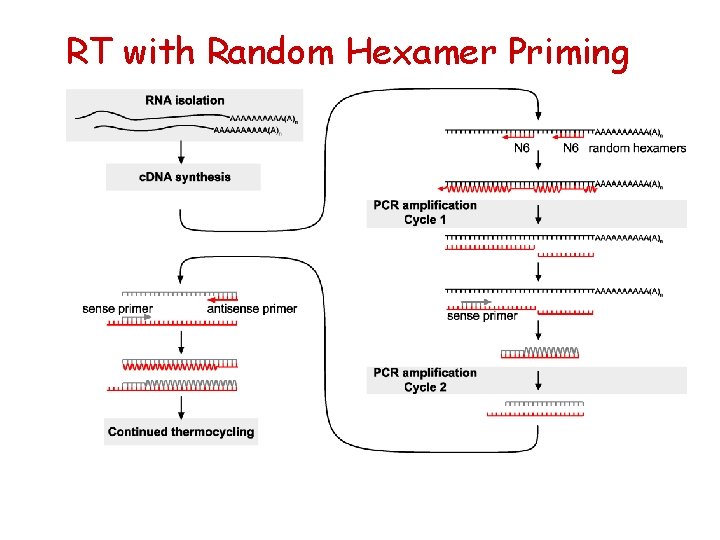
General description of RT reaction Reverse Transcriptases are RNAdependent* DNA polymerases that catalyze first strand DNA synthesis in presence of a suitable primer+ as long as it has a free 3’ OH end. *Can use also single strand DNA as template. + Can be either RNA or DNA.

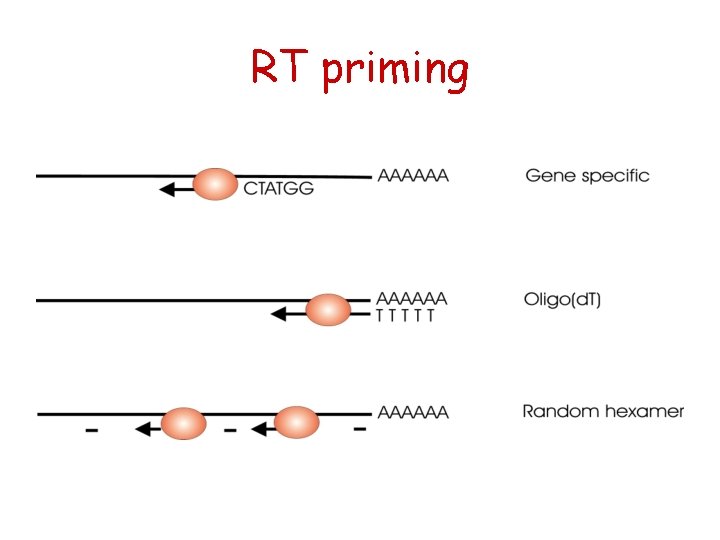
RT priming

RT with Gene-Specific Priming

RT with Oligo(d. T) Priming

RT with Random Hexamer Priming

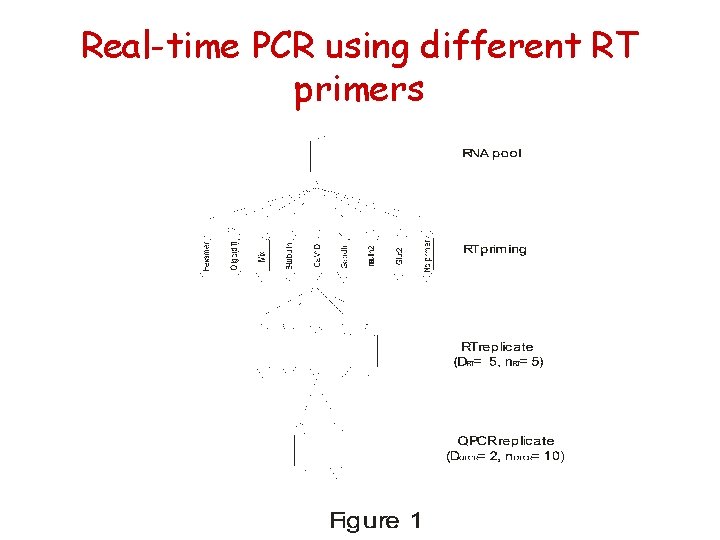
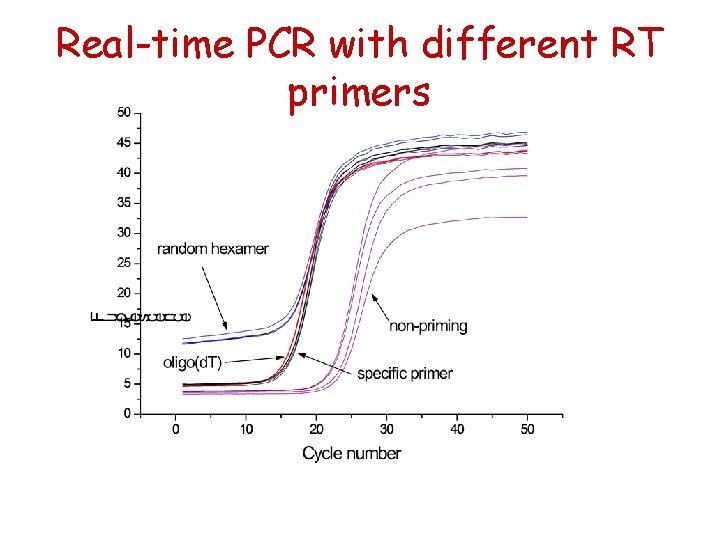
Real-time PCR using different RT primers

Real-time PCR with different RT primers

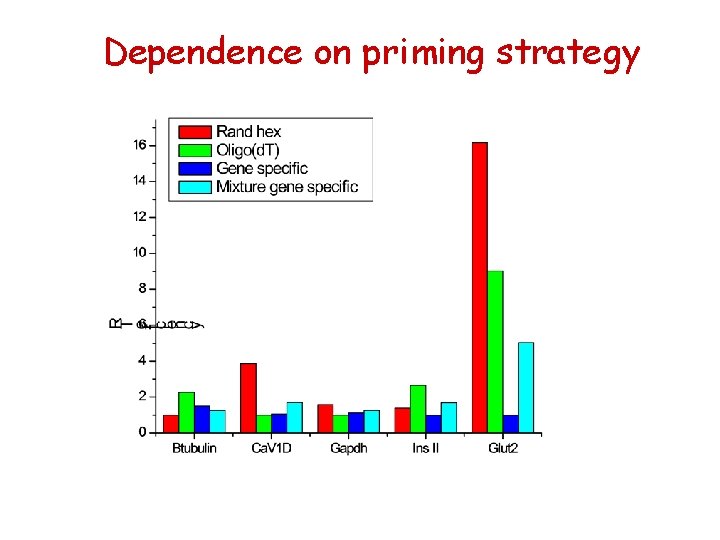
Dependence on priming strategy

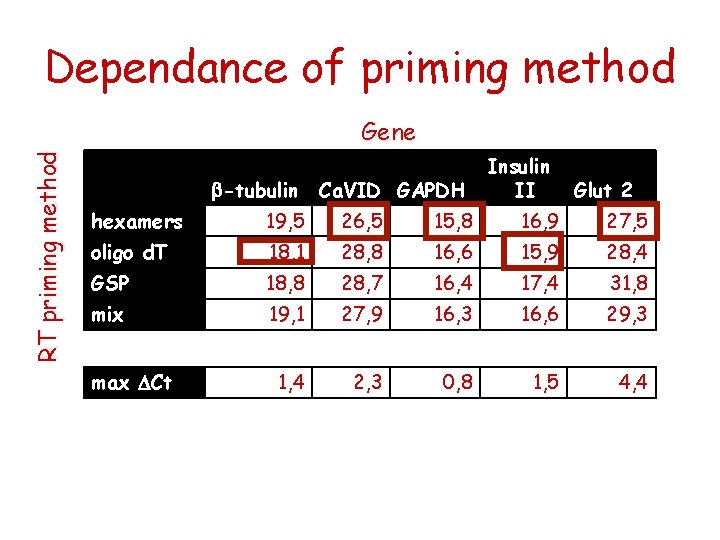
Dependance of priming method RT priming method Gene b-tubulin Ca. VID GAPDH Insulin II Glut 2 hexamers 19, 5 26, 5 15, 8 16, 9 27, 5 oligo d. T 18, 1 28, 8 16, 6 15, 9 28, 4 GSP 18, 8 28, 7 16, 4 17, 4 31, 8 mix 19, 1 27, 9 16, 3 16, 6 29, 3 1, 4 2, 3 0, 8 1, 5 4, 4 max DCt

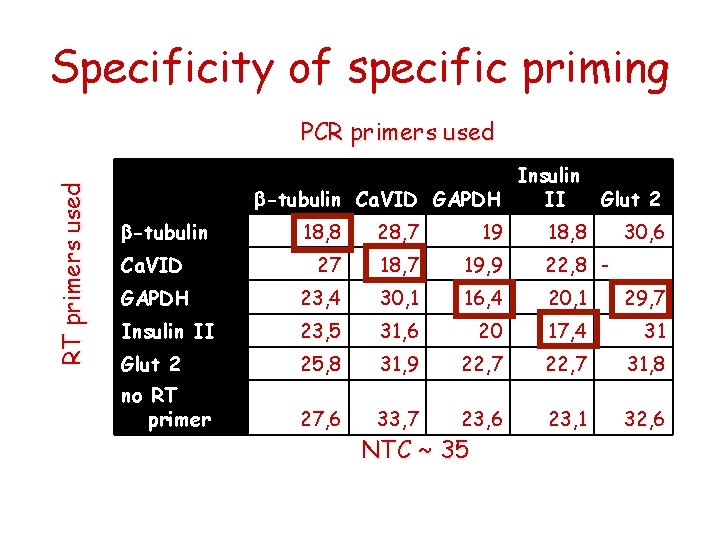
Specificity of specific priming RT primers used PCR primers used Insulin b-tubulin Ca. VID GAPDH II Glut 2 18, 8 28, 7 19 27 18, 7 19, 9 22, 8 - GAPDH 23, 4 30, 1 16, 4 20, 1 29, 7 Insulin II 23, 5 31, 6 20 17, 4 31 Glut 2 25, 8 31, 9 22, 7 31, 8 no RT primer 27, 6 33, 7 23, 6 23, 1 32, 6 b-tubulin Ca. VID NTC ~ 35 18, 8 30, 6

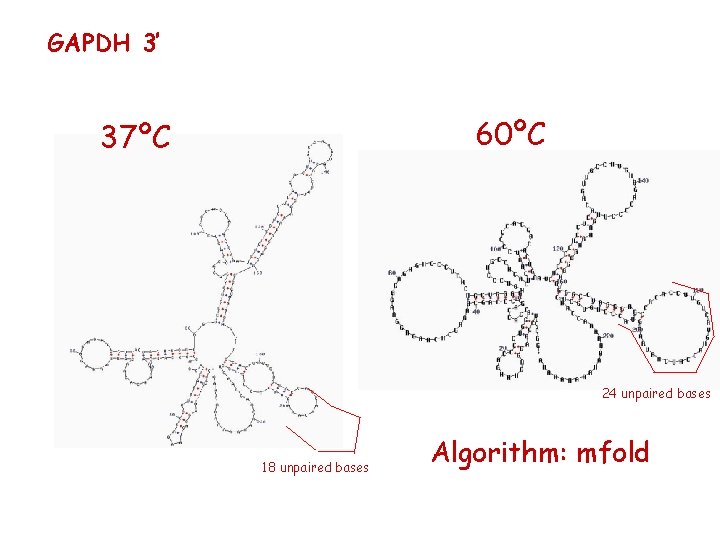
GAPDH 3’ 60ºC 37ºC 24 unpaired bases 18 unpaired bases Algorithm: mfold

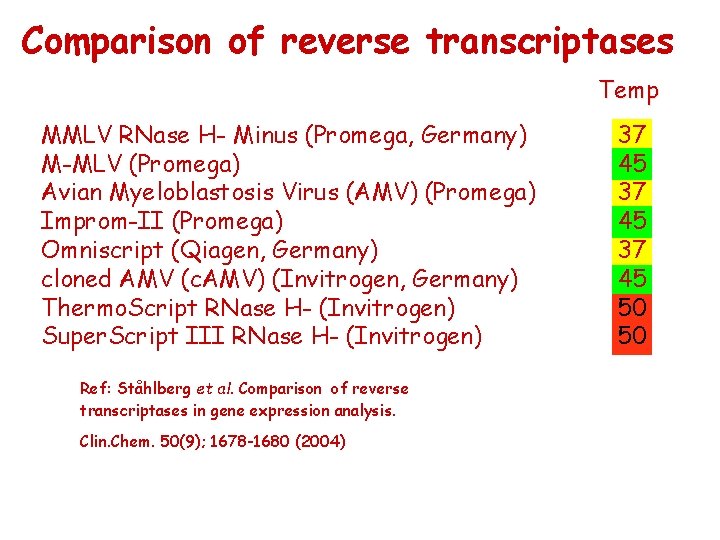
Comparison of reverse transcriptases Temp MMLV RNase H- Minus (Promega, Germany) M-MLV (Promega) Avian Myeloblastosis Virus (AMV) (Promega) Improm-II (Promega) Omniscript (Qiagen, Germany) cloned AMV (c. AMV) (Invitrogen, Germany) Thermo. Script RNase H- (Invitrogen) Super. Script III RNase H- (Invitrogen) Ref: Ståhlberg et al. Comparison of reverse transcriptases in gene expression analysis. Clin. Chem. 50(9); 1678 -1680 (2004) 37 45 50 50

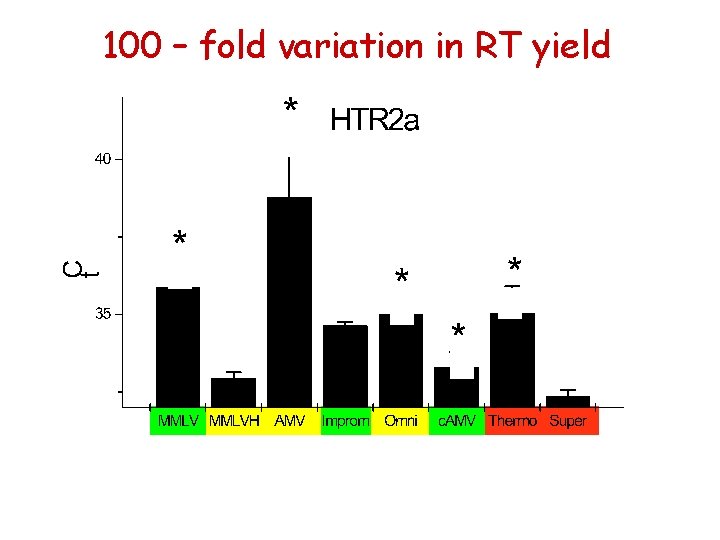
100 – fold variation in RT yield

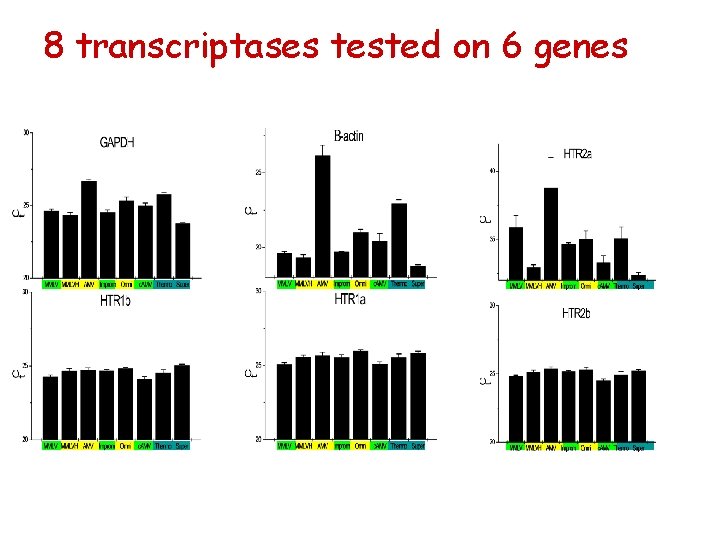
8 transcriptases tested on 6 genes

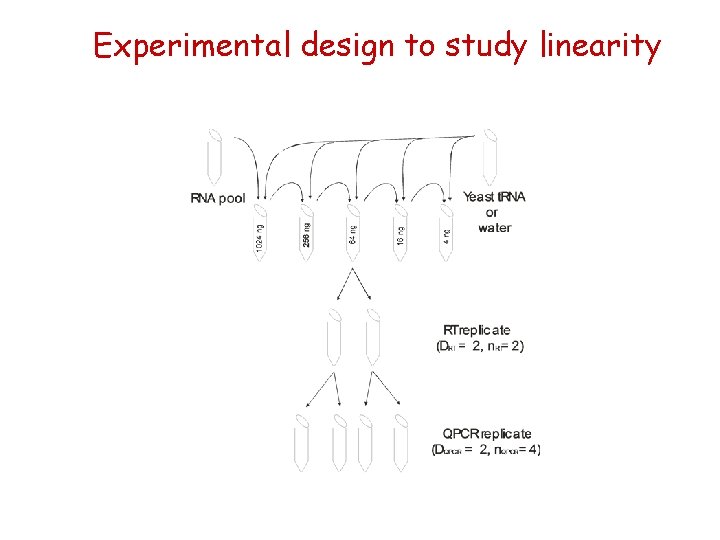
Experimental design to study linearity

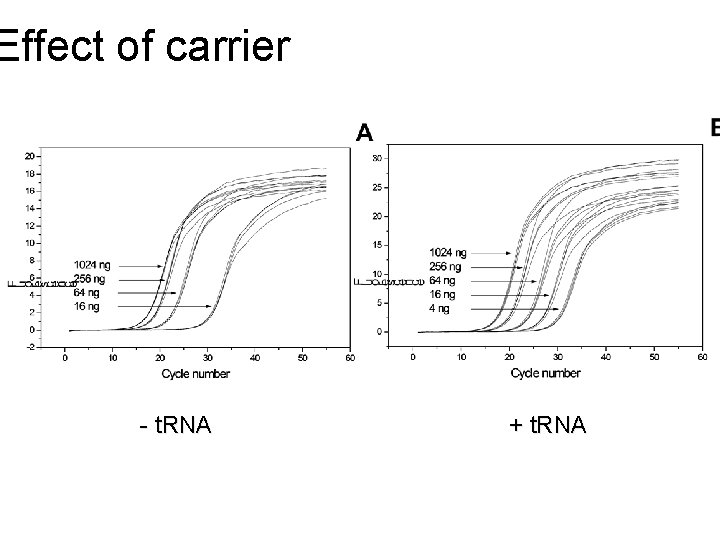
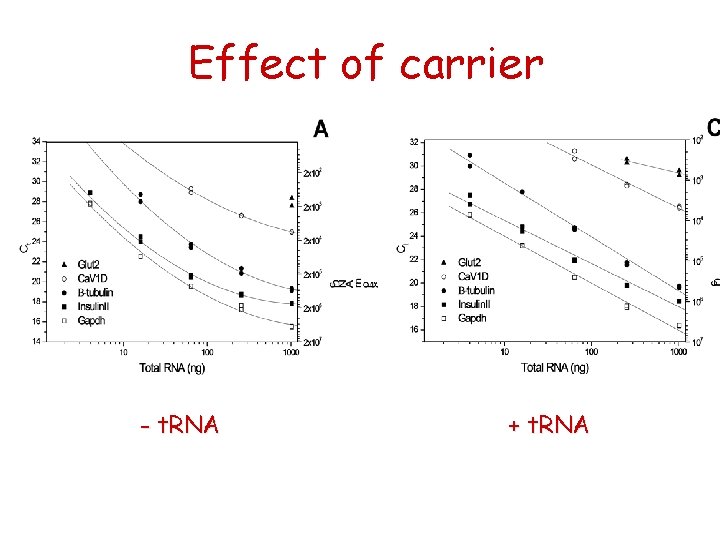
Effect of carrier - t. RNA + t. RNA

Effect of carrier - t. RNA + t. RNA

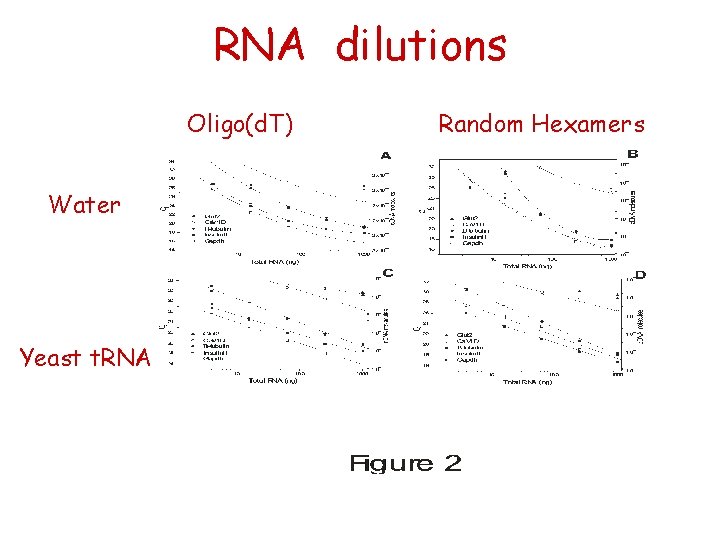
RNA dilutions Oligo(d. T) Water Yeast t. RNA Random Hexamers

Conclusions • • The RT reaction shows higher technical variability than QPCR There is no optimum priming strategy Gene specific primers must target accessible regions The RT yield changes over 100 -fold with the choice of reverse transcriptase The yield variation is gene specific RT yield is proportional to the amount of template in presence of proper carrier Typical RT yield is 10 -50 % RT-QPCR is highly reproducible as long as the same protocol and reaction conditions are used The efficiency of the RT reaction varies from gene to gene and depends on the conditions – run the RT of all samples using exactly the same protocol and reagents under the same conditions
 Rabbi chaim kramer
Rabbi chaim kramer Chaim prinzental
Chaim prinzental Real time software definition
Real time software definition Realtime aps software
Realtime aps software Push notification firebase
Push notification firebase Realtime streaming protocol
Realtime streaming protocol Ecurisa
Ecurisa Market overview real-time interaction management
Market overview real-time interaction management Lightning realtime
Lightning realtime Simple online and realtime tracking
Simple online and realtime tracking Visual rendering
Visual rendering Real time characteristics of embedded operating systems
Real time characteristics of embedded operating systems Realtime communications
Realtime communications Realtime it
Realtime it Realtime it
Realtime it Realtime it
Realtime it Copthorne hotel
Copthorne hotel Cac realtime
Cac realtime Realtime forex
Realtime forex Eva rov
Eva rov Rendering realtime compositing
Rendering realtime compositing Realtime big data
Realtime big data Ad hoc realtime
Ad hoc realtime Rational rose realtime
Rational rose realtime Ams realtime weather maps central
Ams realtime weather maps central Realtime etl
Realtime etl Cos operating system
Cos operating system Realtime
Realtime Realtime optimization
Realtime optimization Realtime diagnostics
Realtime diagnostics Realtime mobile communication
Realtime mobile communication Realtime iep
Realtime iep Real-time messaging protocol
Real-time messaging protocol Alyac realtime service
Alyac realtime service Frankfurt realtime
Frankfurt realtime Realtime interaction
Realtime interaction Realtime networks
Realtime networks Webrtc shim
Webrtc shim Colony pcr yeast
Colony pcr yeast Pcr
Pcr Pcr asimetrica
Pcr asimetrica Rcp extracorpórea
Rcp extracorpórea Poonum patel
Poonum patel Pcr
Pcr Biorad pcr master mix
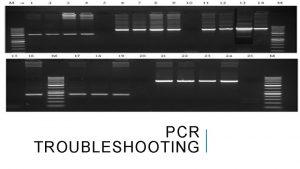
Biorad pcr master mix Pcr troubleshooting multiple bands
Pcr troubleshooting multiple bands Objectives of pcr
Objectives of pcr Fluidigm
Fluidigm Pcr
Pcr Calcul efficacité pcr quantitative
Calcul efficacité pcr quantitative Pcr abi
Pcr abi Molecular lab setup and workflow
Molecular lab setup and workflow Pcr nobel prize
Pcr nobel prize Design change request
Design change request Crystal digital pcr
Crystal digital pcr Missy baker
Missy baker Pcr annealing temperature too high
Pcr annealing temperature too high Pcr biology definition
Pcr biology definition Site:slidetodoc.com
Site:slidetodoc.com Primers dna
Primers dna Pcr
Pcr Pcr animations
Pcr animations Reactantes de fase aguda
Reactantes de fase aguda Pcr file fullprof
Pcr file fullprof Slan real time pcr
Slan real time pcr Basics of microbiology
Basics of microbiology Xét nghiệm pcr
Xét nghiệm pcr Pcr rflp animation
Pcr rflp animation Tail pcr principle
Tail pcr principle Conventional pcr
Conventional pcr