SDS PAGE SDS polyacrylamide gel electrophoresis sodium dodecyl





![Enzyme kinetics (as opposed to simple chemical kinetics) Vo independent of [S] Vo proportional Enzyme kinetics (as opposed to simple chemical kinetics) Vo independent of [S] Vo proportional](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-39.jpg)



![ΔG = Δ G o+ 70 RTln([C][D]/[A][B]) • where A, B, C and D ΔG = Δ G o+ 70 RTln([C][D]/[A][B]) • where A, B, C and D](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-70.jpg)

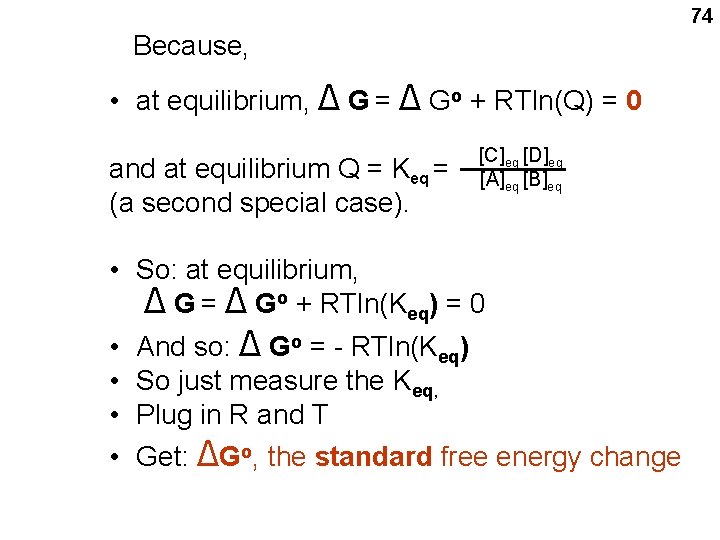


![Exception #2: In the same way, Hydrogen ion concentration, [H+]: 10 -7 M is Exception #2: In the same way, Hydrogen ion concentration, [H+]: 10 -7 M is](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-78.jpg)

- Slides: 79

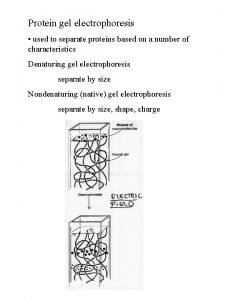
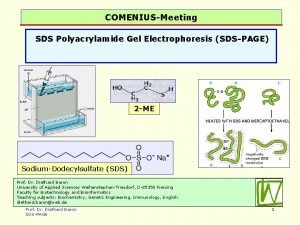
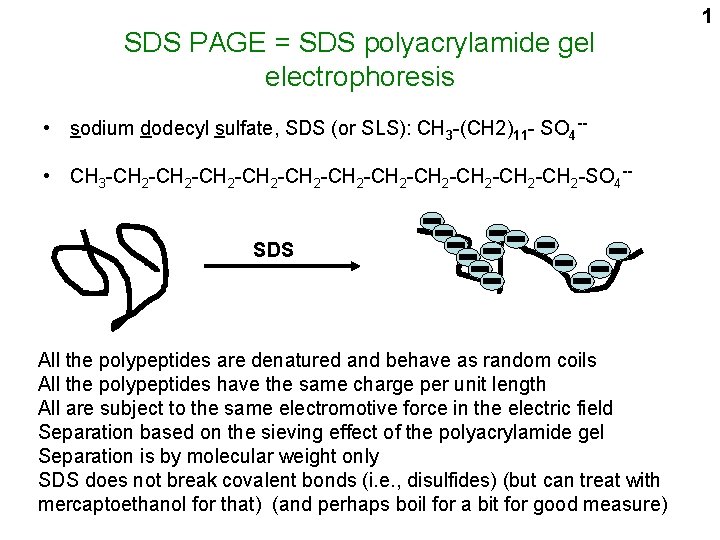
SDS PAGE = SDS polyacrylamide gel electrophoresis • sodium dodecyl sulfate, SDS (or SLS): CH 3 -(CH 2)11 - SO 4 - • CH 3 -CH 2 -CH 2 -CH 2 -CH 2 -SO 4 -SDS All the polypeptides are denatured and behave as random coils All the polypeptides have the same charge per unit length All are subject to the same electromotive force in the electric field Separation based on the sieving effect of the polyacrylamide gel Separation is by molecular weight only SDS does not break covalent bonds (i. e. , disulfides) (but can treat with mercaptoethanol for that) (and perhaps boil for a bit for good measure) 1

2 Disulfides between 2 cysteines can be cleaved in the laboratory by reduction, i. e. , adding 2 Hs (with their electrons) back across the disulfide bond. One adds a reducing agent: mercaptoethanol (HO-CH 2 -SH). In the presence of this reagent, one gets exchange among the disulfides and the sulfhydryls: Protein-CH 2 -S-S-CH 2 -Protein + 2 HO-CH 2 -SH ---> Protein-CH 2 -SH + HS-CH 2 -Protein + HO-CH 2 -S-S-CH 2 -OH The protein's disulfide gets reduced (and the S-S bond cleaved), while the mercaptoethanol gets oxidized, losing electrons and protons and itself forming a disulfide bond.

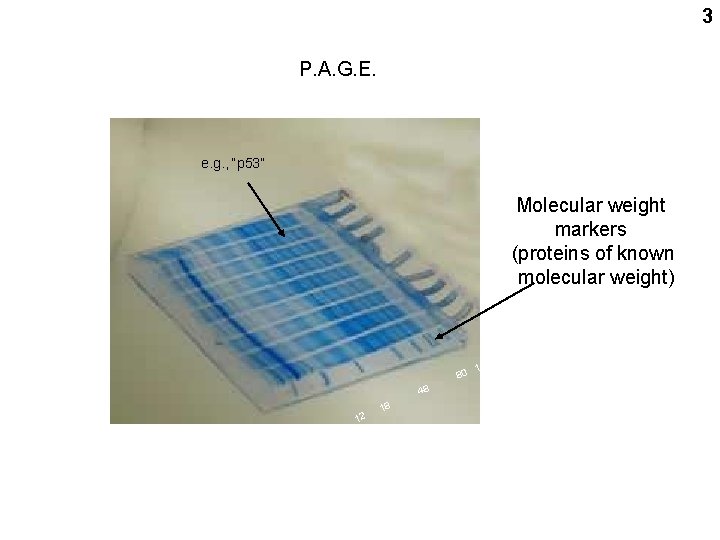
3 P. A. G. E. e. g. , “p 53” Molecular weight markers (proteins of known molecular weight) 160 130 0 0 11 14 80 48 12 18

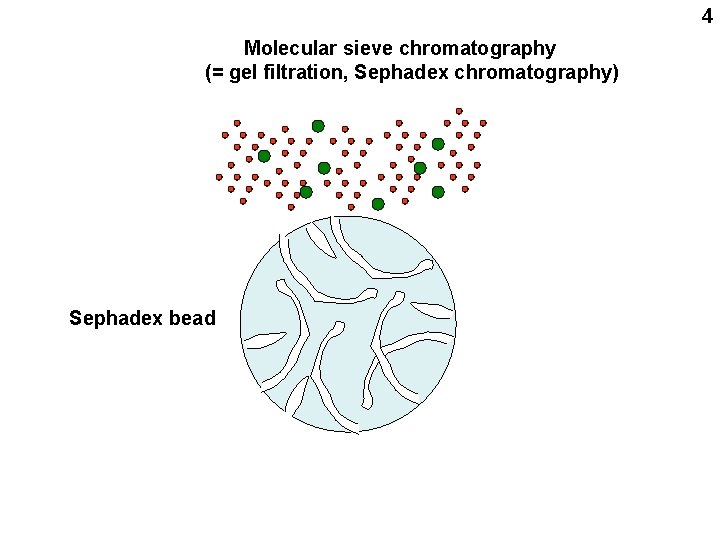
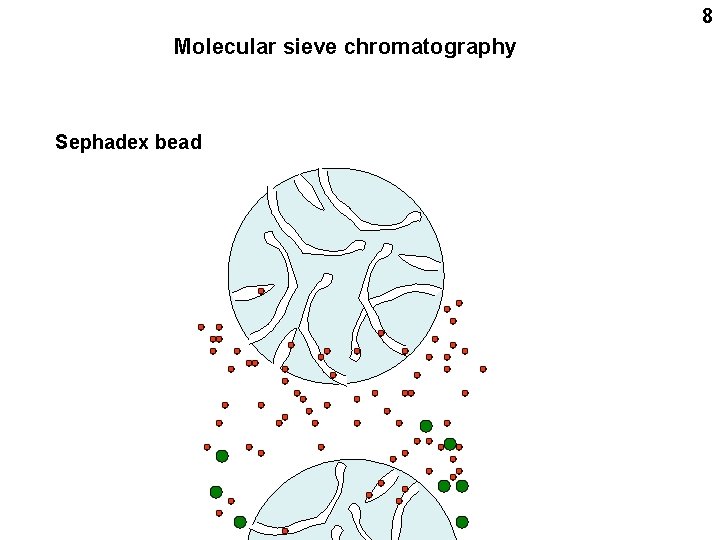
4 Molecular sieve chromatography (= gel filtration, Sephadex chromatography) Sephadex bead

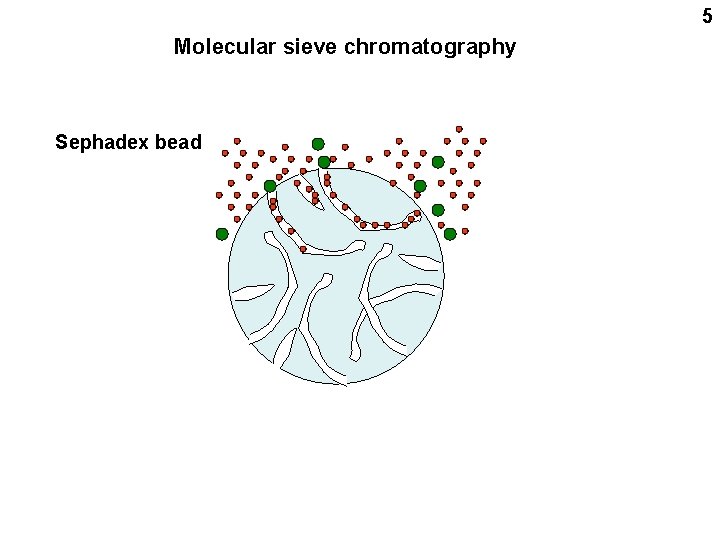
5 Molecular sieve chromatography Sephadex bead

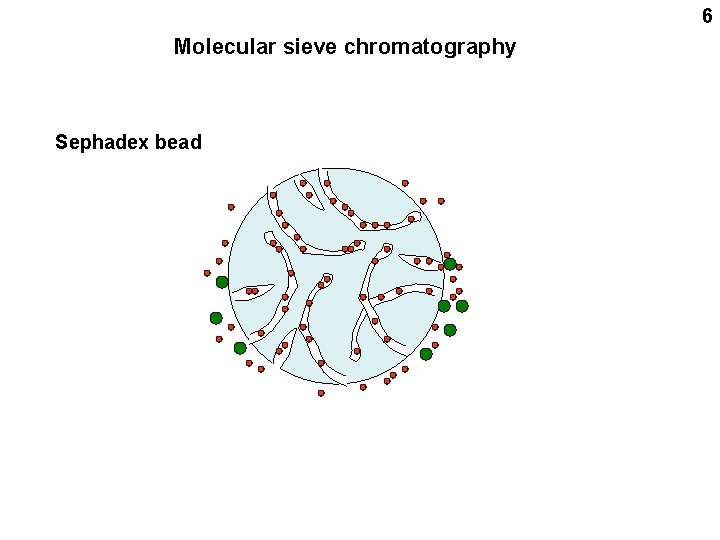
6 Molecular sieve chromatography Sephadex bead

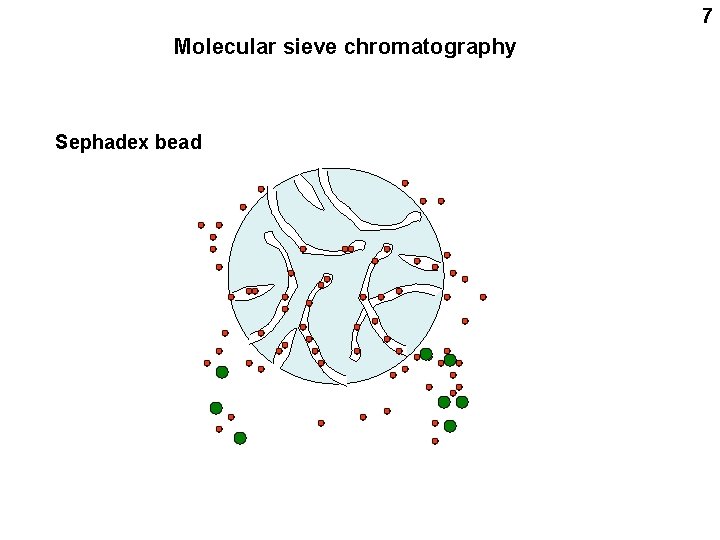
7 Molecular sieve chromatography Sephadex bead

8 Molecular sieve chromatography Sephadex bead

9 Fancy Plain 4 o. C (cold room)

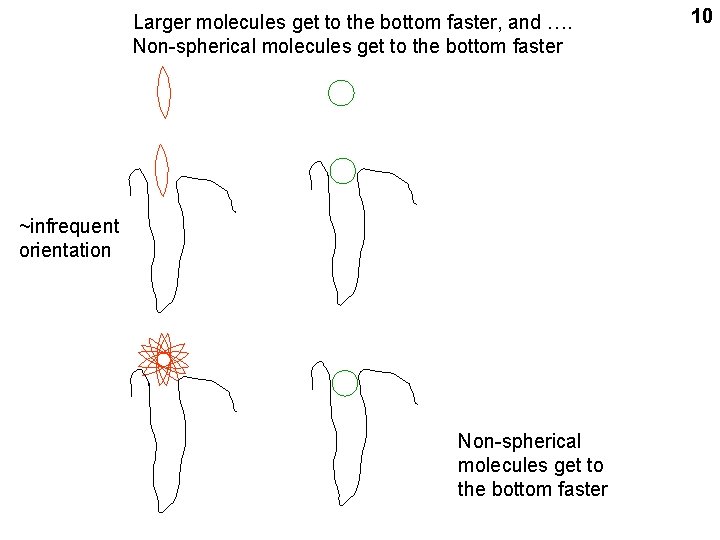
Larger molecules get to the bottom faster, and …. Non-spherical molecules get to the bottom faster ~infrequent orientation Non-spherical molecules get to the bottom faster 10

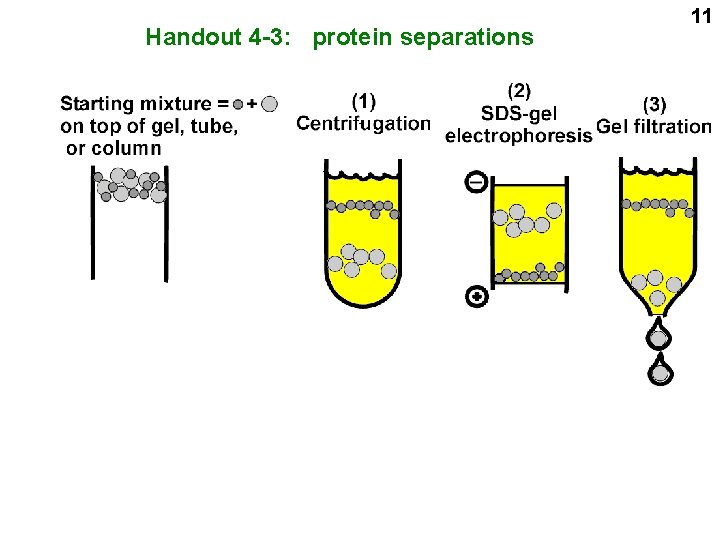
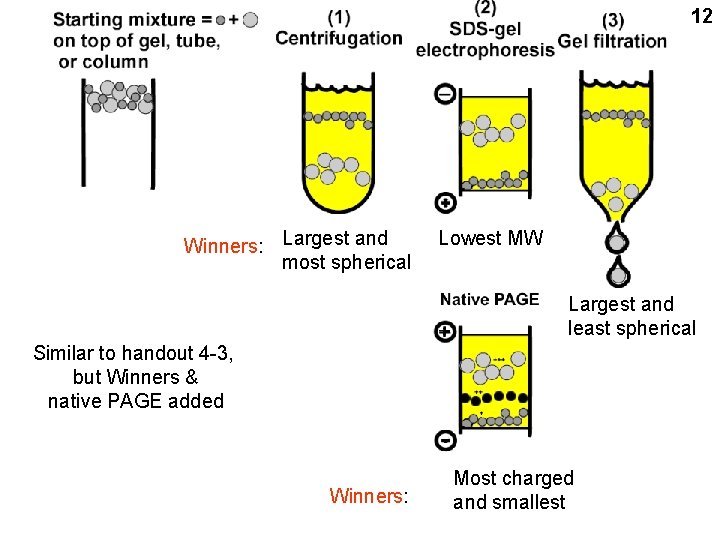
Handout 4 -3: protein separations 11

12 Winners: Largest and most spherical Lowest MW Largest and least spherical Similar to handout 4 -3, but Winners & native PAGE added Winners: Most charged and smallest

13 Enzymes = protein catalysts

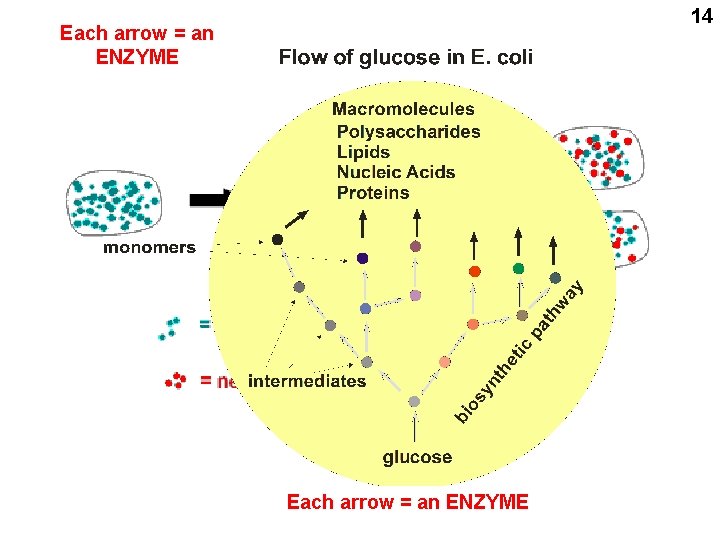
14 Each arrow = an ENZYME

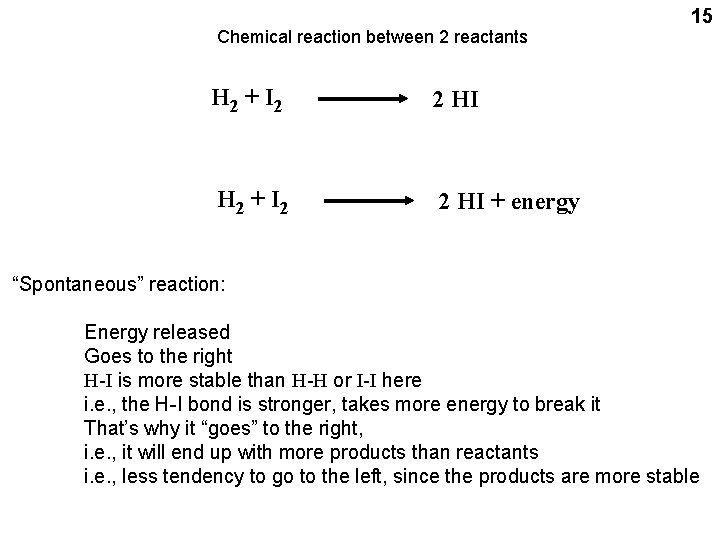
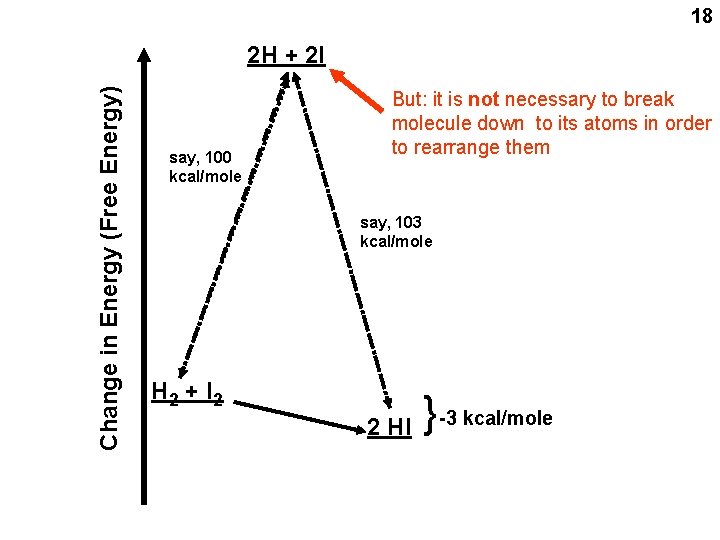
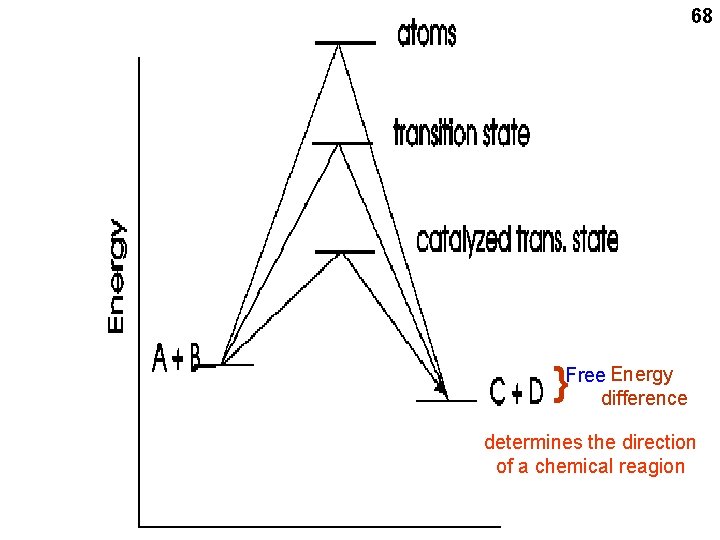
Chemical reaction between 2 reactants H 2 + I 2 2 HI + energy 15 “Spontaneous” reaction: Energy released Goes to the right H-I is more stable than H-H or I-I here i. e. , the H-I bond is stronger, takes more energy to break it That’s why it “goes” to the right, i. e. , it will end up with more products than reactants i. e. , less tendency to go to the left, since the products are more stable

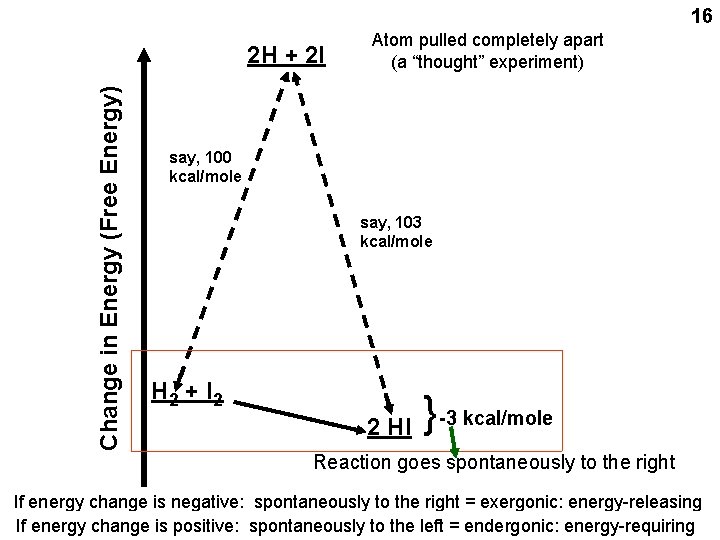
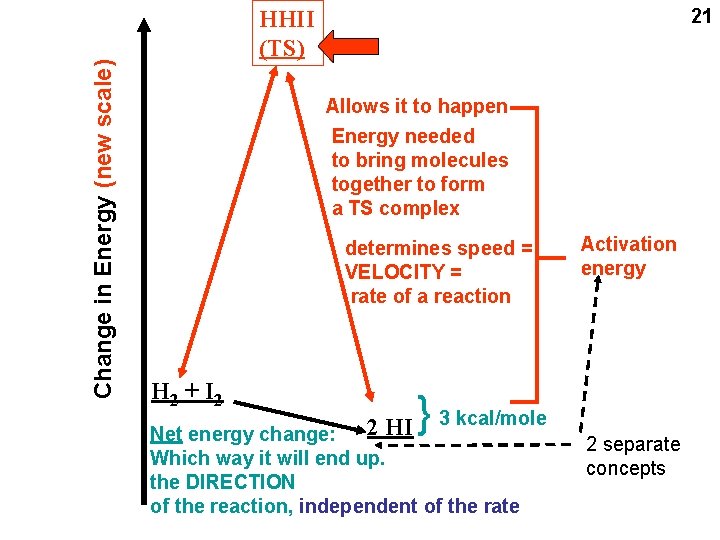
16 say, 100 kcal/mole say, 103 kcal/mole H 2 + I 2 2 HI { Change in Energy (Free Energy) 2 H + 2 I Atom pulled completely apart (a “thought” experiment) -3 kcal/mole Reaction goes spontaneously to the right If energy change is negative: spontaneously to the right = exergonic: energy-releasing If energy change is positive: spontaneously to the left = endergonic: energy-requiring

17 Different ways of writing chemical reactions H 2 + I 2 2 HI H 2 + I 2 2 HI

18 say, 100 kcal/mole But: it is not necessary to break molecule down to its atoms in order to rearrange them say, 103 kcal/mole H 2 + I 2 2 HI { Change in Energy (Free Energy) 2 H + 2 I -3 kcal/mole

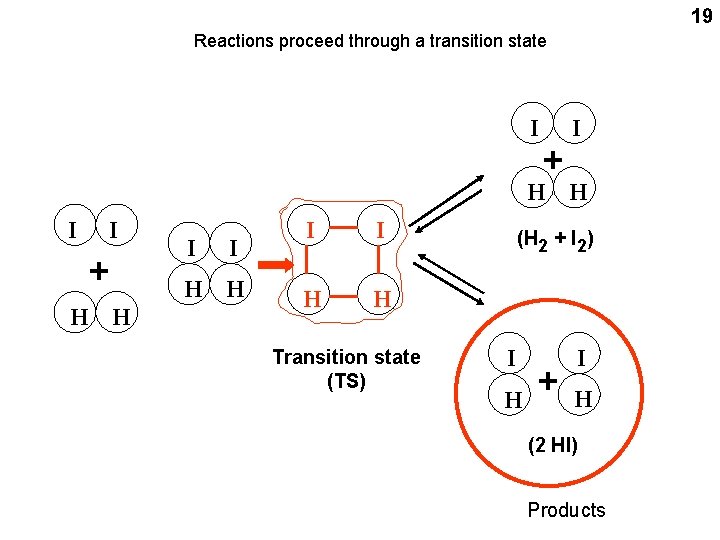
19 Reactions proceed through a transition state I + I H H I I + H H I I H H Transition state (TS) (H 2 + I 2) I H + I H (2 HI) Products

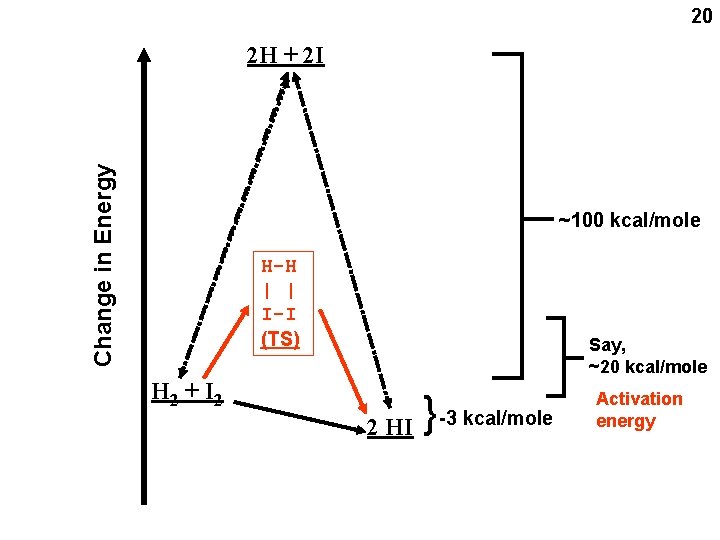
20 Change in Energy 2 H + 2 I ~100 kcal/mole H-H | | I-I (TS) Say, ~20 kcal/mole 2 HI { H 2 + I 2 -3 kcal/mole Activation energy

Allows it to happen Energy needed to bring molecules together to form a TS complex determines speed = VELOCITY = rate of a reaction H 2 + I 2 2 HI { Change in Energy (new scale) 21 HHII (TS) Activation energy 3 kcal/mole Net energy change: Which way it will end up. the DIRECTION of the reaction, independent of the rate 2 separate concepts

22 Concerns about the cell’s chemical reactions • Direction – We need it to go in the direction we want • Speed – We need it to go fast enough to have the cell double in one generation

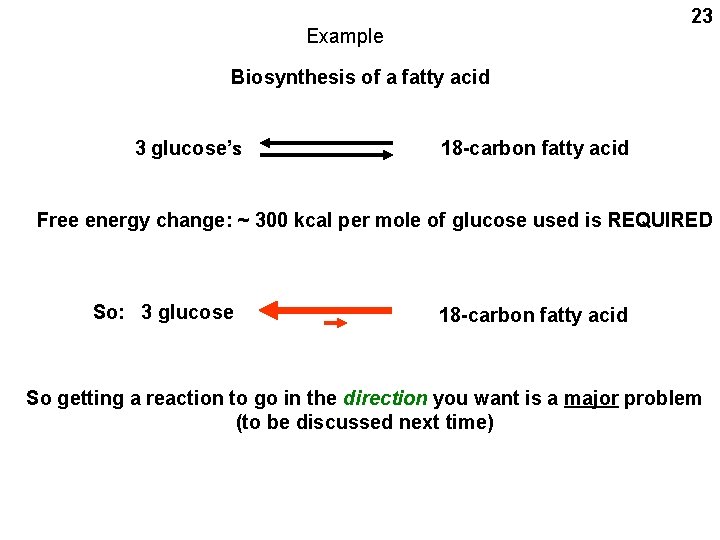
23 Example Biosynthesis of a fatty acid 3 glucose’s 18 -carbon fatty acid Free energy change: ~ 300 kcal per mole of glucose used is REQUIRED So: 3 glucose 18 -carbon fatty acid So getting a reaction to go in the direction you want is a major problem (to be discussed next time)

24 Concerns about the cell’s chemical reactions • Direction – We need it to go in the direction we want • Speed – We need it to go fast enough to have the cell double in one generation – Catalysts deal with this second problem, which we will now consider

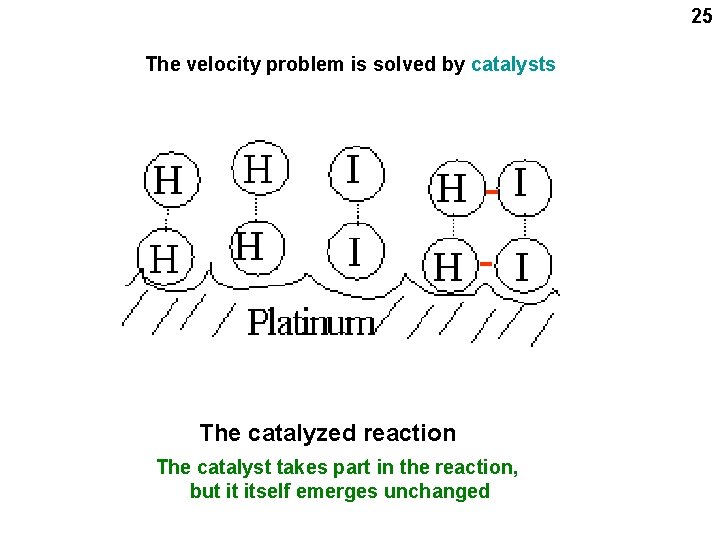
25 The velocity problem is solved by catalysts The catalyzed reaction The catalyst takes part in the reaction, but it itself emerges unchanged

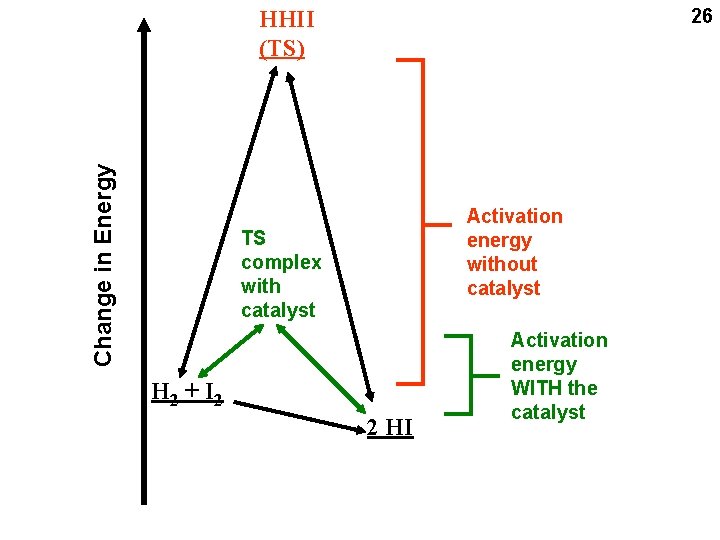
26 Change in Energy HHII (TS) Activation energy without catalyst TS complex with catalyst H 2 + I 2 2 HI Activation energy WITH the catalyst

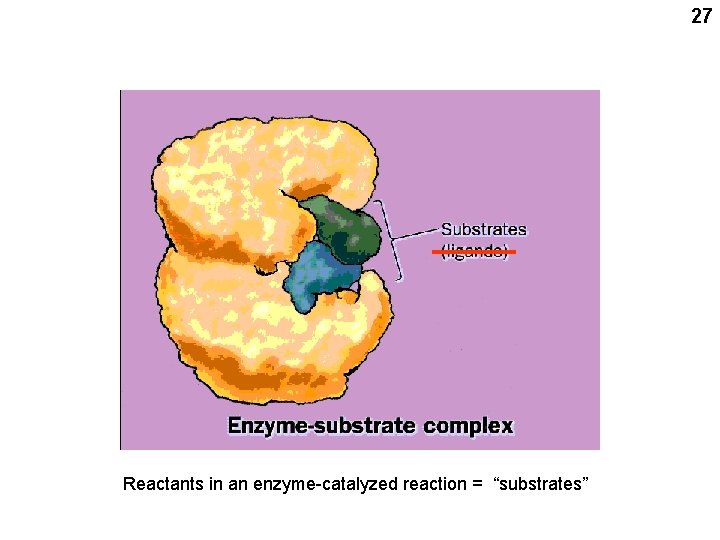
27 Reactants in an enzyme-catalyzed reaction = “substrates”

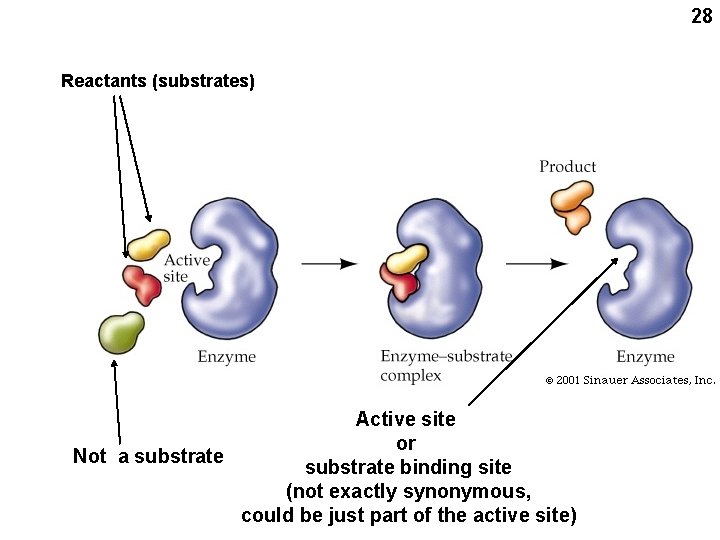
28 Reactants (substrates) Active site or Not a substrate binding site (not exactly synonymous, could be just part of the active site)

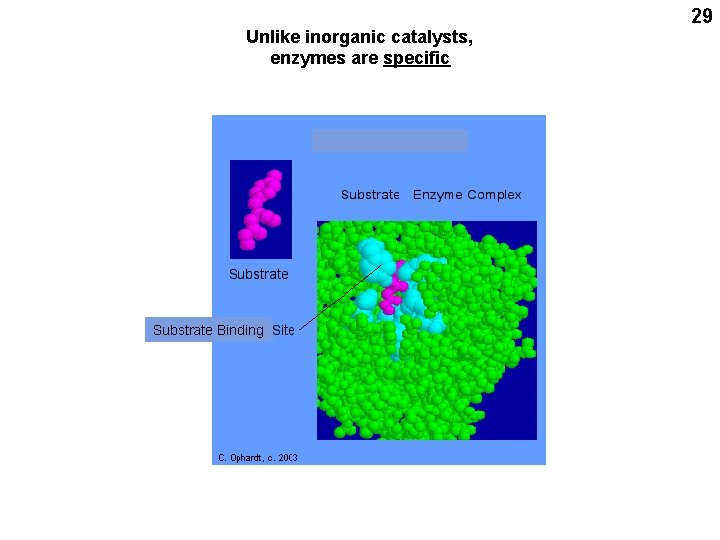
Unlike inorganic catalysts, enzymes are specific Substrate Binding 29

30 Small molecules bind with great specificity to pockets on ENZYME surfaces Too far

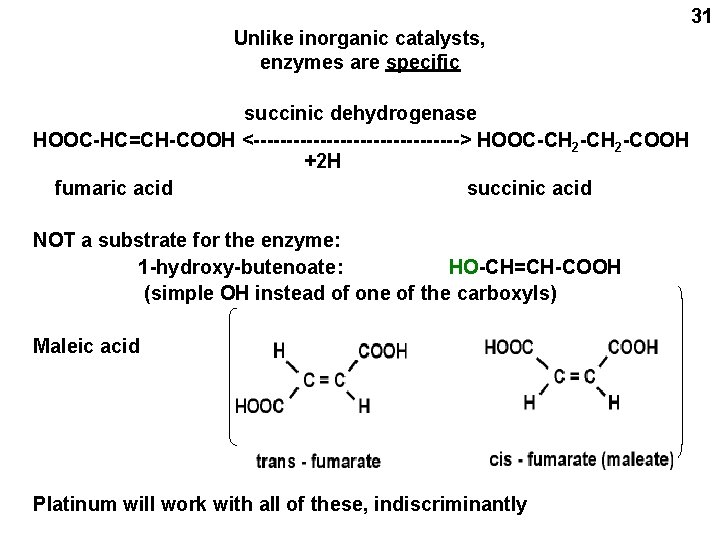
Unlike inorganic catalysts, enzymes are specific succinic dehydrogenase HOOC-HC=CH-COOH <----------------> HOOC-CH 2 -COOH +2 H fumaric acid succinic acid NOT a substrate for the enzyme: 1 -hydroxy-butenoate: HO-CH=CH-COOH (simple OH instead of one of the carboxyls) Maleic acid Platinum will work with all of these, indiscriminantly 31

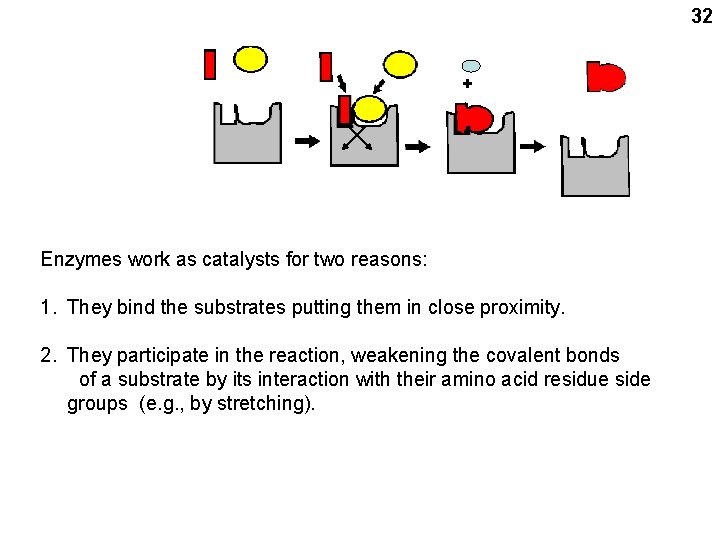
32 + Enzymes work as catalysts for two reasons: 1. They bind the substrates putting them in close proximity. 2. They participate in the reaction, weakening the covalent bonds of a substrate by its interaction with their amino acid residue side groups (e. g. , by stretching).

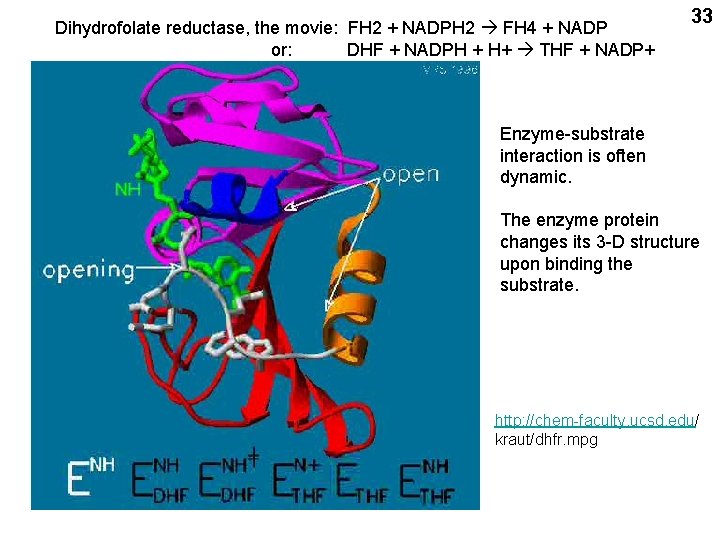
Dihydrofolate reductase, the movie: FH 2 + NADPH 2 FH 4 + NADP or: DHF + NADPH + H+ THF + NADP+ 33 Enzyme-substrate interaction is often dynamic. The enzyme protein changes its 3 -D structure upon binding the substrate. http: //chem-faculty. ucsd. edu/ kraut/dhfr. mpg

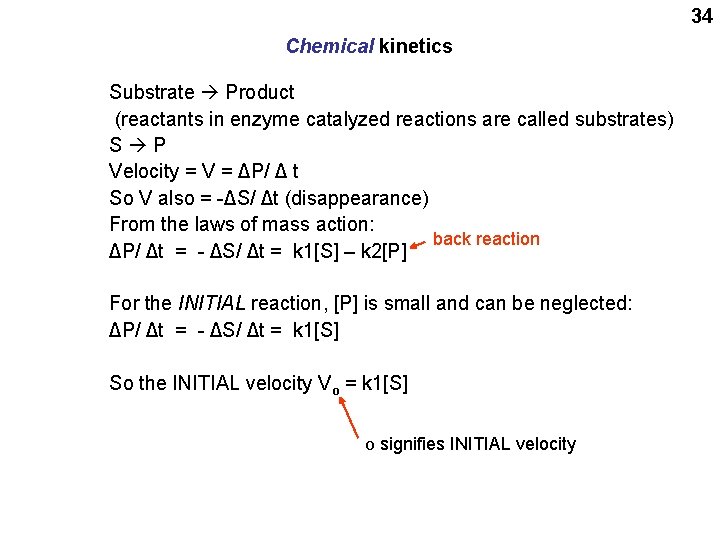
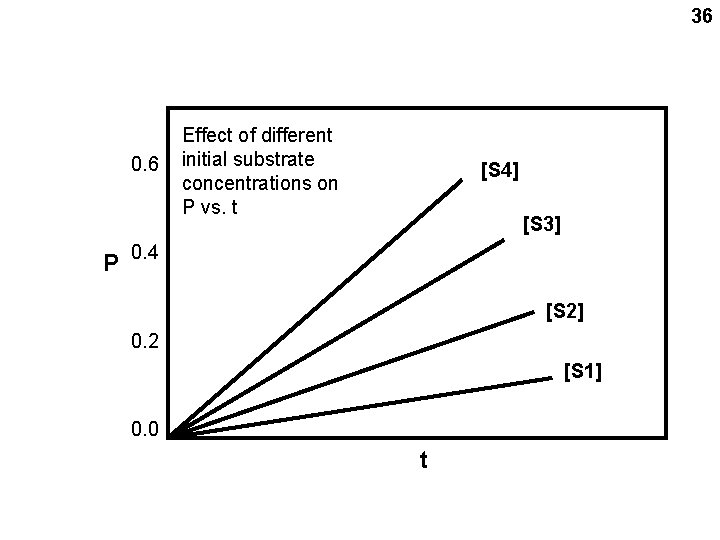
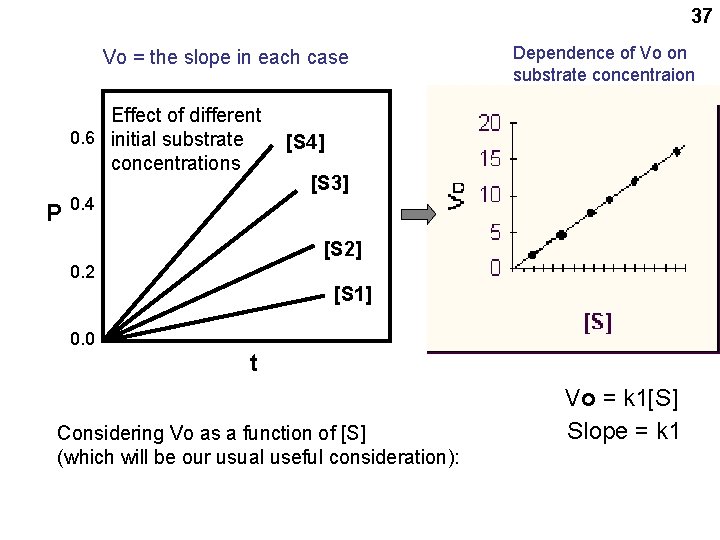
34 Chemical kinetics Substrate Product (reactants in enzyme catalyzed reactions are called substrates) S P Velocity = V = ΔP/ Δ t So V also = -ΔS/ Δt (disappearance) From the laws of mass action: back reaction ΔP/ Δt = - ΔS/ Δt = k 1[S] – k 2[P] For the INITIAL reaction, [P] is small and can be neglected: ΔP/ Δt = - ΔS/ Δt = k 1[S] So the INITIAL velocity Vo = k 1[S] O signifies INITIAL velocity

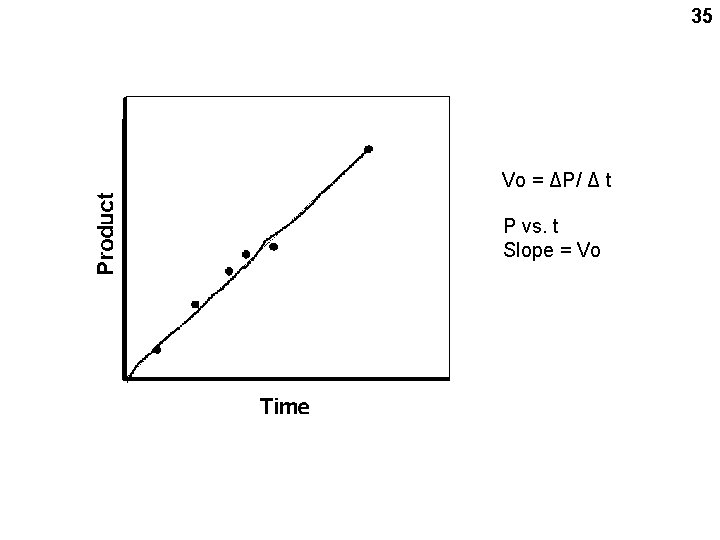
35 Vo = ΔP/ Δ t P vs. t Slope = Vo

36 0. 6 P Effect of different initial substrate concentrations on P vs. t [S 4] [S 3] 0. 4 [S 2] 0. 2 [S 1] 0. 0 t

37 Vo = the slope in each case Effect of different 0. 6 initial substrate concentrations P Dependence of Vo on substrate concentraion [S 4] [S 3] 0. 4 [S 2] 0. 2 [S 1] 0. 0 t Considering Vo as a function of [S] (which will be our usual useful consideration): Vo = k 1[S] Slope = k 1

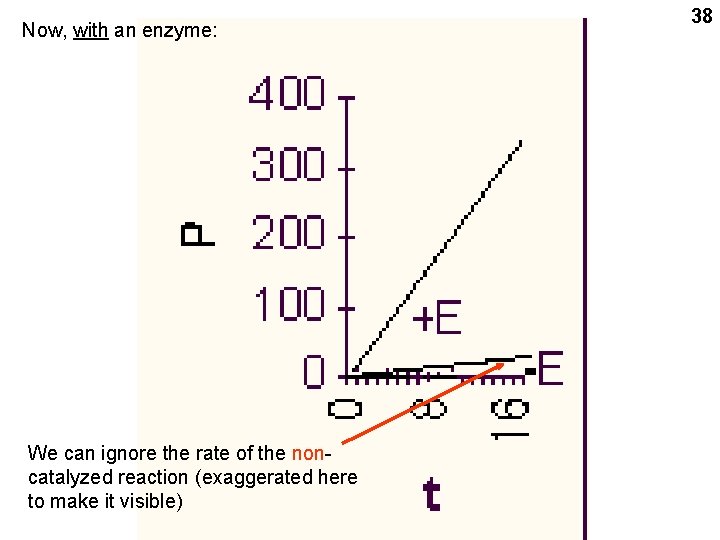
Now, with an enzyme: We can ignore the rate of the noncatalyzed reaction (exaggerated here to make it visible) 38
![Enzyme kinetics as opposed to simple chemical kinetics Vo independent of S Vo proportional Enzyme kinetics (as opposed to simple chemical kinetics) Vo independent of [S] Vo proportional](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-39.jpg)
Enzyme kinetics (as opposed to simple chemical kinetics) Vo independent of [S] Vo proportional to [S] Can we understand this curve? 39

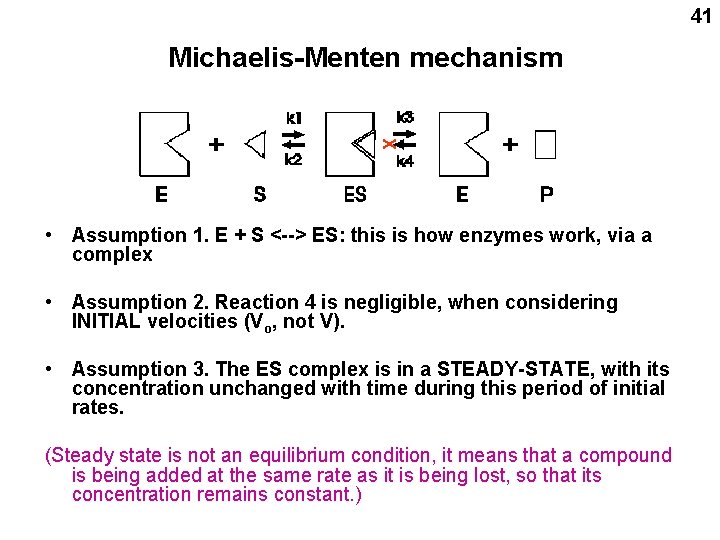
40 Michaelis and Menten mechanism for the action of enzymes (1913)

41 Michaelis-Menten mechanism X • Assumption 1. E + S <--> ES: this is how enzymes work, via a complex • Assumption 2. Reaction 4 is negligible, when considering INITIAL velocities (Vo, not V). • Assumption 3. The ES complex is in a STEADY-STATE, with its concentration unchanged with time during this period of initial rates. (Steady state is not an equilibrium condition, it means that a compound is being added at the same rate as it is being lost, so that its concentration remains constant. )

42

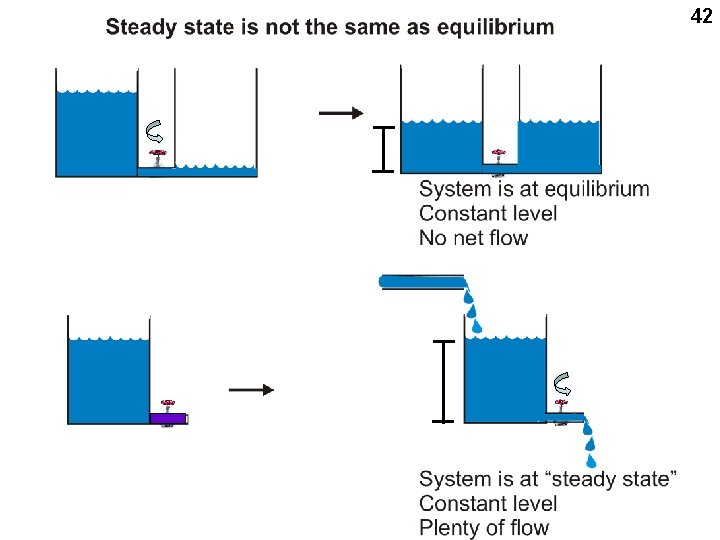
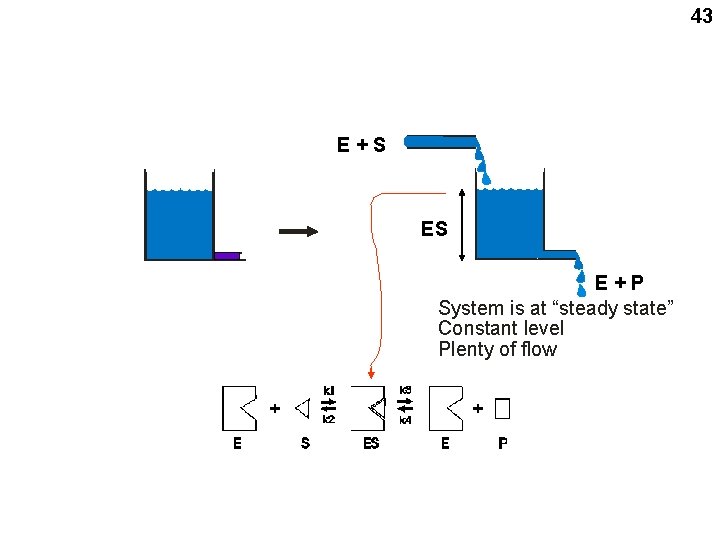
43 System is at equilibrium Constant level No net flow E+S ES E+P System is at “steady state” Constant level Plenty of flow

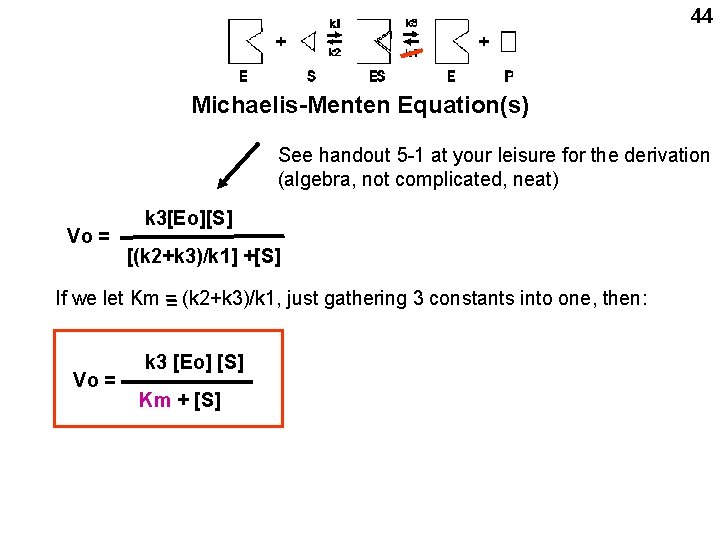
44 Michaelis-Menten Equation(s) See handout 5 -1 at your leisure for the derivation (algebra, not complicated, neat) Vo = k 3[Eo][S] [(k 2+k 3)/k 1] +[S] If we let Km = = (k 2+k 3)/k 1, just gathering 3 constants into one, then: Vo = k 3 [Eo] [S] Km + [S]

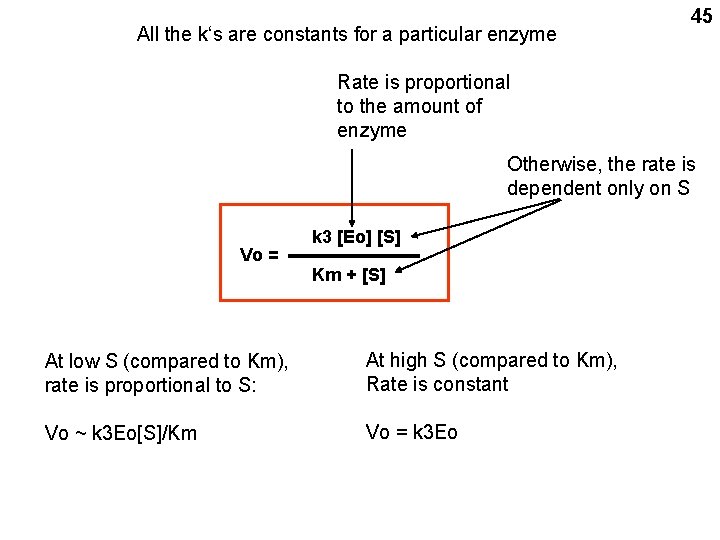
All the k‘s are constants for a particular enzyme 45 Rate is proportional to the amount of enzyme Otherwise, the rate is dependent only on S Vo = k 3 [Eo] [S] Km + [S] At low S (compared to Km), rate is proportional to S: At high S (compared to Km), Rate is constant Vo ~ k 3 Eo[S]/Km Vo = k 3 Eo

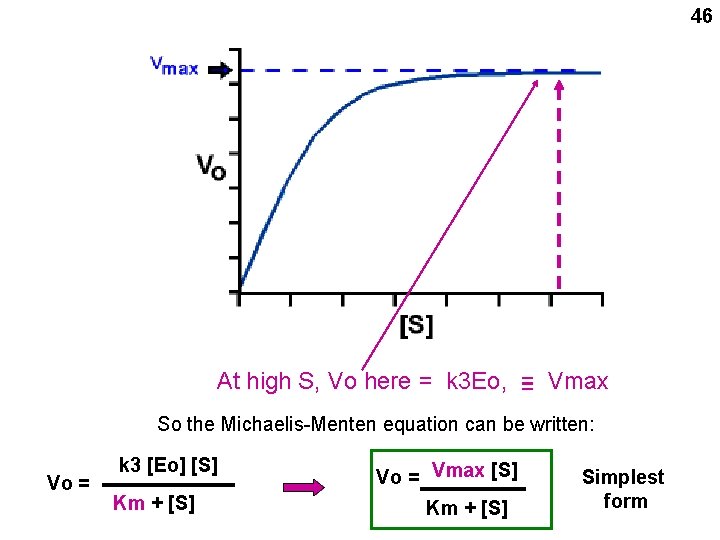
46 At high S, Vo here = k 3 Eo, = = Vmax So the Michaelis-Menten equation can be written: Vo = k 3 [Eo] [S] Km + [S] Vo = Vmax [S] Km + [S] Simplest form

47 Understanding Vmax: ( the maximum intital velocity achievable with a given amount of enzyme ) Now, Vmax = k 3 Eo So: k 3 = Vmax/Eo = the maximum (d. P/dt)/Eo, = the maximum (-d. S/dt)/Eo k 3 = the TURNOVER NUMBER • the maximum number of moles of substrate converted to product per mole of enzyme per second; • the maximum number of molecules of substrate converted to product per molecule of enzyme per second • Turnover number (k 3) then is: a measure of the enzyme's catalytic power.

48 Some turnover numbers (per second) • Succinic dehydrogenase: • Most enzymes: • The winner: Carbonic anhydrase (CO 2 +H 20 19 (below average) 100 -1000 H 2 CO 3) 600, 000 That’s 600, 000 molecules of substrate, per molecule of enzyme, per second. Picture it! You can’t.

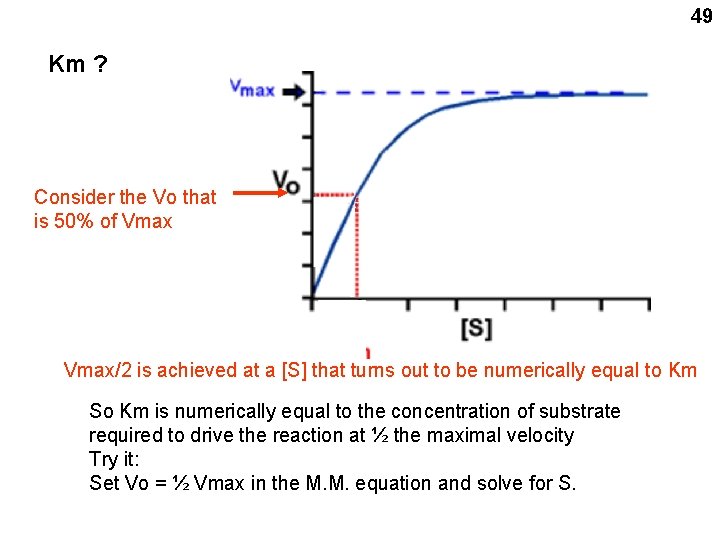
49 Km ? Consider the Vo that is 50% of Vmax/2 is achieved at a [S] that turns out to be numerically equal to Km So Km is numerically equal to the concentration of substrate required to drive the reaction at ½ the maximal velocity Try it: Set Vo = ½ Vmax in the M. M. equation and solve for S.

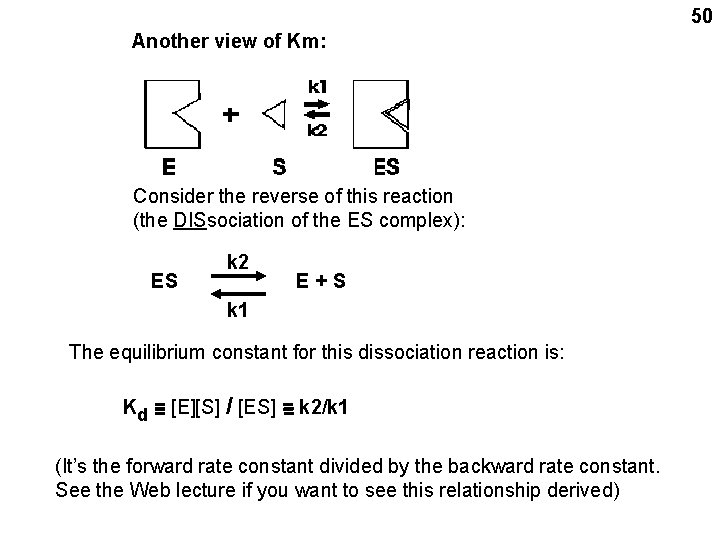
50 Another view of Km: Consider the reverse of this reaction (the DISsociation of the ES complex): ES k 2 E+S k 1 The equilibrium constant for this dissociation reaction is: Kd = = [E][S] / [ES] = = k 2/k 1 (It’s the forward rate constant divided by the backward rate constant. See the Web lecture if you want to see this relationship derived)

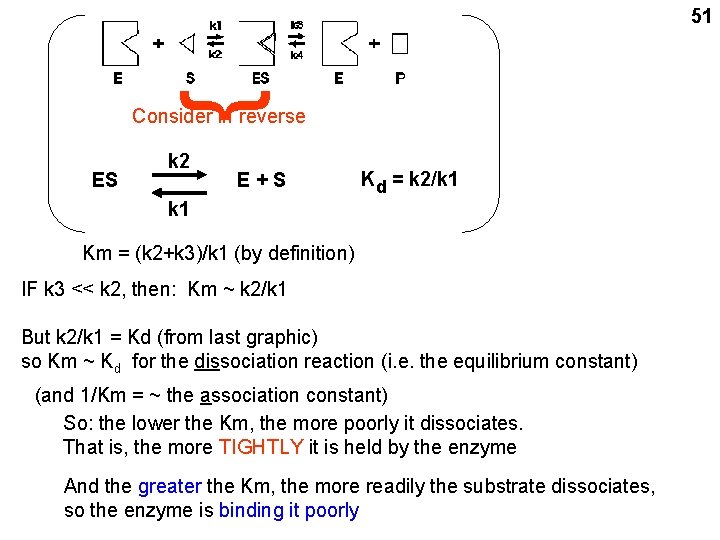
{ 51 Consider in reverse ES k 2 E+S Kd = k 2/k 1 Km = (k 2+k 3)/k 1 (by definition) IF k 3 << k 2, then: Km ~ k 2/k 1 But k 2/k 1 = Kd (from last graphic) so Km ~ Kd for the dissociation reaction (i. e. the equilibrium constant) (and 1/Km = ~ the association constant) So: the lower the Km, the more poorly it dissociates. That is, the more TIGHTLY it is held by the enzyme And the greater the Km, the more readily the substrate dissociates, so the enzyme is binding it poorly

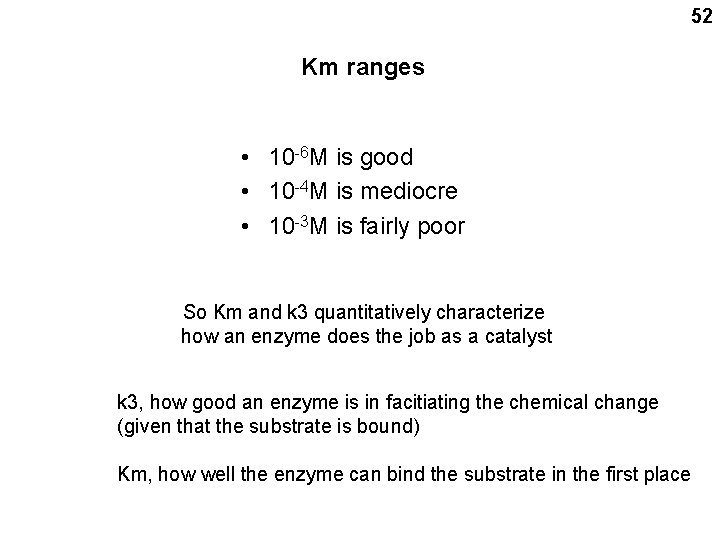
52 Km ranges • 10 -6 M is good • 10 -4 M is mediocre • 10 -3 M is fairly poor So Km and k 3 quantitatively characterize how an enzyme does the job as a catalyst k 3, how good an enzyme is in facitiating the chemical change (given that the substrate is bound) Km, how well the enzyme can bind the substrate in the first place

53 Got this far Exam one material ends at this point.

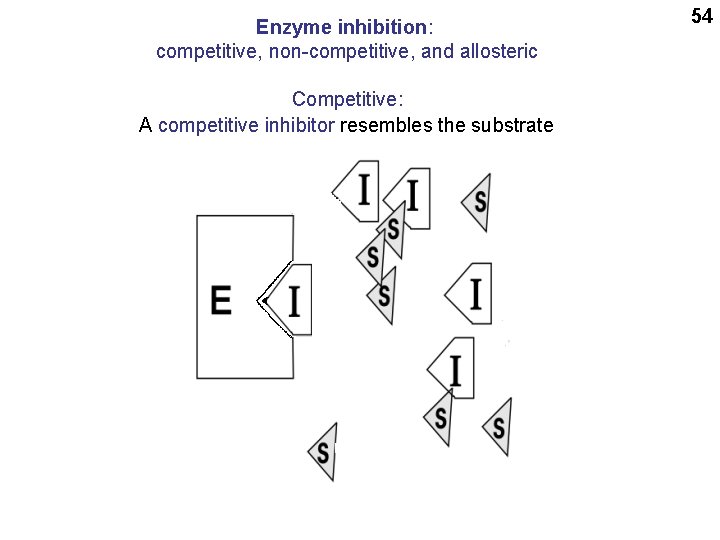
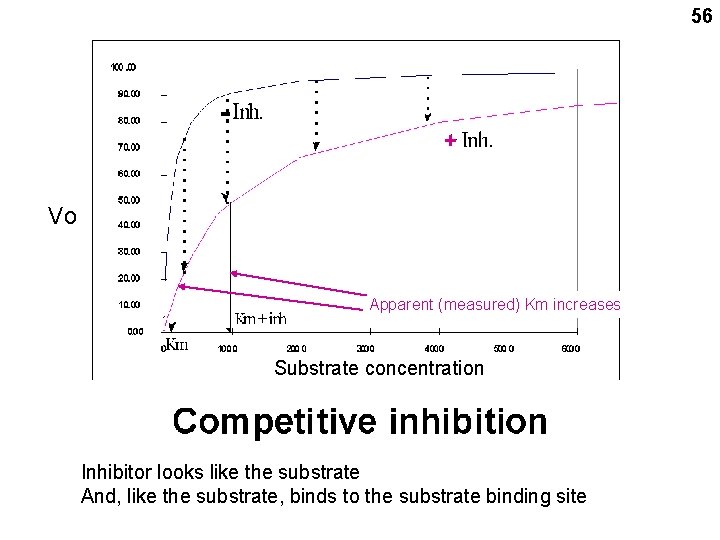
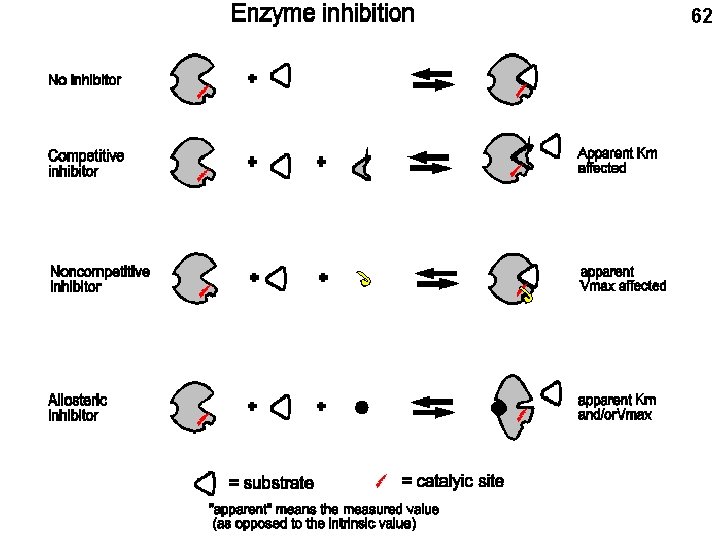
Enzyme inhibition: competitive, non-competitive, and allosteric Competitive: A competitive inhibitor resembles the substrate 54

55 A competitive inhibitor can be swamped out at high substrate concentrations Handout 5 -3 b

56 - + Vo Apparent (measured) Km increases Substrate concentration Inhibitor looks like the substrate And, like the substrate, binds to the substrate binding site

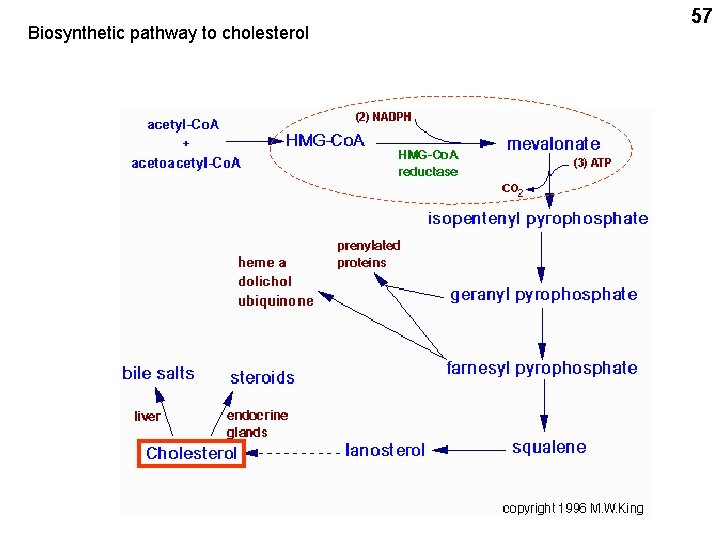
Biosynthetic pathway to cholesterol 57

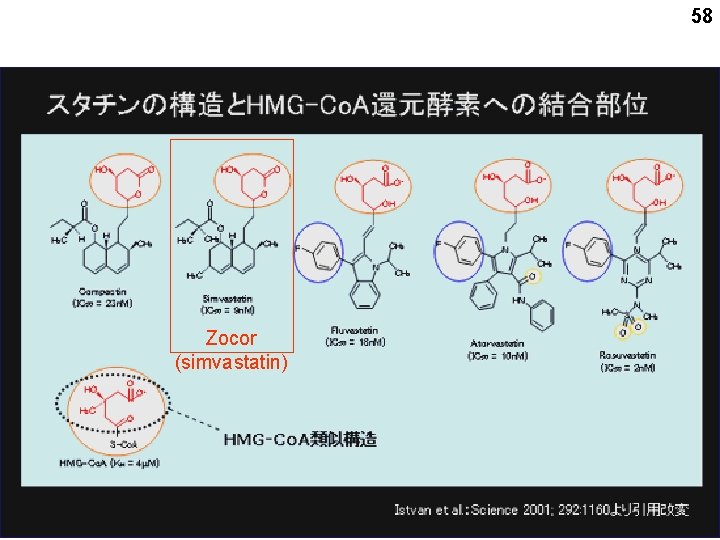
58 Zocor (simvastatin)

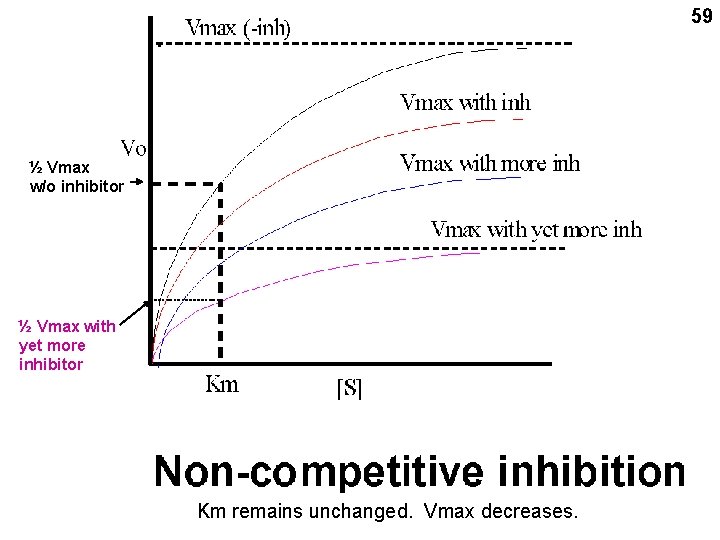
59 ½ Vmax w/o inhibitor ½ Vmax with yet more inhibitor Km remains unchanged. Vmax decreases.

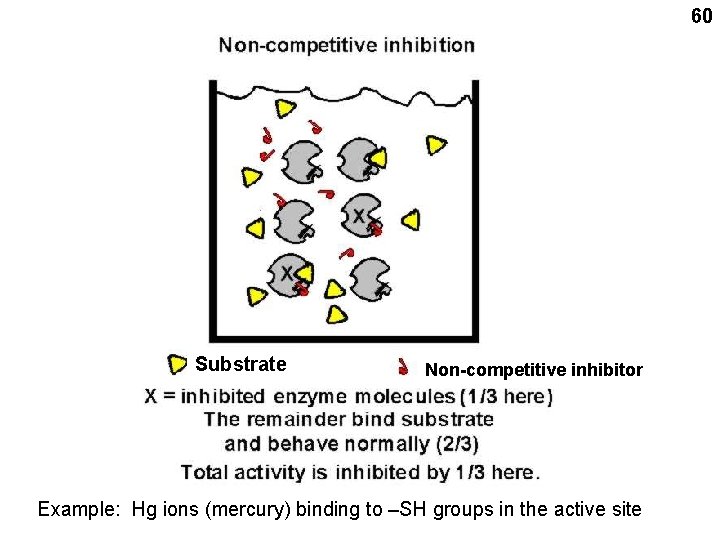
60 Substrate Non-competitive inhibitor Example: Hg ions (mercury) binding to –SH groups in the active site

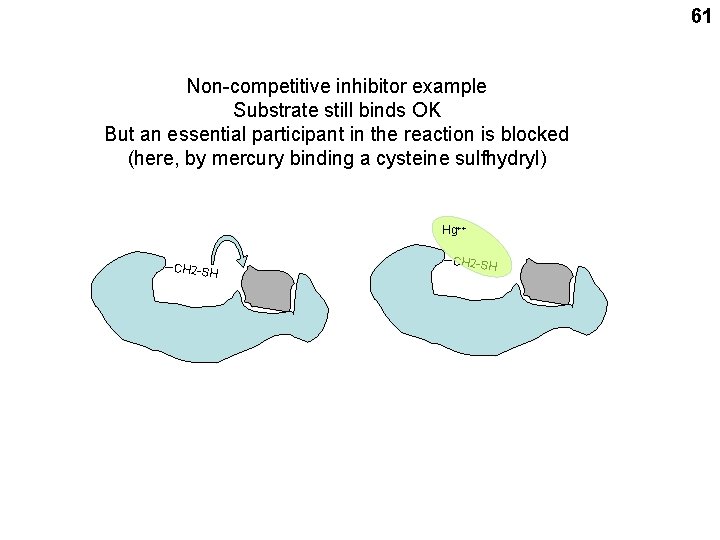
61 Non-competitive inhibitor example Substrate still binds OK But an essential participant in the reaction is blocked (here, by mercury binding a cysteine sulfhydryl) Hg++ --CH 2 -S H

62

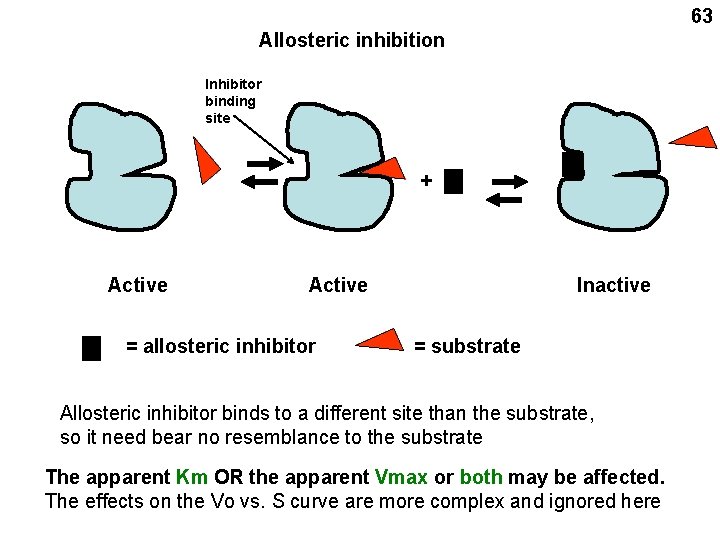
63 Allosteric inhibition Inhibitor binding site + Active = allosteric inhibitor Inactive = substrate Allosteric inhibitor binds to a different site than the substrate, so it need bear no resemblance to the substrate The apparent Km OR the apparent Vmax or both may be affected. The effects on the Vo vs. S curve are more complex and ignored here

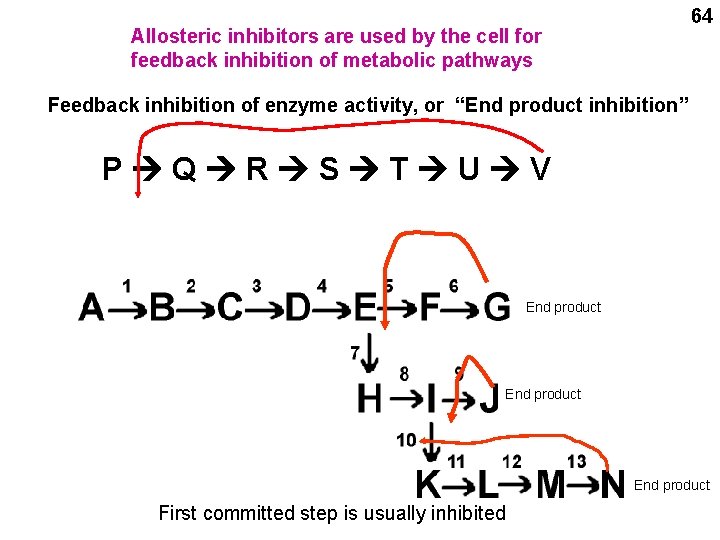
64 Allosteric inhibitors are used by the cell for feedback inhibition of metabolic pathways Feedback inhibition of enzyme activity, or “End product inhibition” P Q R S T U V End product First committed step is usually inhibited

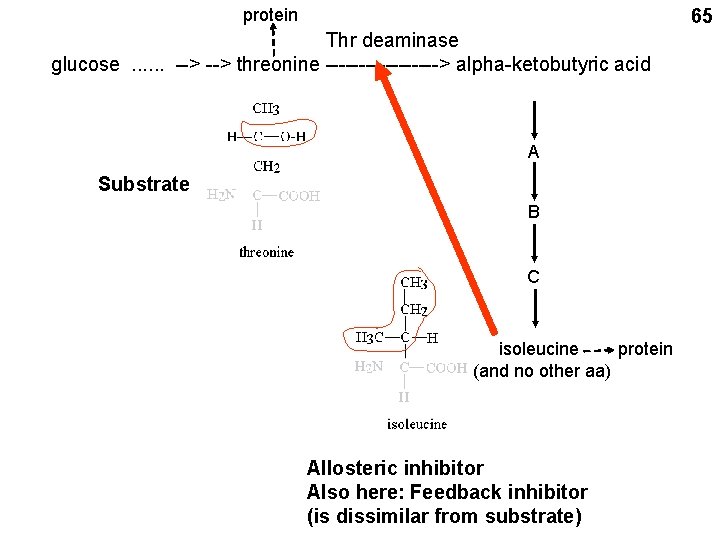
protein 65 Thr deaminase glucose. . . --> threonine ---------> alpha-ketobutyric acid A Substrate B C protein isoleucine (and no other aa) Allosteric inhibitor Also here: Feedback inhibitor (is dissimilar from substrate)

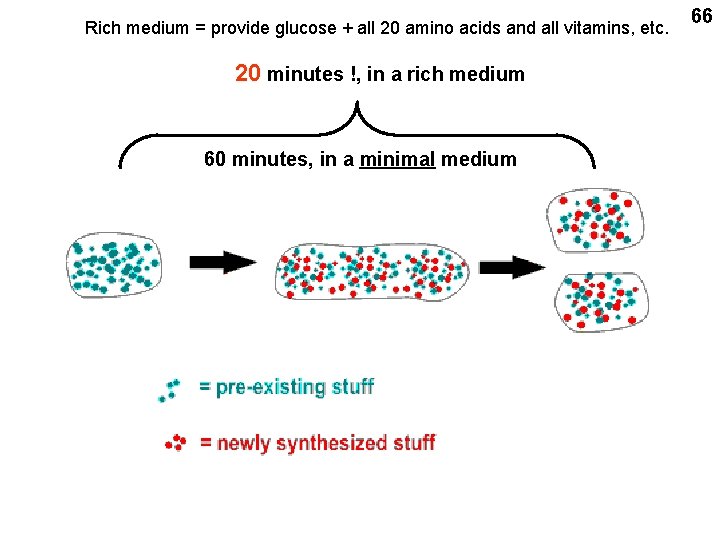
Rich medium = provide glucose + all 20 amino acids and all vitamins, etc. 20 minutes !, in a rich medium 60 minutes, in a minimal medium 66

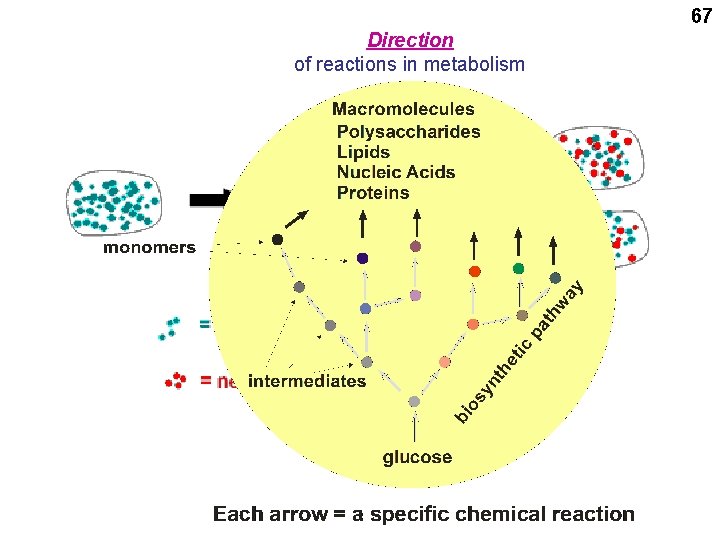
67 Direction of reactions in metabolism

68 Energy }Freedifference determines the direction of a chemical reagion

69 For the model reaction A + B C + D, written in the left-to-right direction indicated: Consider the quantity called the change in free energy associated with a chemical reaction, or: Δ G Such that: • IF Δ G IS <0: THEN A AND B WILL TEND TO PRODUCE C AND D (i. e. , tends to go to the right). • IF Δ G IS >0: THEN C AND D WILL TEND TO PRODUCE A AND B. (i. e. , tends to go to the left) • IF Δ G IS = 0: THEN THE REACTION WILL BE AT EQUILIBRIUM: NOT TENDING TO GO IN EITHER DIRECTION IN A NET WAY.
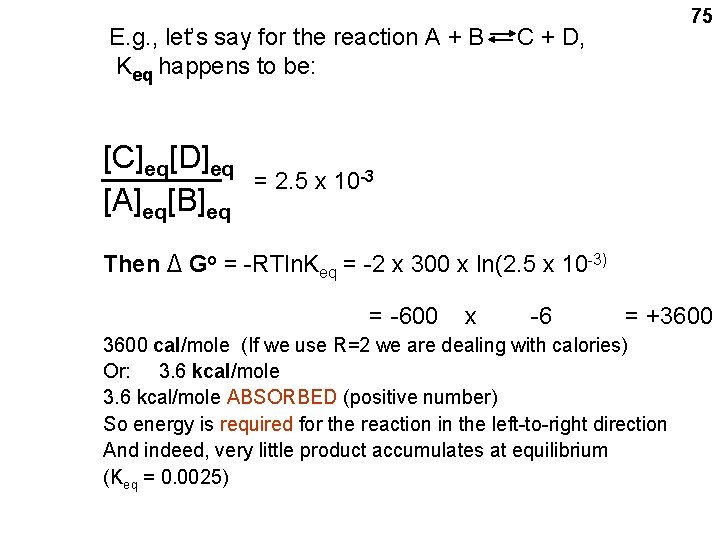
![ΔG Δ G o 70 RTlnCDAB where A B C and D ΔG = Δ G o+ 70 RTln([C][D]/[A][B]) • where A, B, C and D](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-70.jpg)
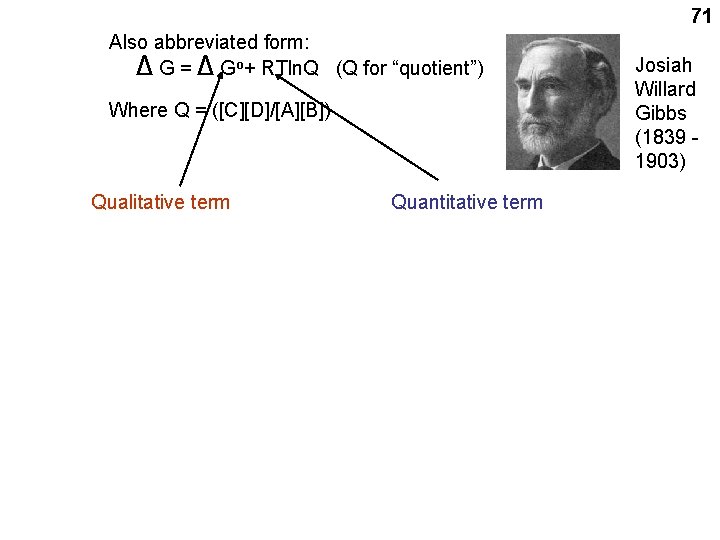
ΔG = Δ G o+ 70 RTln([C][D]/[A][B]) • where A, B, C and D are the concentrations of the reactants and the products AT THE MOMENT BEING CONSIDERED. (i. e. , these A, B, C, D’s here are not the equilibrium concentrations) • R = UNIVERSAL GAS CONSTANT = 1. 98 CAL / DEG K MOLE (R =~2) • T = ABSOLUTE TEMP ( o. K ) 0 o. C = 273 o. K; Room temp = 25 o C = 298 o K (T =~ 300) • ln = NATURAL LOG • Δ Go = a CONSTANT: a quantity related to the INTRINSIC properties of A, B, C, and D

71 Also abbreviated form: Δ G = Δ Go+ RTln. Q (Q for “quotient”) Where Q = ([C][D]/[A][B]) Qualitative term Quantitative term Josiah Willard Gibbs (1839 1903)

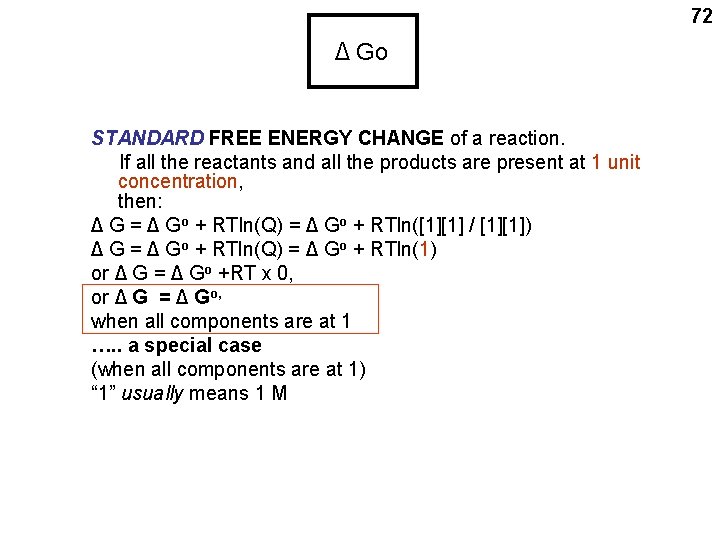
72 Δ Go STANDARD FREE ENERGY CHANGE of a reaction. If all the reactants and all the products are present at 1 unit concentration, then: Δ G = Δ Go + RTln(Q) = Δ Go + RTln([1][1] / [1][1]) Δ G = Δ Go + RTln(Q) = Δ Go + RTln(1) or Δ G = Δ Go +RT x 0, or Δ G = Δ Go, when all components are at 1 …. . a special case (when all components are at 1) “ 1” usually means 1 M

So Δ G and Δ Go are quite different, and not to be confused with each other. Δ Go allows us to compare all reactions under the same standard reaction conditions that we all agree to, independent of concentrations. So it allows a comparison of the stabilities of the bonds in the reactants vs. the products. It is useful. AND, It is easily measured. 73
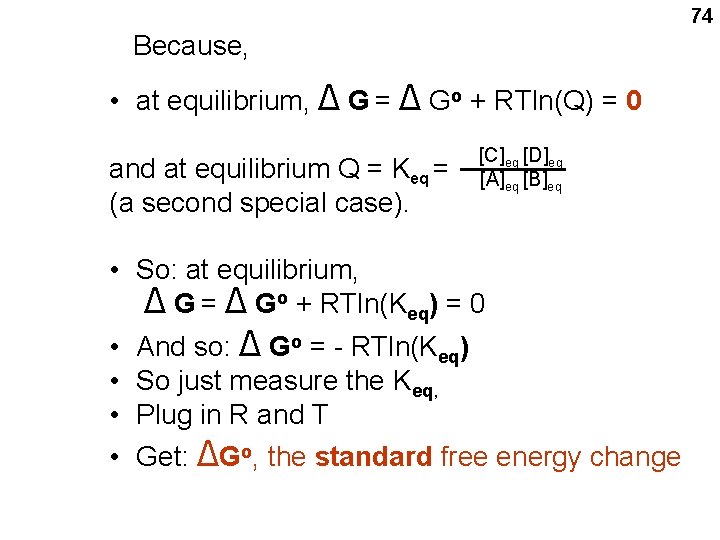
74 Because, • at equilibrium, Δ G = Δ Go + RTln(Q) = 0 and at equilibrium Q = Keq = (a second special case). [C]eq [D]eq [A]eq [B]eq • So: at equilibrium, Δ G = Δ Go + RTln(Keq) = 0 • • And so: Δ Go = - RTln(Keq) So just measure the Keq, Plug in R and T Get: ΔGo, the standard free energy change

E. g. , let’s say for the reaction A + B Keq happens to be: [C]eq[D]eq [A]eq[B]eq 75 C + D, = 2. 5 x 10 -3 Then Δ Go = -RTln. Keq = -2 x 300 x ln(2. 5 x 10 -3) = -600 x -6 = +3600 cal/mole (If we use R=2 we are dealing with calories) Or: 3. 6 kcal/mole ABSORBED (positive number) So energy is required for the reaction in the left-to-right direction And indeed, very little product accumulates at equilibrium (Keq = 0. 0025)

Note: If ΔGo = +3. 6 for the reaction A + B < --- >C + D Then ΔGo = -3. 6 for the reaction C + D <--- > A + B (Reverse the reaction: switch the sign) And: For reactions of more than simple 1 to 1 stoichiometries: a. A + b. B <--> c. C + d. D, ΔG = ΔGo + RT ln [C]c[D]d [A]a[B]b 76

Some exceptions to the 1 M standard condition: Exception #1: • 1) Water: 55 M (pure water) is considered the “unit” concentration instead of 1 M The concentration of water rarely changes during the course of an aqueous reaction, since water is at such a high concentration. • So when calulating Go, instead of writing in “ 55” when water participates in a reaction (e. g. , a hydrolysis) we write “ 1. ” • This is not cheating; we are in charge of what is a “standard” condition, and we all agree to this: 55 M H 20 is unit (“ 1”) concentration for the purpose of defining Go. 77
![Exception 2 In the same way Hydrogen ion concentration H 10 7 M is Exception #2: In the same way, Hydrogen ion concentration, [H+]: 10 -7 M is](https://slidetodoc.com/presentation_image_h2/2ff3dc88418247698c9123c0d0cbde8c/image-78.jpg)
Exception #2: In the same way, Hydrogen ion concentration, [H+]: 10 -7 M is taken as unit concentration, by biochemists. since p. H 7 is maintained in most parts of the cell despite a reaction that may produce acid or base. This definition of the standard free energy change requires the designation ΔGo’ However, I will not bother. But it should be understood we are always talking about ΔGo’ in this course. 78

Summary 79 ΔG = Go + RTln(Q) This combination of one qualitative and one quantitative (driving) term tell the direction of a chemical reaction in any particular circumstance ΔGo = - RTln(Keq) The ΔGo for any reaction is a constant that can be looked up in a book.
 Polyacrylamide gel electrophoresis (page)
Polyacrylamide gel electrophoresis (page) Electrolysis gel
Electrolysis gel Agarose gel
Agarose gel Agarose gel electrophoresis vs sds page
Agarose gel electrophoresis vs sds page Zone electrophoresis definition
Zone electrophoresis definition Lambda electrolysis
Lambda electrolysis Gel electrophoresis definition
Gel electrophoresis definition Gel electrophoresis separates dna by
Gel electrophoresis separates dna by Process of gel electrophoresis
Process of gel electrophoresis Gel electrophoresis definition
Gel electrophoresis definition Gel electrophoresis separates dna by
Gel electrophoresis separates dna by Pipette measure
Pipette measure Translate image
Translate image Gel electrophoresis result
Gel electrophoresis result Parts of electrophoresis apparatus
Parts of electrophoresis apparatus Gel electrophoresis lab ap bio
Gel electrophoresis lab ap bio Selective breeding definition biology
Selective breeding definition biology Disadvantages of agarose gel electrophoresis
Disadvantages of agarose gel electrophoresis Gel electrophoresis advantages
Gel electrophoresis advantages Gel electrophoresis definition
Gel electrophoresis definition Gel kim olursan ol yine gel
Gel kim olursan ol yine gel Sodium oxidanide
Sodium oxidanide Sodium carbonate and calcium chloride balanced equation
Sodium carbonate and calcium chloride balanced equation Oxygen bleach
Oxygen bleach Sodium carbonate and sodium bicarbonate reaction
Sodium carbonate and sodium bicarbonate reaction What is the iupac name of the base naoh?
What is the iupac name of the base naoh? Sodium thiosulfate and sodium hypochlorite reaction
Sodium thiosulfate and sodium hypochlorite reaction Sds page
Sds page Icat
Icat Dizilab4
Dizilab4 Gel page
Gel page Apa title format
Apa title format Stability of sols
Stability of sols Hemoglobin electrophoresis
Hemoglobin electrophoresis Polyclonal gammopathy electrophoresis pattern
Polyclonal gammopathy electrophoresis pattern Instrumentation of electrophoresis
Instrumentation of electrophoresis Paper electrophoresis
Paper electrophoresis Glimpase
Glimpase Haemoglobin normal range
Haemoglobin normal range Automated+electrophoresis+system
Automated+electrophoresis+system Formula of electrophoretic mobility
Formula of electrophoretic mobility Capillary electrophoresis disadvantages
Capillary electrophoresis disadvantages Hemoglobin electrophoresis
Hemoglobin electrophoresis Electrophoresis
Electrophoresis Zonal electrophoresis
Zonal electrophoresis Electrophoresis separation technique
Electrophoresis separation technique Hemoglobin electrophoresis
Hemoglobin electrophoresis Laser doppler electrophoresis
Laser doppler electrophoresis Factors affecting electrophoresis
Factors affecting electrophoresis Paper electrophoresis
Paper electrophoresis Paiweb nacional
Paiweb nacional Two signal words used on whmis 2015 labels
Two signal words used on whmis 2015 labels Victor buczkowski
Victor buczkowski Sulfuric acid sds
Sulfuric acid sds Stylistic devices and expressive means
Stylistic devices and expressive means Skular voc
Skular voc What does sds stand for whmis
What does sds stand for whmis Hazard identification section of the sds
Hazard identification section of the sds Sds umbc
Sds umbc John l. holland
John l. holland Calendrier sds unige
Calendrier sds unige Msds stand for
Msds stand for Sulfuric acid sds
Sulfuric acid sds Honeywell sds protocol
Honeywell sds protocol Instruksi tes holland
Instruksi tes holland Examples of mos and sds
Examples of mos and sds Section 8 of sds sheet
Section 8 of sds sheet Sds meta skills
Sds meta skills Sds security services
Sds security services Holland sds
Holland sds Explorador de carreras y ocupaciones
Explorador de carreras y ocupaciones Hhps
Hhps What does ghs stand for in whmis
What does ghs stand for in whmis Sulfuric acid sds
Sulfuric acid sds Lembar epps
Lembar epps Sds 2 connection design
Sds 2 connection design Student disability services (sds) office
Student disability services (sds) office Jst umn
Jst umn Sds occupations finder
Sds occupations finder Sd pantai indah cilincing
Sd pantai indah cilincing