The applications of advanced proteomic and genomic techniques










- Slides: 37

The applications of advanced proteomic and genomic techniques in cancer Seminar by Dr. Ayat Al-Ghafari Thursday 21 st November 2013

Outlines Overview (cancer) § Protein synthesis § What does proteomic mean? 1. Two-dimensional gel electrophoresis 2. Mass spectrometry (MS) § What does genomic mean? 1. Gene expression microarray §

Overview (cancer) � Cancer � It is a life threatening disease. is a heterogeneous and multifactorial disease. � Although many advances have been made to treat cancer, the survival rate for patients with some types of cancer remains poor. �A main reason behind this is the limitation in understanding the biology of cancer.

Overview (cancer) � Recently, advanced proteomic and genomic approaches have been used to understand cancers and to improve the response of patients to currently used drugs. � In this seminar, I will focused on two major proteomic and one major genomic techniques that are widely used to elucidate the differences in protein and gene expressions, respectively and their correlation with cancer. � Before that I will briefly mentioned how proteins are formed in our cells.

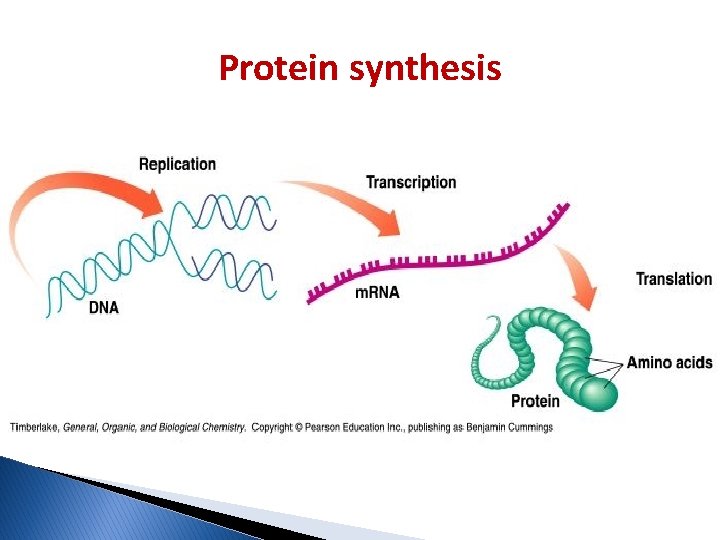
Protein synthesis � The two main processes involved in protein synthesis are: 1. The (transcription) of genomic information from DNA to m. RNA in the same way as the DNA is replicated during cell division. This process takes place in the nucleus. 2. The conversion of m. RNA to protein at the ribosome (translation) which takes place in the cytoplasm.

Protein synthesis

So…… What does proteomic mean? ? ?

Overview (Proteomic) � The analysis and identification of thousands of proteins in a cell or tissue mixture has become easier since the introduction of two-dimensional gel electrophoresis in 1975 and protein mass spectrometry in 1985. � Proteomic is the study of proteome (the collection of all proteins in the cell). � It was first invented in 1997 by Marc Wilkins when he was working on the concept as a Ph. D student.

Aims of Proteomic � Detect the different proteins expressed by tissue, cell culture, or organism using 2 -Dimensional Gel Electrophoresis. � Store those information in a database. � Compare expression profiles between a healthy cell vs. a diseased cell. � The data comparison can then be used for testing and rational drug design.

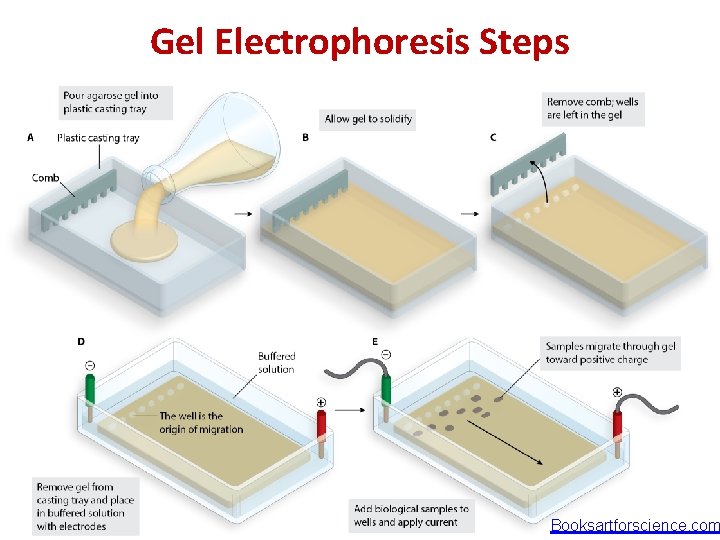
Gel Electrophoresis � Is a separation technique based on the motion of charged molecules in an electric field. � Usually used a porous matrix (Agarose or polyacrylamide). � For protein separation: Polyacrylamide gel provides a porous matrix (PAGE – Polyacrylamide Gel Electrophoresis) � Sample is stained with comassie blue or bromophenol blue to make it visible in the gel when we load the sample on the gel and during the run.

Gel Electrophoresis Steps Booksartforscience. com

� Separation 1 -Dimensional (1 -D) Gel electrophoresis in only 1 dimension based on the molecular weight. � Smaller molecules travel further through the gel. � Electric field across gel separates molecules based on the charge of the molecules. � Proteins are treated with the denaturing detergent SDS (sodium dodecyl sulfate) which coats the protein with negative charges, hence SDS-PAGE.

Two-dimensional (2 -D) gel electrophoresis � It is a technique used to separate proteins based on two properties: 1. In the first dimension, proteins are separated according to their isoelectric point (p. I) in a process known as isoelectric focusing (IEF) along a strip that has an immobilized p. H gradient range. 2. In the second dimension, proteins are separated in a vertical direction according to their denatured molecular weight through a gradient polyacrylamide gel.

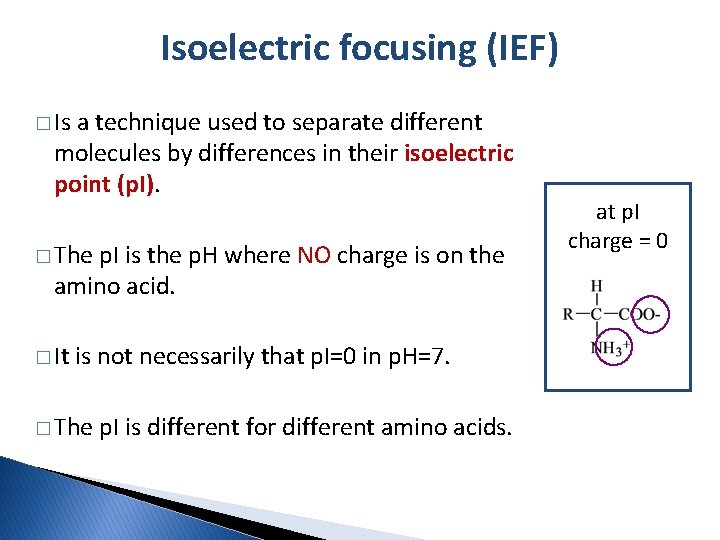
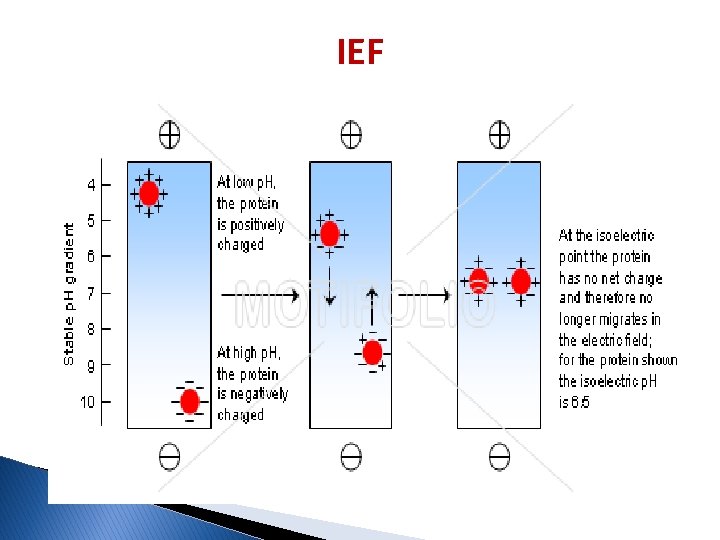
Isoelectric focusing (IEF) � Is a technique used to separate different molecules by differences in their isoelectric point (p. I). � The p. I is the p. H where NO charge is on the amino acid. � It is not necessarily that p. I=0 in p. H=7. � The p. I is different for different amino acids. at p. I charge = 0

IEF


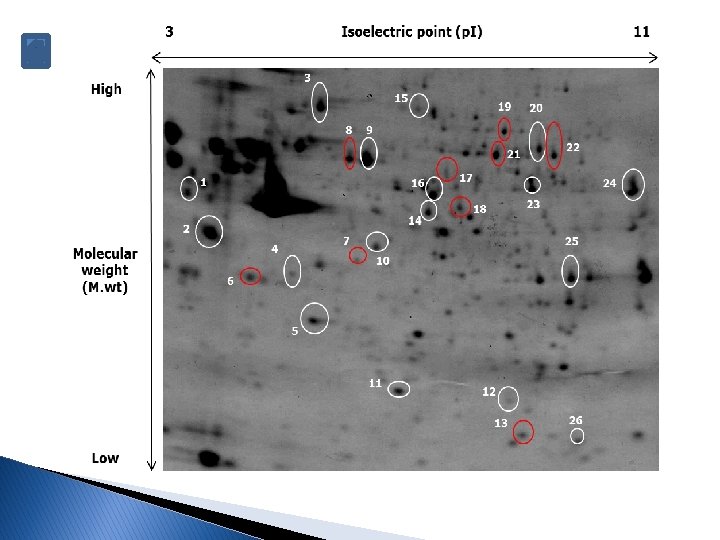
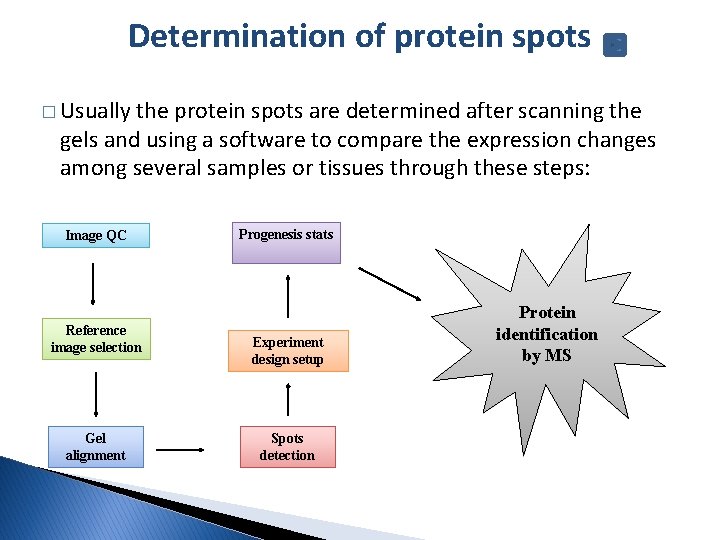
Determination of protein spots � Usually the protein spots are determined after scanning the gels and using a software to compare the expression changes among several samples or tissues through these steps: Image QC Reference image selection Gel alignment Progenesis stats Experiment design setup Spots detection Protein identification by MS

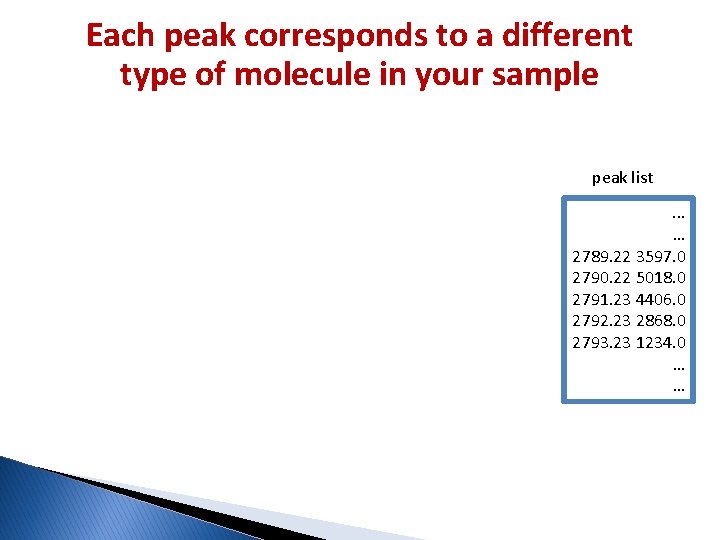
Mass Spectrometry (MS) � MS is an analytical technique that produces spectra of the masses of the atoms or molecules comprising a sample. � If there is a mixture of different molecules in a sample, all the masses are measured simultaneously. So you get a spectrum. � The mass spectrum is a plot of the (relative) abundances of the ions at each m/z ratio.

Each peak corresponds to a different type of molecule in your sample peak list. . . … 2789. 22 3597. 0 2790. 22 5018. 0 2791. 23 4406. 0 2792. 23 2868. 0 2793. 23 1234. 0 … …

Components of MS 1. Ionisation-Gaseous atoms of a particular element are bombarded with electrons fired from an electron gun. These electron particles will give the gaseous atoms of the specific element a charge. This charge may vary; or some atoms may have no charge at all. 2. Acceleration-The atoms, now charged, are called ions and because these ions are charged they can be accelerated by an electric field.

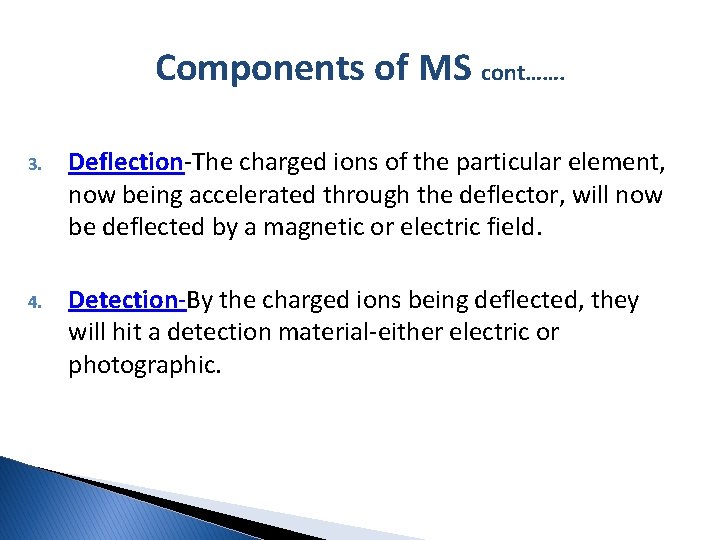
Components of MS cont……. 3. Deflection-The charged ions of the particular element, now being accelerated through the deflector, will now be deflected by a magnetic or electric field. 4. Detection-By the charged ions being deflected, they will hit a detection material-either electric or photographic.

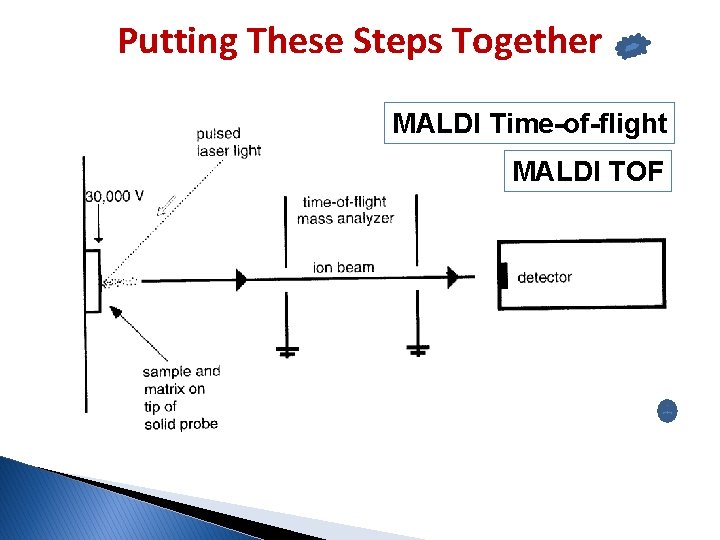
Putting These Steps Together MALDI Time-of-flight MALDI TOF Drift region (D)

Final result = the spectrum www. csd. uwo. ca

Final step in Protein “identification” with MS � The final step is the detection of the protein of interest in the database and this can be done through the following steps: 1. The mass of the protein in the sample is detected by spectrometer. 2. Then the unknown masses in our sample were compared to other known masses in database to determine the protein type.

Next, we will talk about genomic applications

Overview (Genomic) � Before understanding what genomics mean, we need to know what is genome? ? �A genome is “all the DNA contained in an organism or a cell, which includes both the chromosomes within the nucleus and the DNA in mitochondria… all our genes together [Source: National Human Genome Research Institute]. � Genomics is “the study of functions and interactions of all the genes in the genome, including their interactions with environmental factors” [Collins, F. and Guttmacher, A. “Genomic Medicine—A Primer, ” NEJM, Vol. 347: 1512 -1520].

Overview (Genomic) Cont…. . � Genetic variations are important factors in cancer. � Understanding how these genes alterations can increase the risk of cancer and affect therapies is important to draw a comprehensive picture about it and help to design a new therapy regimen.

Aims of genomics � Genetic testing ◦ To detect mutations. � Pharmacogenomics ◦ The development of drugs tailored to specific subpopulations based on genes. ◦ Pharmacogenomics has the potential to: �Decrease side effects of drugs. �Increase drug effectiveness. �Make drug development faster and less costly.

Aims of genomics � Recent ◦ ◦ ◦ research in genomics includes Learning more about the genetic underpinnings of chronic diseases Developing mouse models of human genes Developing genetic fingerprinting for childhood cancer Conducting stem cell research Identifying tumor suppressor genes

Gene expression microarray (DNA Microarray) � One of the major genomic applications which plays an important role in understanding cancer biology is gene expression microarray. � In a gene expression experiment, the expression levels of thousands of genes are simultaneously monitored to study the effects of certain treatments, diseases, and developmental stages on gene expression by comparing gene expression in infected to that in uninfected cells or tissues.

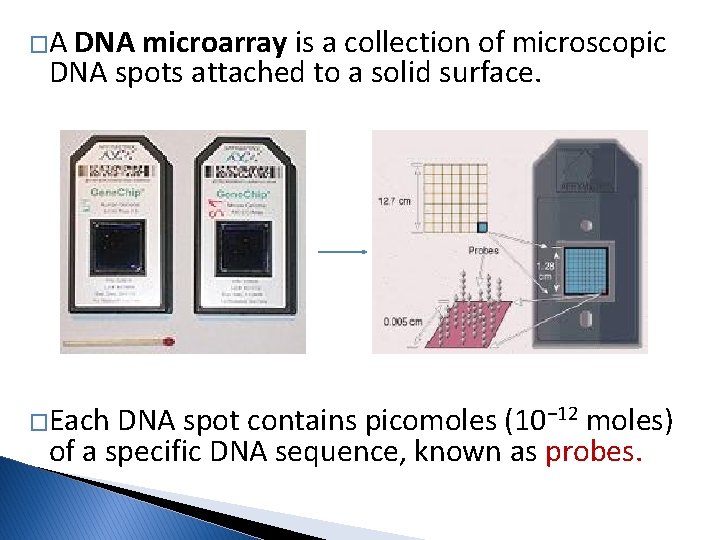
�A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. �Each DNA spot contains picomoles (10− 12 moles) of a specific DNA sequence, known as probes.

� These probes can be a short section of a gene or other DNA element that are used to hybridize a complimentary DNA (c. DNA) sample under high-stringency conditions. � Probe-target hybridization is usually detected and quantified by detection of fluorescent or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target.

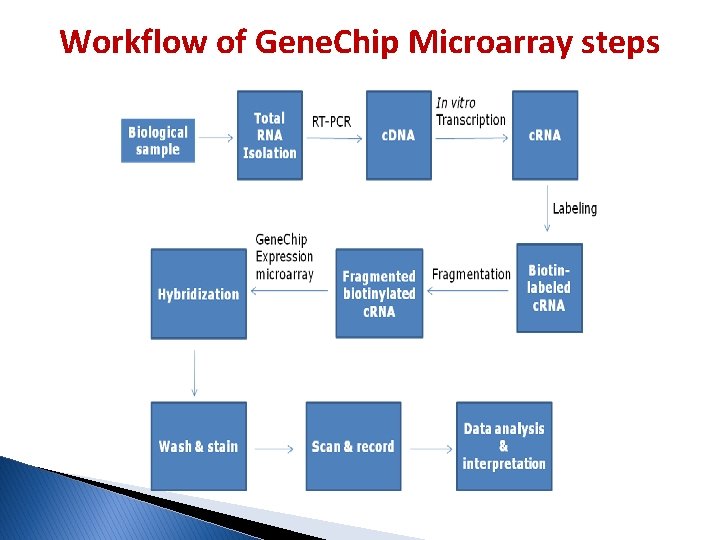
Workflow of Gene. Chip Microarray steps

Final result will be presented in different colors. Each color represent different expression pattern for the genes. These colors known as probes intensities. The black features represent no intensity. The intensity level from lowest to highest by color is: Dark blue -> Blue -> Light Blue -> Green -> Yellow -> Orange -> Red - > White.

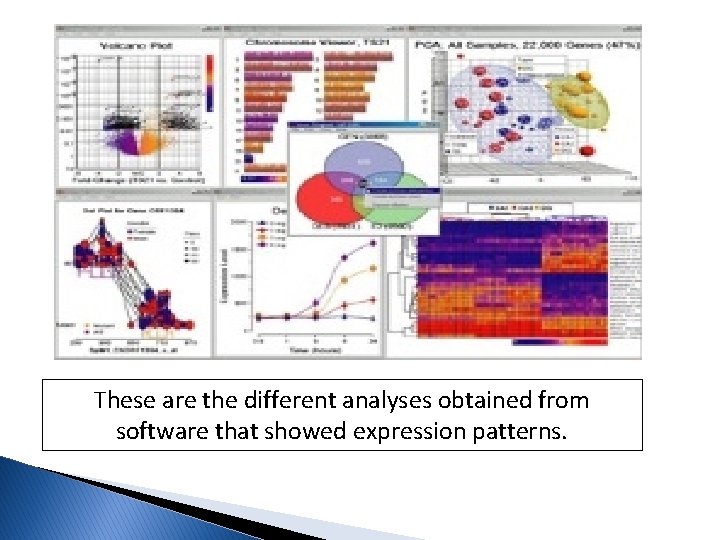
These are the different analyses obtained from software that showed expression patterns.

References � � HANAHAN, D. & WEINBERG, R. 2000. The hallmarks of cancer. Cell, 100, 57 -70. O'FARRELL, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. The Journal of Biological Chemistry, 250, 4007 -4021. WHITEHOUSE, C. M. , DREYER, R. N. , YAMASHITA, M. & FENN, J. B. 1985. Electrospray interface for liquid chromatographs and mass spectrometers. Analytical Chemistry, 57, 675 -679. GRESHOCK, J. , NATHANSON, K. , MARTIN, A. M. , ZHANG, L. , COUKOS, G. , WEBER, B. L. & ZAKS, T. Z. 2007. Cancer cell lines as genetic models of their parent histology: analyses based on array comparative genomic hybridization. Cancer Research, 67, 3594 -3600

Thanks for your attention ………
 Genomic equivalence definition
Genomic equivalence definition Genomic england
Genomic england Genomic england
Genomic england Genomic england
Genomic england Genomic instability
Genomic instability Genomic
Genomic Genomic imprinting definition
Genomic imprinting definition Genomic signal processing
Genomic signal processing Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic equivalence definition
Genomic equivalence definition Counting techniques in discrete mathematics
Counting techniques in discrete mathematics Advanced business applications
Advanced business applications Advanced business applications
Advanced business applications Advanced sketching techniques
Advanced sketching techniques Advanced evasion techniques
Advanced evasion techniques Advanced counting techniques
Advanced counting techniques Advanced interviewing techniques
Advanced interviewing techniques Advanced image processing techniques
Advanced image processing techniques Advanced counting techniques in discrete mathematics
Advanced counting techniques in discrete mathematics Advanced data visualization techniques
Advanced data visualization techniques Preengineered building
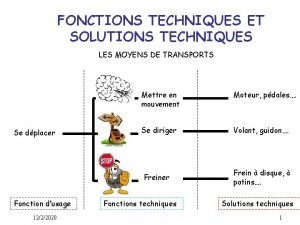
Preengineered building Fonctions techniques et solutions techniques
Fonctions techniques et solutions techniques Hình ảnh bộ gõ cơ thể búng tay
Hình ảnh bộ gõ cơ thể búng tay Ng-html
Ng-html Bổ thể
Bổ thể Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Voi kéo gỗ như thế nào
Voi kéo gỗ như thế nào Glasgow thang điểm
Glasgow thang điểm Hát lên người ơi alleluia
Hát lên người ơi alleluia Môn thể thao bắt đầu bằng từ đua
Môn thể thao bắt đầu bằng từ đua Thế nào là hệ số cao nhất
Thế nào là hệ số cao nhất Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới Công thức tính thế năng
Công thức tính thế năng Trời xanh đây là của chúng ta thể thơ
Trời xanh đây là của chúng ta thể thơ Cách giải mật thư tọa độ
Cách giải mật thư tọa độ 101012 bằng
101012 bằng Phản ứng thế ankan
Phản ứng thế ankan Các châu lục và đại dương trên thế giới
Các châu lục và đại dương trên thế giới