Sodium dodecyl sulfate polyacrylamide gel electrophoresis SDSPAGE Provides













- Slides: 33

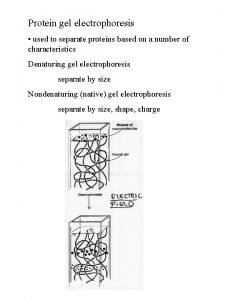
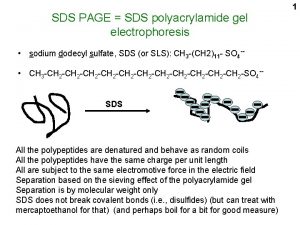
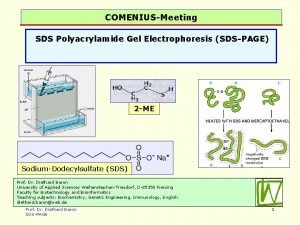
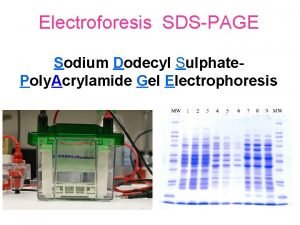
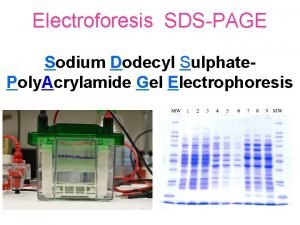
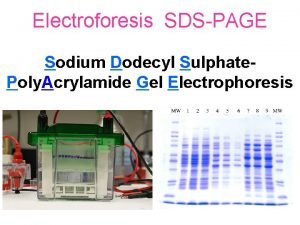
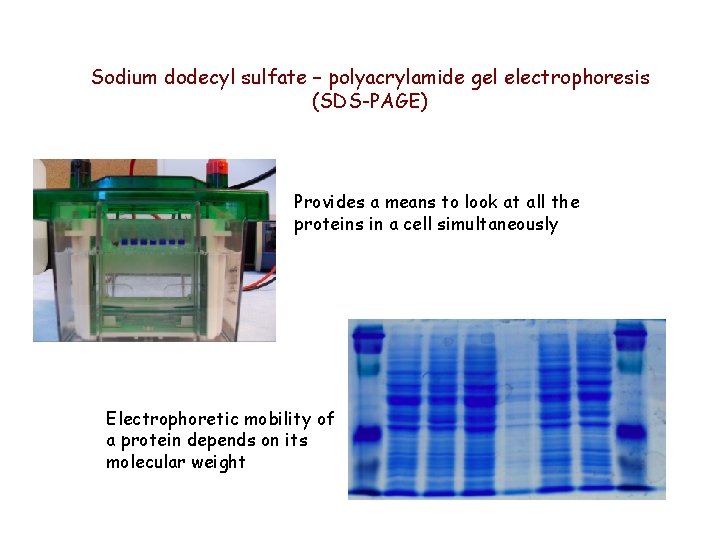
Sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS-PAGE) Provides a means to look at all the proteins in a cell simultaneously Electrophoretic mobility of a protein depends on its molecular weight

What principles govern the separation of proteins by SDS -PAGE? How does one prepare and run SDS-PAGE gels? How are SDS-PAGE gels analyzed?

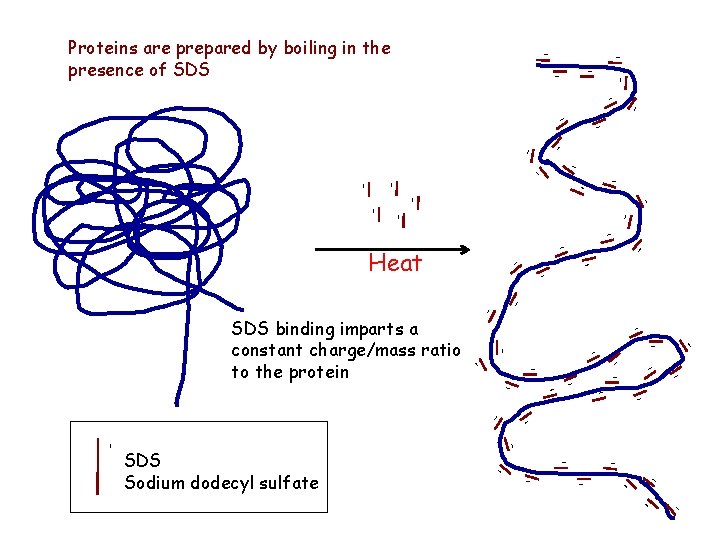
- - Proteins are prepared by boiling in the presence of SDS - - - - - - Heat - - - - - - - SDS Sodium dodecyl sulfate - - - SDS binding imparts a constant charge/mass ratio to the protein

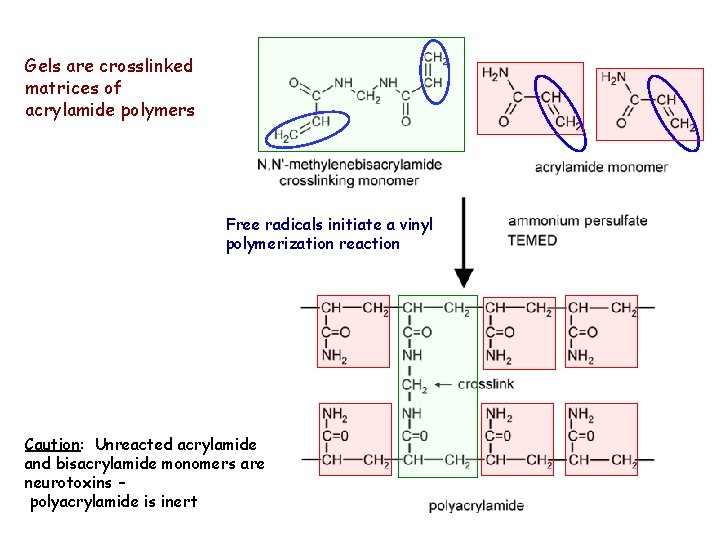
Gels are crosslinked matrices of acrylamide polymers Free radicals initiate a vinyl polymerization reaction Caution: Unreacted acrylamide and bisacrylamide monomers are neurotoxins – polyacrylamide is inert

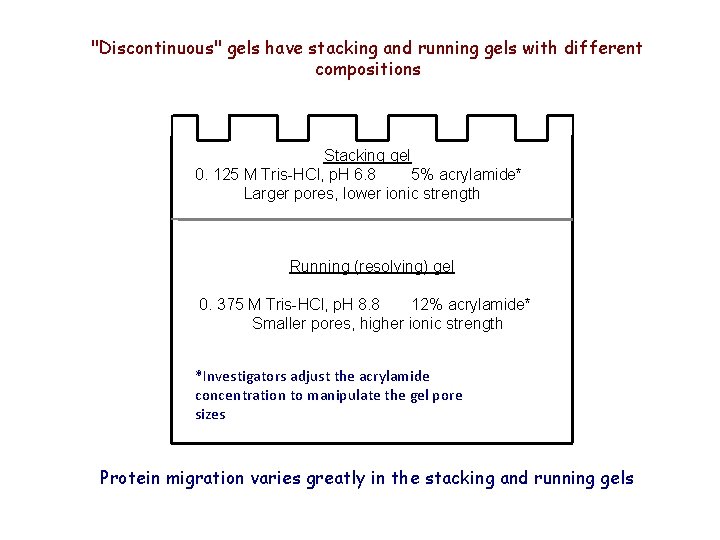
"Discontinuous" gels have stacking and running gels with different compositions Stacking gel 0. 125 M Tris-HCl, p. H 6. 8 5% acrylamide* Larger pores, lower ionic strength Running (resolving) gel 0. 375 M Tris-HCl, p. H 8. 8 12% acrylamide* Smaller pores, higher ionic strength *Investigators adjust the acrylamide concentration to manipulate the gel pore sizes Protein migration varies greatly in the stacking and running gels

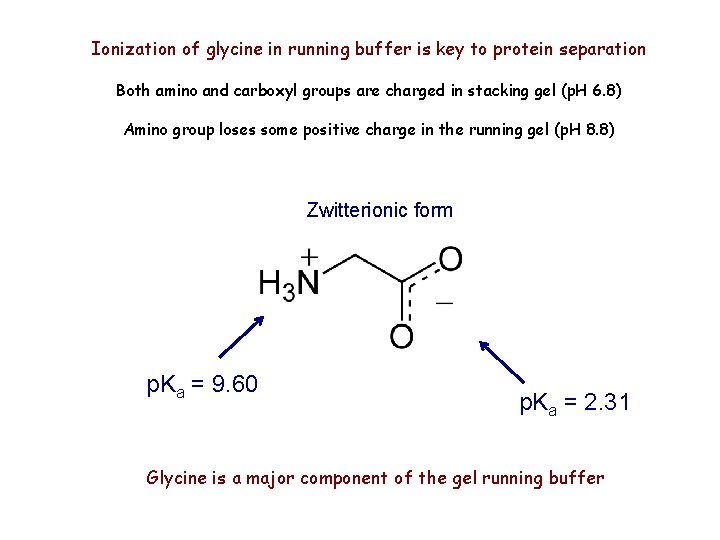
Ionization of glycine in running buffer is key to protein separation Both amino and carboxyl groups are charged in stacking gel (p. H 6. 8) Amino group loses some positive charge in the running gel (p. H 8. 8) Zwitterionic form p. Ka = 9. 60 p. Ka = 2. 31 Glycine is a major component of the gel running buffer

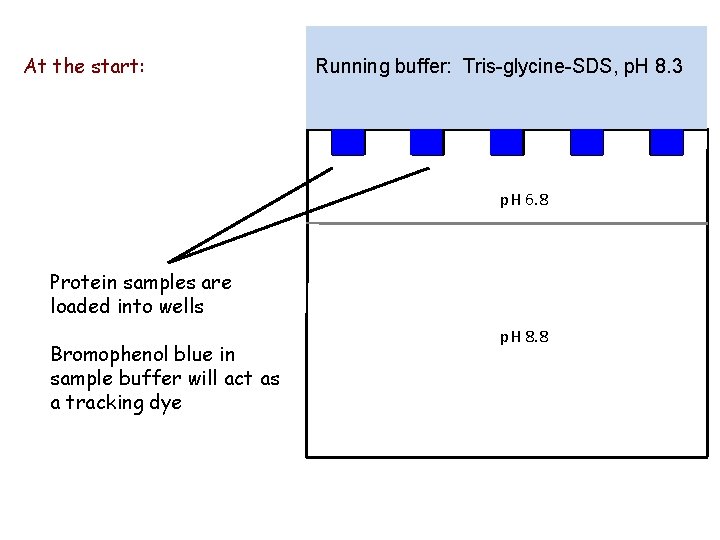
At the start: Running buffer: Tris-glycine-SDS, p. H 8. 3 p. H 6. 8 Protein samples are loaded into wells Bromophenol blue in sample buffer will act as a tracking dye p. H 8. 8

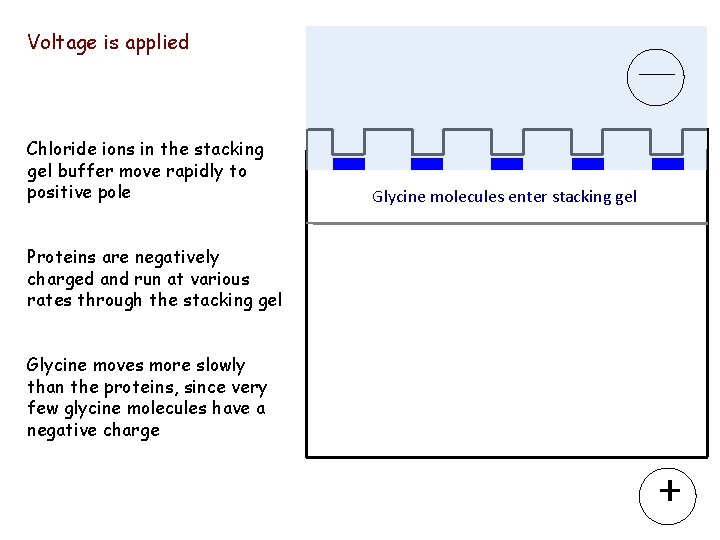
Voltage is applied ____ Chloride ions in the stacking gel buffer move rapidly to positive pole Glycine molecules enter stacking gel Proteins are negatively charged and run at various rates through the stacking gel Glycine moves more slowly than the proteins, since very few glycine molecules have a negative charge +

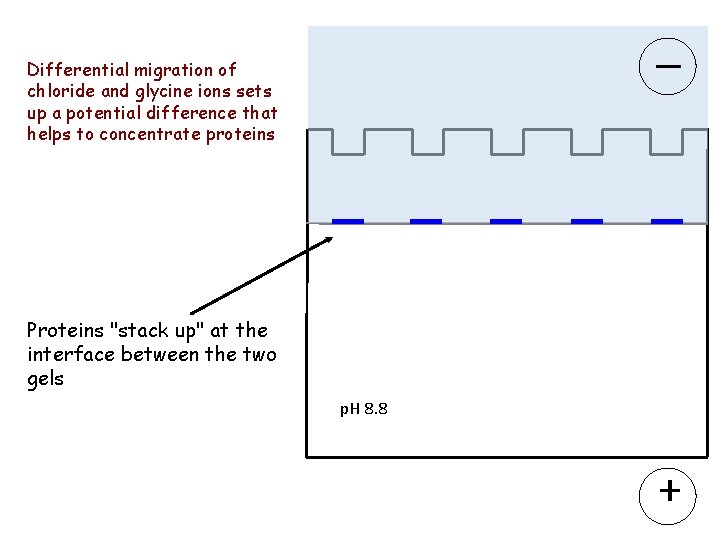
Differential migration of chloride and glycine ions sets up a potential difference that helps to concentrate proteins Proteins "stack up" at the interface between the two gels p. H 8. 8 +

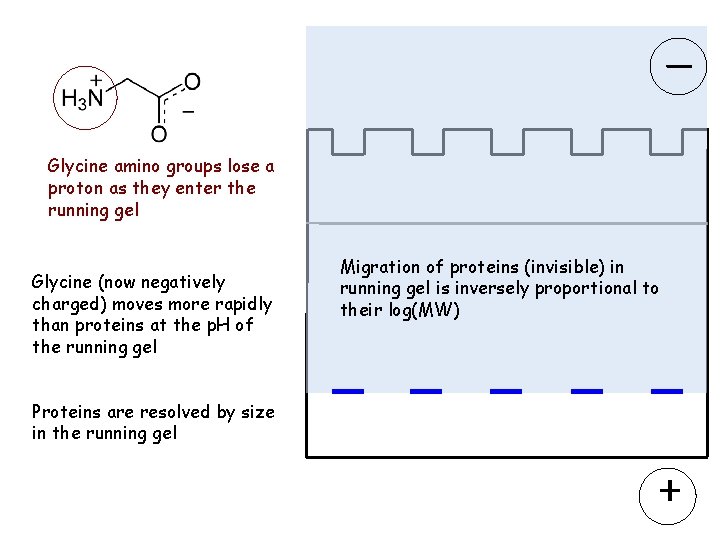
Glycine amino groups lose a proton as they enter the running gel Glycine (now negatively charged) moves more rapidly than proteins at the p. H of the running gel Migration of proteins (invisible) in running gel is inversely proportional to their log(MW) Proteins are resolved by size in the running gel +

What principles govern the separation of proteins by SDS -PAGE? How does one prepare and run SDS-PAGE gels? How are SDS-PAGE gels analyzed?

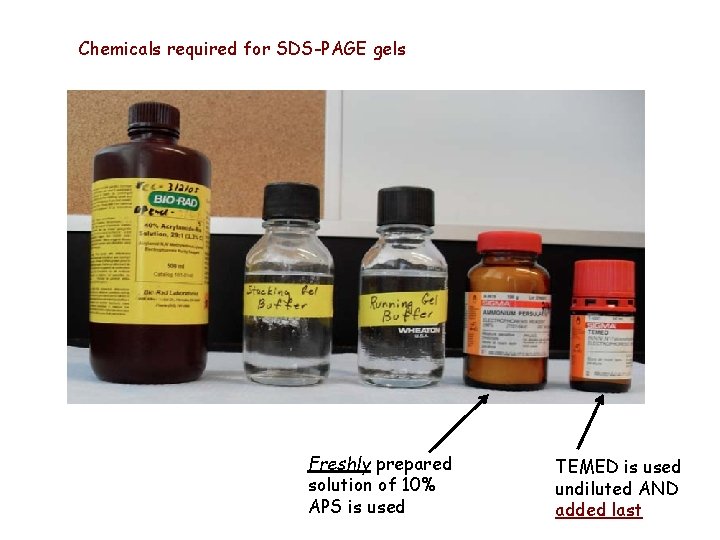
Chemicals required for SDS-PAGE gels Freshly prepared solution of 10% APS is used TEMED is used undiluted AND added last

Gel will be formed between two glass plates Larger plate has precisely milled spacers at either edge that will generate a 1 mm separation between the assembled plates Second plate is shorter, but has the same width as the spacer plate Plate is VERY fragile! Make sure plates are CLEAN before proceeding!

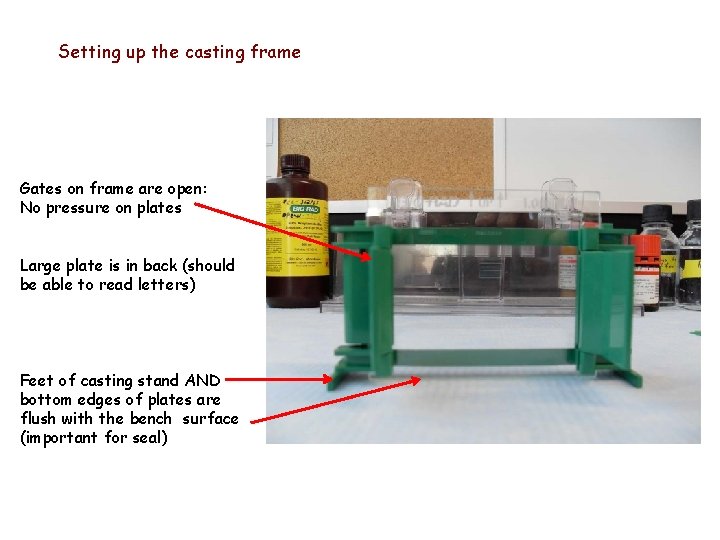
Setting up the casting frame Gates on frame are open: No pressure on plates Large plate is in back (should be able to read letters) Feet of casting stand AND bottom edges of plates are flush with the bench surface (important for seal)

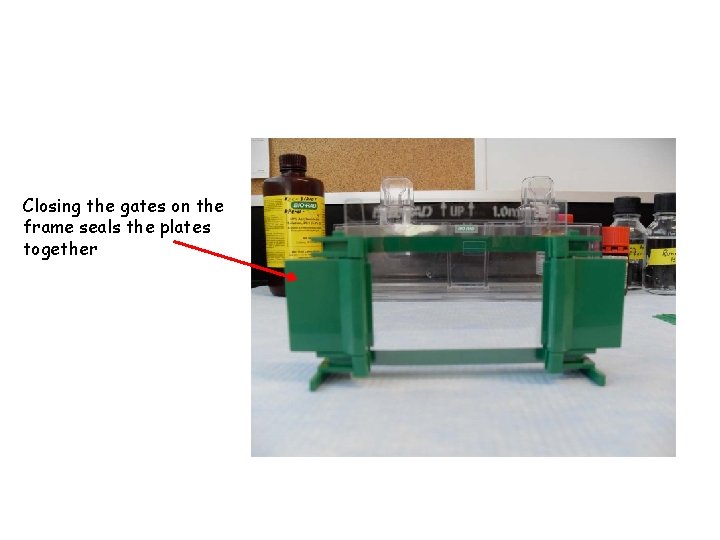
Closing the gates on the frame seals the plates together

Clip the assembled casting frame into the casting stand (can accommodate two gels) Spongy pad at the bottom of the stand contributes to a tight seal

Test for potential leaks with deionized water Pour the water out after you have finished the test

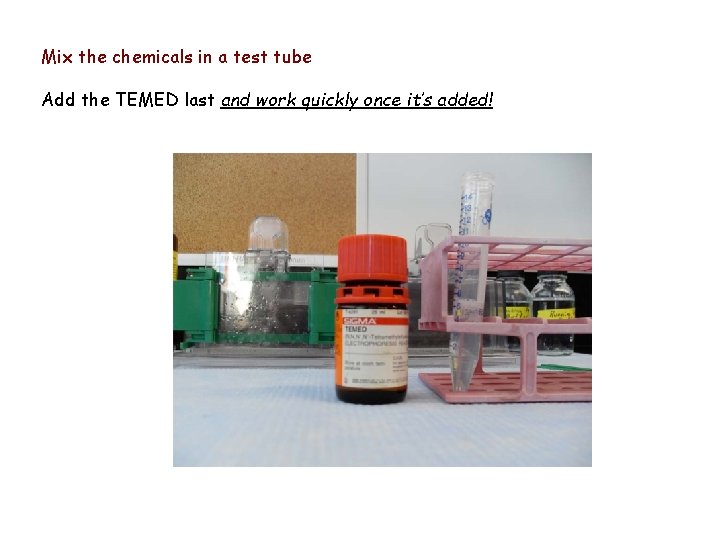
Mix the chemicals in a test tube Add the TEMED last and work quickly once it’s added!

Pipet acrylamide mixture to the top of the frame gates Avoid air bubbles as much as possible (O 2 inhibits polymerization) Overlay the acrylamide with a thin layer of deionized water The interface between the running gel and water will disappear as the gel polymerizes, but it will reappear as a sharp boundary when polymerization is complete

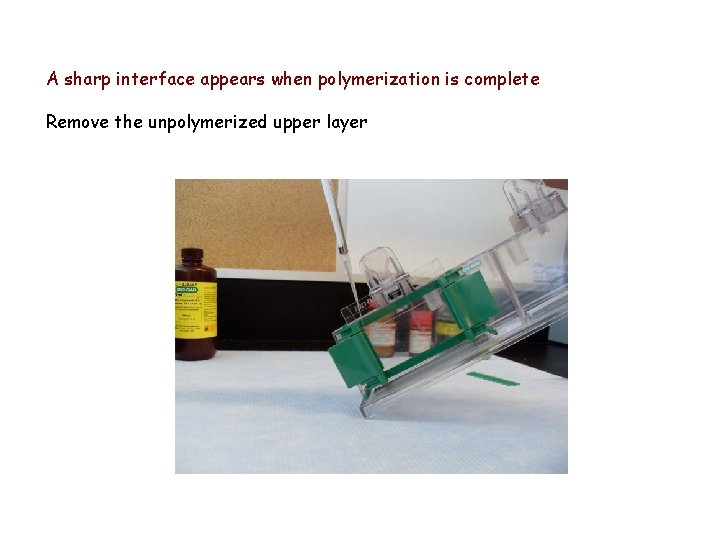
A sharp interface appears when polymerization is complete Remove the unpolymerized upper layer

Combine the components of the stacking gel, adding catalysts last Pipette the solution on top of the running gel, leaving some room for the sample comb – BE QUICK!

Insert the sample comb into the stacking gel solution Be careful to avoid trapping bubbles! (may need to reposition the comb)

Time to run the gel! Remove the gel from the casting frame

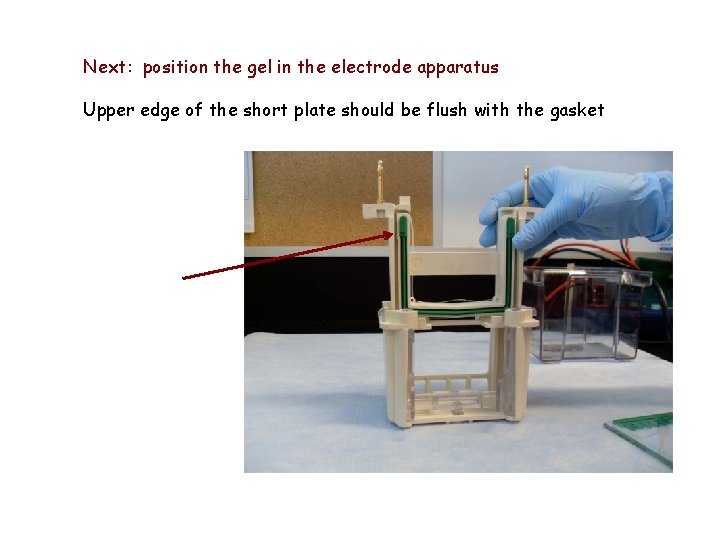
Next: position the gel in the electrode apparatus Upper edge of the short plate should be flush with the gasket

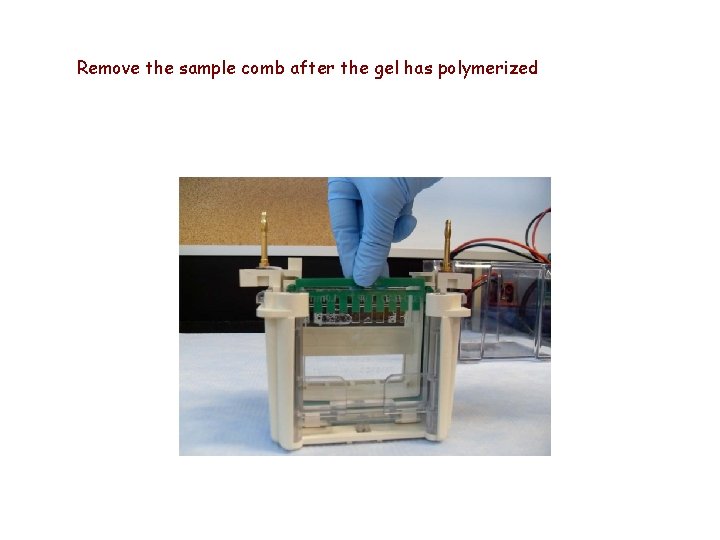
Remove the sample comb after the gel has polymerized

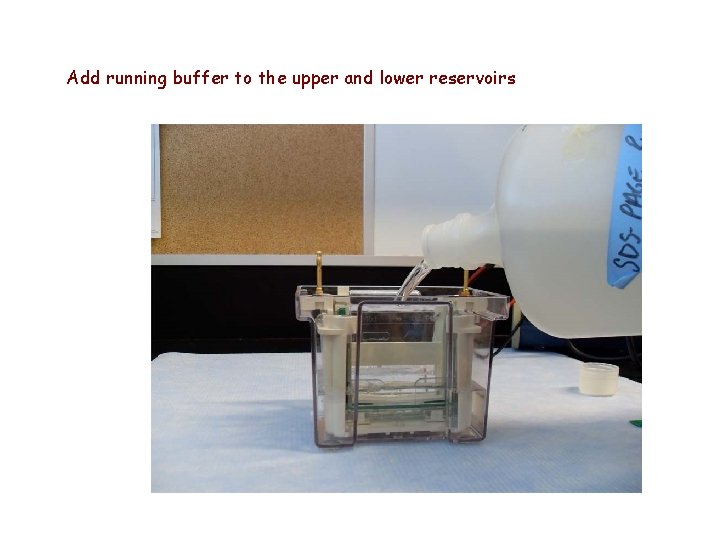
Add running buffer to the upper and lower reservoirs

Load the samples Place a loaded gel loading tip at the bottom of a sample well Slowly expel the sample, avoiding air bubbles

Connect the gel to the power supply

Monitor the progress of the tracking dye Turn off the power before the tracking dye runs off the gel!

What principles govern the separation of proteins by SDS -PAGE? How does one prepare and run SDS-PAGE gels? How are SDS-PAGE gels analyzed?

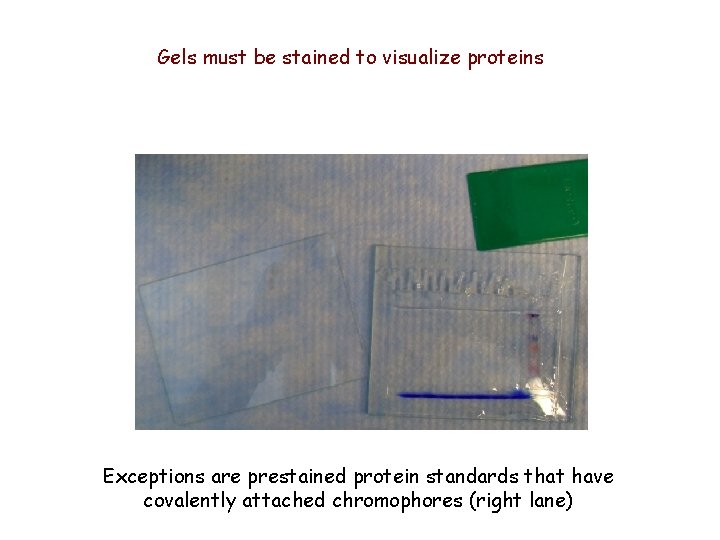
Gels must be stained to visualize proteins Exceptions are prestained protein standards that have covalently attached chromophores (right lane)

Proteins are visualized with Coomassie Brilliant Blue G-250 Gels are rinsed with water several times to remove gel chemicals and are then incubated with Simply Blue® a colloidal suspension of Coomassie Blue G-250

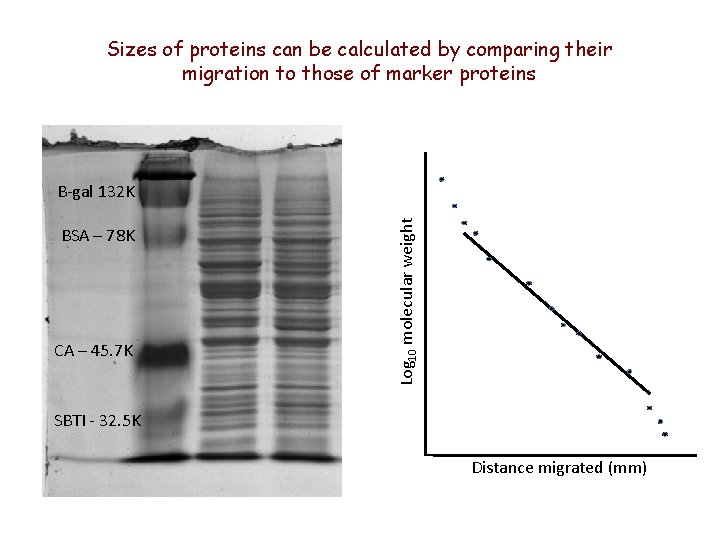
Sizes of proteins can be calculated by comparing their migration to those of marker proteins BSA – 78 K CA – 45. 7 K Log 10 molecular weight B-gal 132 K SBTI - 32. 5 K Distance migrated (mm)
 Electrolysis gel
Electrolysis gel Polyacrylamide gel electrophoresis (page)
Polyacrylamide gel electrophoresis (page) Agarose gel electrophoresis vs sds page
Agarose gel electrophoresis vs sds page Agarose gel vs polyacrylamide gel
Agarose gel vs polyacrylamide gel Spectator ions
Spectator ions Calcium carbonate nitric acid reaction
Calcium carbonate nitric acid reaction Gel electrophoresis separates dna by
Gel electrophoresis separates dna by Process of gel electrophoresis
Process of gel electrophoresis Electrophoresis definition
Electrophoresis definition Gel electrophoresis why do smaller fragments move faster
Gel electrophoresis why do smaller fragments move faster Gel electrophoresis lab ap bio
Gel electrophoresis lab ap bio P-20 micropipette
P-20 micropipette Translate
Translate Gel electrophoresis result
Gel electrophoresis result Endosmosis
Endosmosis Is gel electrophoresis a biotechnology
Is gel electrophoresis a biotechnology Ashanthi desilva
Ashanthi desilva Disadvantages of agarose gel electrophoresis
Disadvantages of agarose gel electrophoresis Gel electrophoresis advantages
Gel electrophoresis advantages Zone electrophoresis definition
Zone electrophoresis definition Lambda electrolysis
Lambda electrolysis Anode cathode gel electrophoresis
Anode cathode gel electrophoresis Gel kim olursan ol yine gel
Gel kim olursan ol yine gel Balance the following chemical equations
Balance the following chemical equations Oxygen bleach
Oxygen bleach Sodium carbonate hydrochloric acid
Sodium carbonate hydrochloric acid What is the iupac name of the base naoh?
What is the iupac name of the base naoh? Redox volumetric analysis
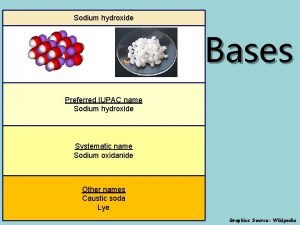
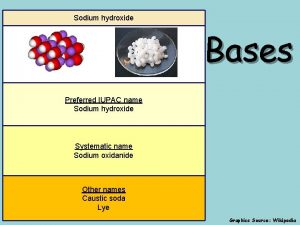
Redox volumetric analysis Sodium hydroxide iupac id sodium oxidanide
Sodium hydroxide iupac id sodium oxidanide Copper sulfate and potassium iodide
Copper sulfate and potassium iodide Magnesium sulfate and urine output
Magnesium sulfate and urine output Mgso4 side effects
Mgso4 side effects Ammonium sulfate precipitation
Ammonium sulfate precipitation Asthma complications
Asthma complications