Protein Tertiary Structure Comparison Dong Xu Computer Science



















- Slides: 61

Protein Tertiary Structure Comparison Dong Xu Computer Science Department 271 C Life Sciences Center 1201 East Rollins Road University of Missouri-Columbia, MO 65211 -2060 E-mail: xudong@missouri. edu 573 -882 -7064 (O) http: //digbio. missouri. edu

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

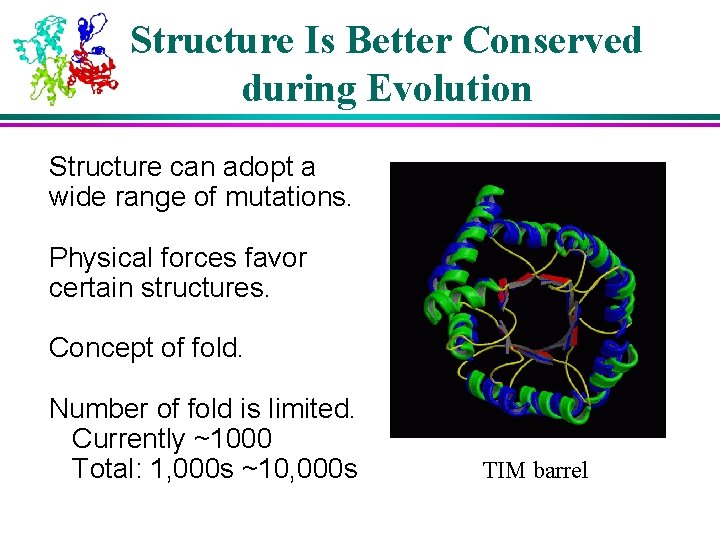
Structure Is Better Conserved during Evolution Structure can adopt a wide range of mutations. Physical forces favor certain structures. Concept of fold. Number of fold is limited. Currently ~1000 Total: 1, 000 s ~10, 000 s TIM barrel

Alignment of Protein Structure l Three-dimensional structure of one protein compared against three-dimensional structure of second protein l Atoms (protein backbones) fit together as closely as possible to minimize the average deviation

Why Align Structures? (1) Additional measure of protein similarity l Structure generally preserved better than sequence over the course of evolution l l Provide more information on the relationship between proteins than what sequence alignment can offer l Allows classification of proteins based on structural similarities

Why Align Structures? (2) l l Basis for protein fold identification (prediction) Sometimes sequence similarity between two proteins exists, but is not strong enough to produce an unambiguous alignment (gold standard for sequence comparison). Pinpoint the active sites more accurately. Allows identification of common substructures of interest

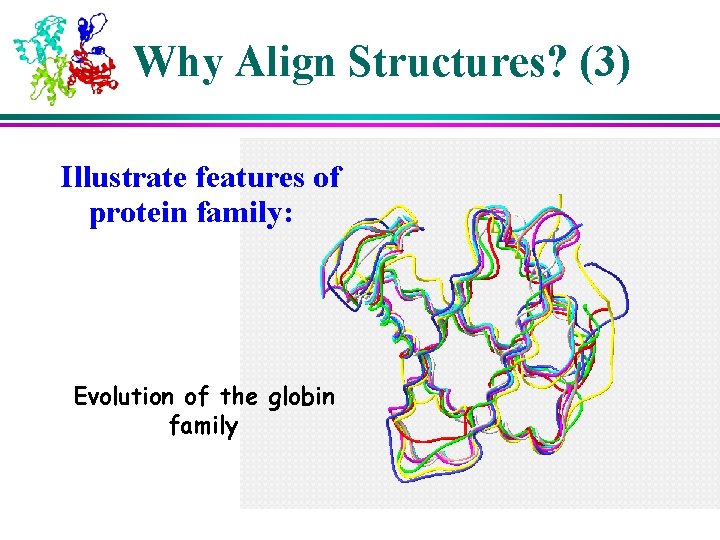
Why Align Structures? (3) Illustrate features of protein family: Evolution of the globin family

Why Align Structures? (4) Illustrate interesting evolutionary/functional relationship between proteins: Two ferredoxins, 1 DOI and 1 AWD, are aligned structurally, showing an insertion in 1 DOI that contains potassium-ion binding sites. This may be the result of adaptations to the high salt environment of the Dead Sea.

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

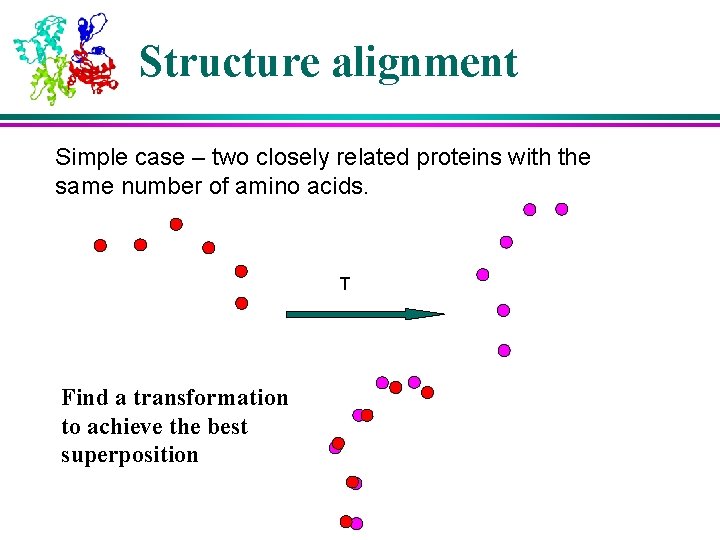
Structure alignment Simple case – two closely related proteins with the same number of amino acids. T Find a transformation to achieve the best superposition

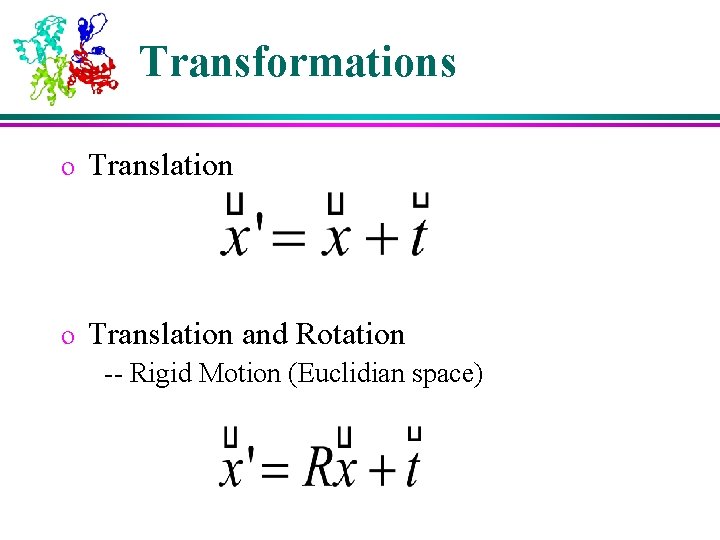
Transformations o Translation and Rotation -- Rigid Motion (Euclidian space)

Types of Structure Comparison o Sequence-dependent vs. sequenceindependent structural alignment o Global vs. local structural alignment o Pairwise vs. multiple structural alignment

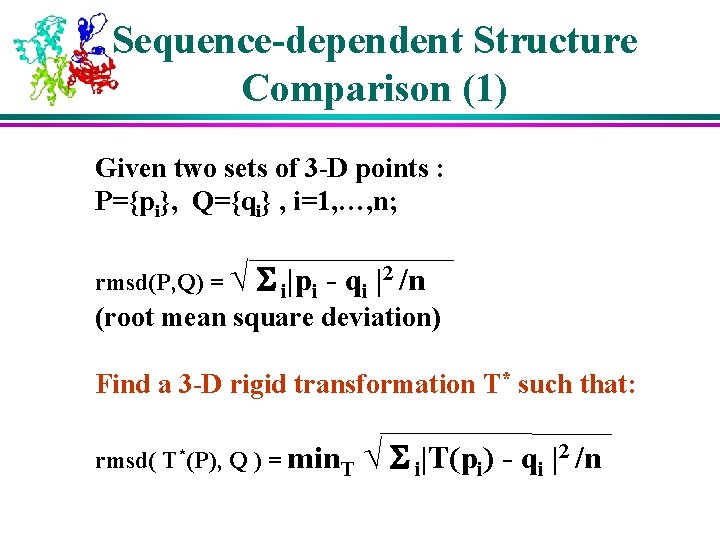
Sequence-dependent Structure Comparison (1) Given two sets of 3 -D points : P={pi}, Q={qi} , i=1, …, n; rmsd(P, Q) = √ S i|pi - qi |2 /n (root mean square deviation) Find a 3 -D rigid transformation T* such that: rmsd( T*(P), Q ) = min. T √ S i|T(pi) - qi |2 /n

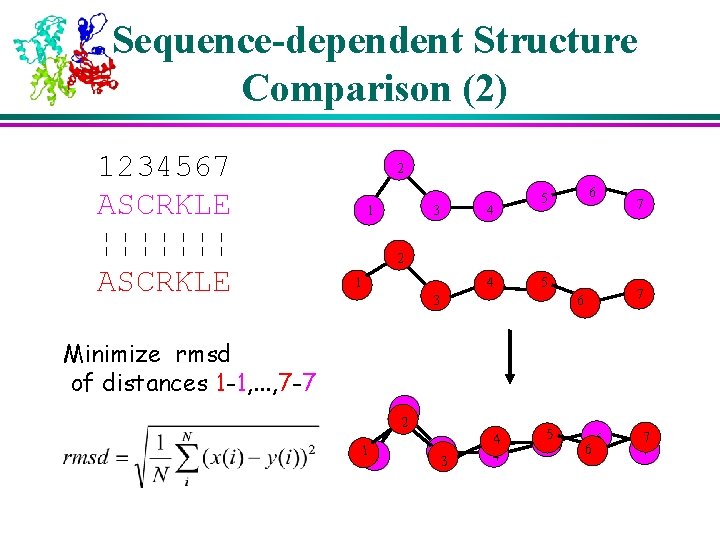
Sequence-dependent Structure Comparison (2) 1234567 ASCRKLE ¦¦¦¦¦¦¦ ASCRKLE 2 1 3 4 6 5 7 2 1 4 5 3 7 6 Minimize rmsd of distances 1 -1, . . . , 7 -7 2 2 11 33 4 4 5 5 6 6 7 7

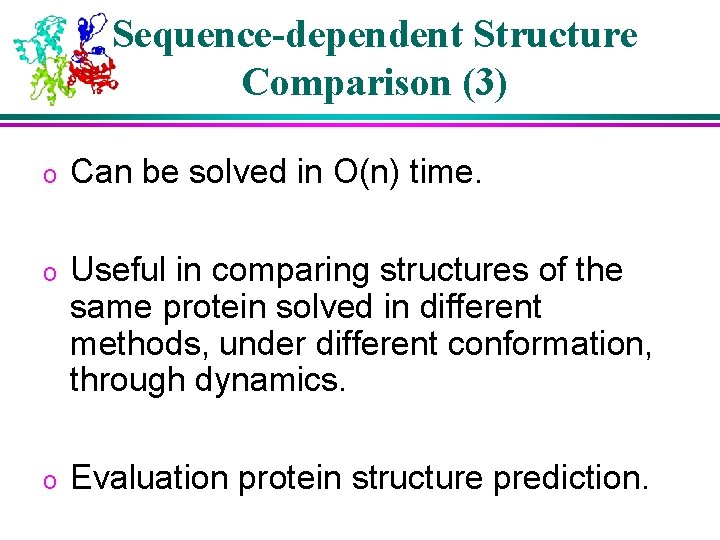
Sequence-dependent Structure Comparison (3) o Can be solved in O(n) time. o Useful in comparing structures of the same protein solved in different methods, under different conformation, through dynamics. o Evaluation protein structure prediction.

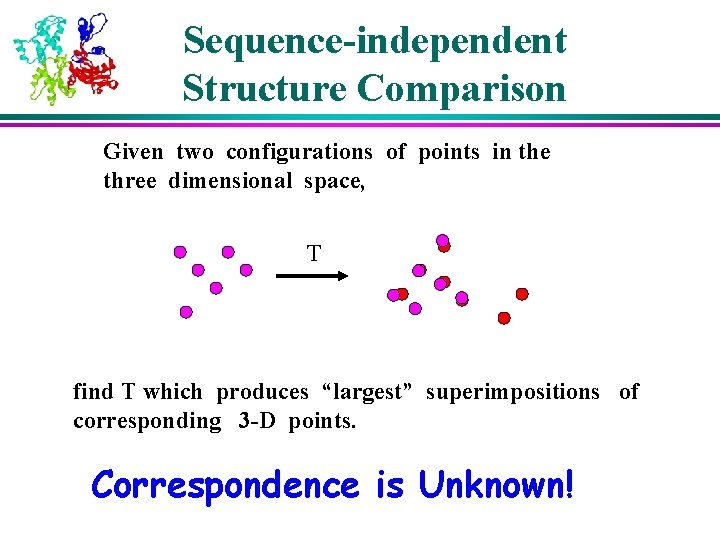
Sequence-independent Structure Comparison Given two configurations of points in the three dimensional space, T find T which produces “largest” superimpositions of corresponding 3 -D points. Correspondence is Unknown!

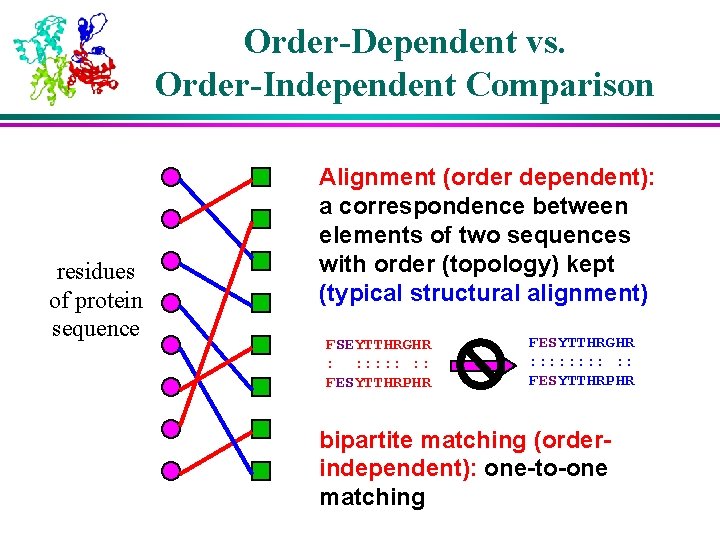
Order-Dependent vs. Order-Independent Comparison residues of protein sequence Alignment (order dependent): a correspondence between elements of two sequences with order (topology) kept (typical structural alignment) FSEYTTHRGHR : : : : FESYTTHRPHR FESYTTHRGHR : : : : : FESYTTHRPHR bipartite matching (orderindependent): one-to-one matching

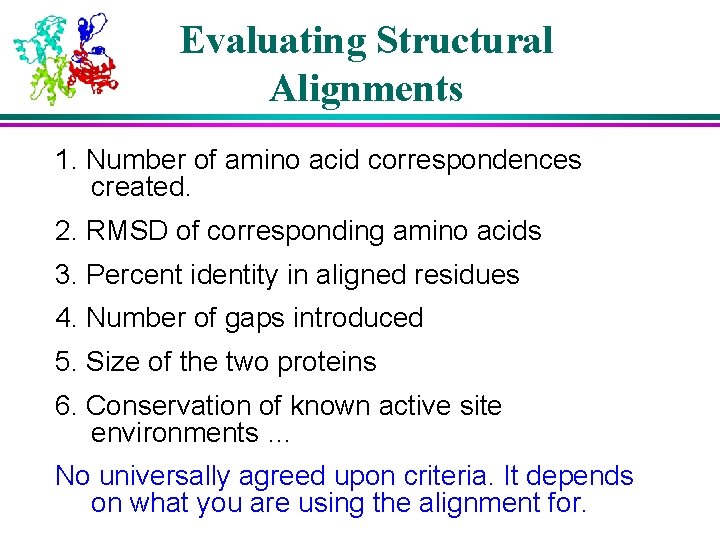
Evaluating Structural Alignments 1. Number of amino acid correspondences created. 2. RMSD of corresponding amino acids 3. Percent identity in aligned residues 4. Number of gaps introduced 5. Size of the two proteins 6. Conservation of known active site environments … No universally agreed upon criteria. It depends on what you are using the alignment for.

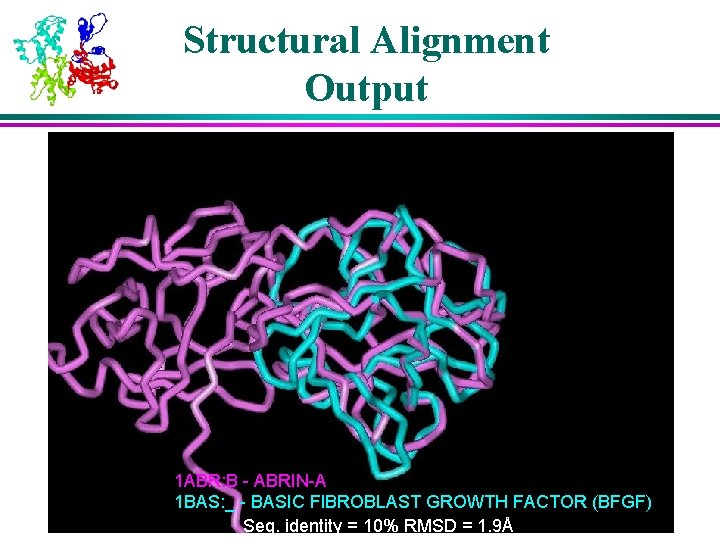
Structural Alignment Output 1 ABR: B - ABRIN-A 1 BAS: _ - BASIC FIBROBLAST GROWTH FACTOR (BFGF) Seq. identity = 10% RMSD = 1. 9Å

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

How to recognize structural similarities 1. By eye (SCOP) 2. Algorithmically o point-based methods use properties of points (distances) to establish correspondence Ø Dynamic programming (SSAP) Ø Distance matrix (DALI) o secondary structure-based methods use vectors representing secondary structures to establish correspondences (LOCK). o Image processing based method.

Structural Comparison Algorithms l Due to the high compute complexity, practical algorithms rely on heuristics l Fully automated structure analysis has not been as successful as analyses with human intervention in taking in to account the biological implications

SSAP l SSAP: Secondary Structure Alignment Program l Incorporates double dynamic programming to produce a structural alignment between two proteins

Basic Ideas of SSAP The similarity between residue i in molecule A and residue k in molecule B is characterised in terms of their structural surroundings This similarity can be quantified into a score, Sik Based on this similarity score and some specified gap penalty, dynamic programming is used to find the optimal structural alignment

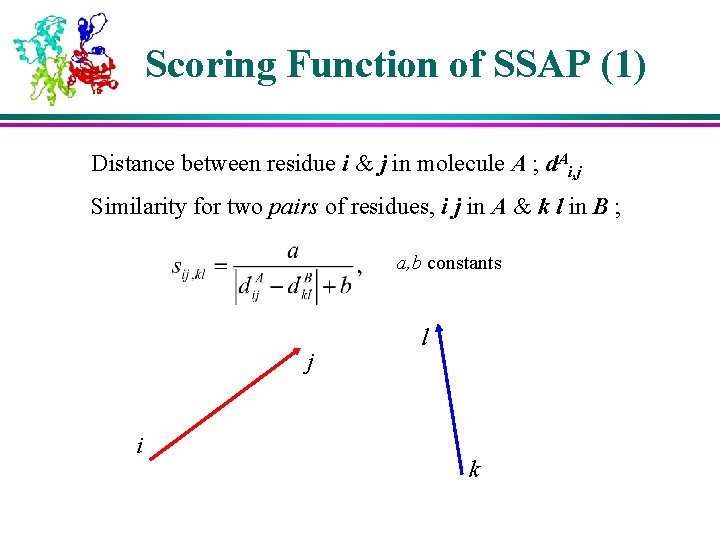
Scoring Function of SSAP (1) Distance between residue i & j in molecule A ; d. Ai, j Similarity for two pairs of residues, i j in A & k l in B ; a, b constants j i l k

Scoring Function of SSAP (2) Similarity between residue i in A and residue k in B ; Si, k is big if the distances from residue i in A to the 2 n nearest neighbours are similar to the corresponding distances around k in B

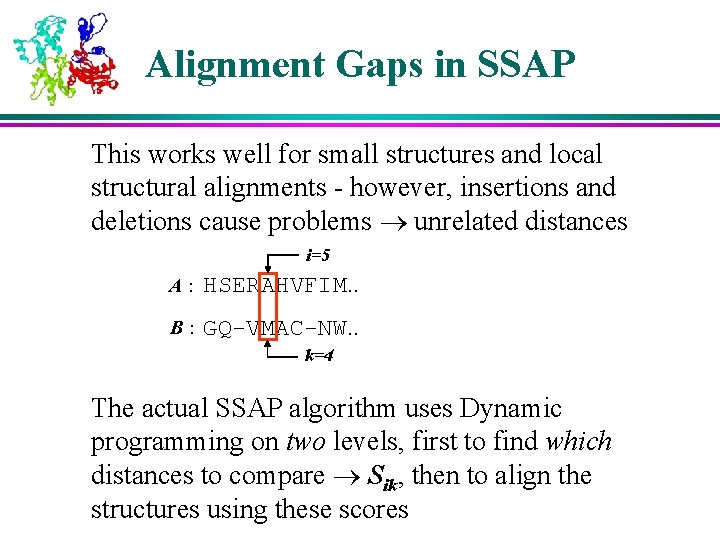
Alignment Gaps in SSAP This works well for small structures and local structural alignments - however, insertions and deletions cause problems unrelated distances i=5 A: HSERAHVFIM. . B: GQ-VMAC-NW. . k=4 The actual SSAP algorithm uses Dynamic programming on two levels, first to find which distances to compare Sik, then to align the structures using these scores

Steps in SSAP (1) l 1) Calculate vectors from C of one amino acid to set of nearby amino acids å Vectors from two separate proteins compared å Difference (expressed as an angle) calculated, and converted to score l 2) Matrix for scores of vector differences from one protein to the next is computed.

Steps in SSAP (2) l 3) Optimal alignment found using global dynamic programming, with a constant gap penalty l 4) Next amino acid residue considered, optimal path to align this amino acid to the second sequence computed

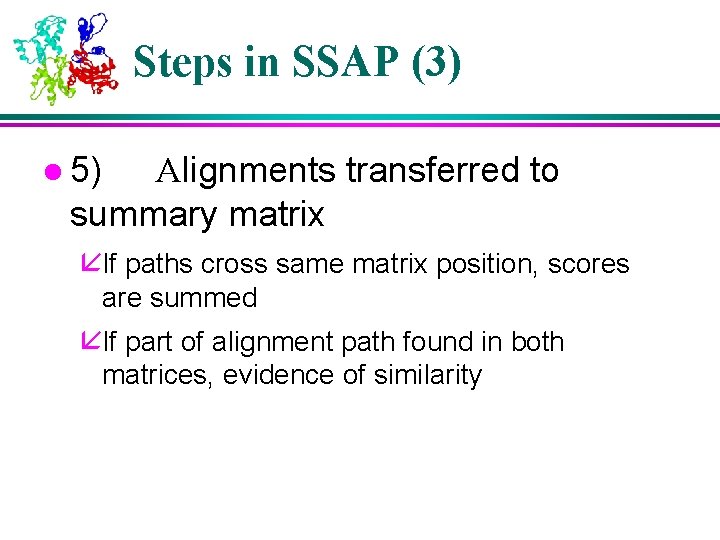
Steps in SSAP (3) l 5) Alignments transferred to summary matrix åIf paths cross same matrix position, scores are summed åIf part of alignment path found in both matrices, evidence of similarity

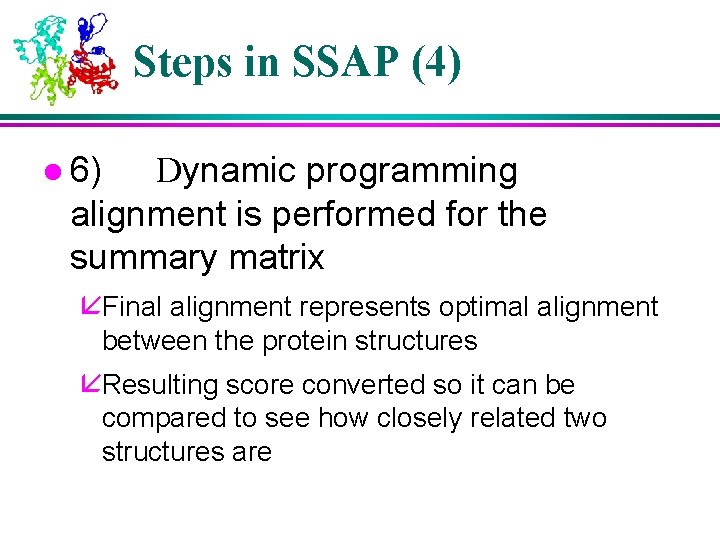
Steps in SSAP (4) l 6) Dynamic programming alignment is performed for the summary matrix åFinal alignment represents optimal alignment between the protein structures åResulting score converted so it can be compared to see how closely related two structures are

Summary of SSAP

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

Distance Matrix Approach l Uses graphical procedure similar to dot plots l Identifies residues that lie most closely together in three-dimensional structure l Two sequences with similar structure can have dot plots superimposed

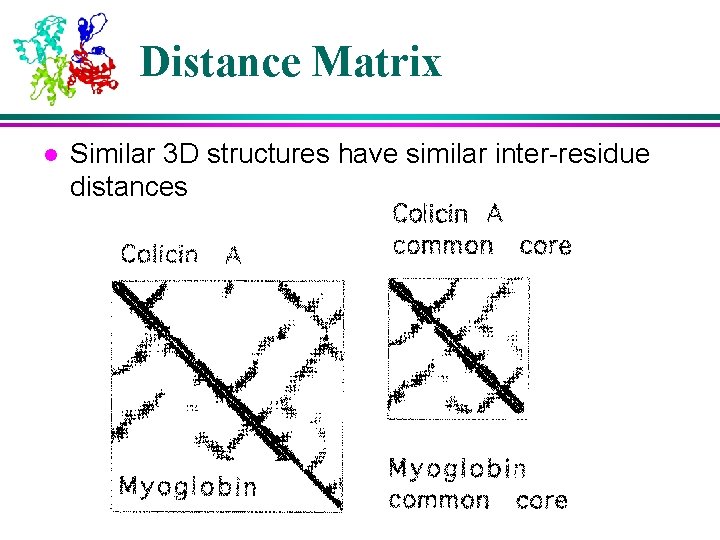
Distance Matrix l Similar 3 D structures have similar inter-residue distances

DALI l Distance Alignment Tool (DALI) l Uses distance matrix method to align protein structures l Assembly step uses Monte Carlo simulation to find submatrices that can be aligned

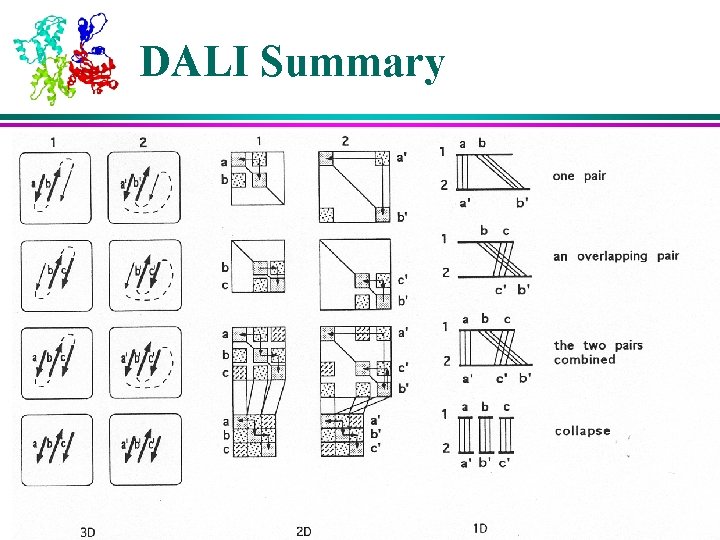
DALI Summary

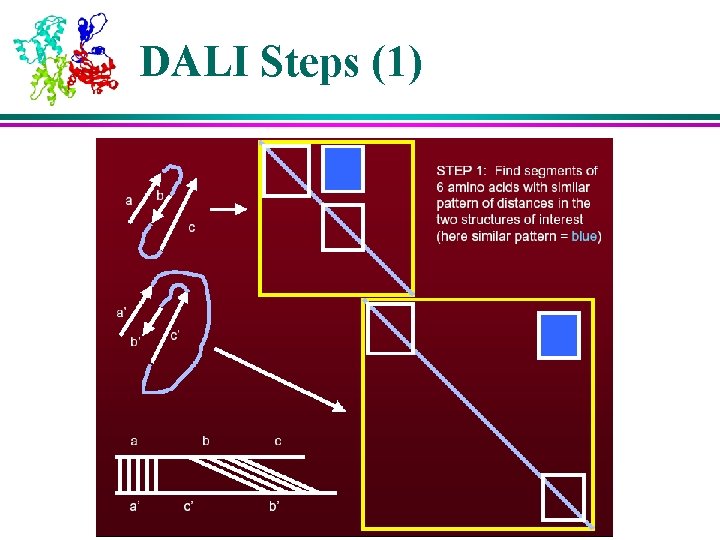
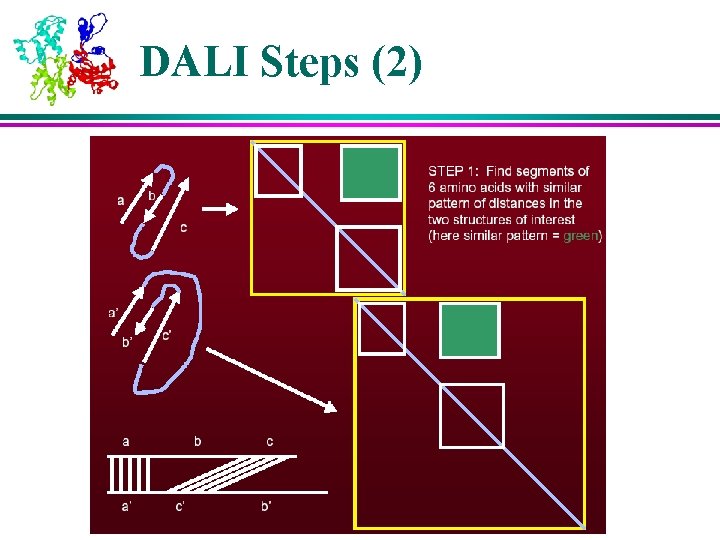
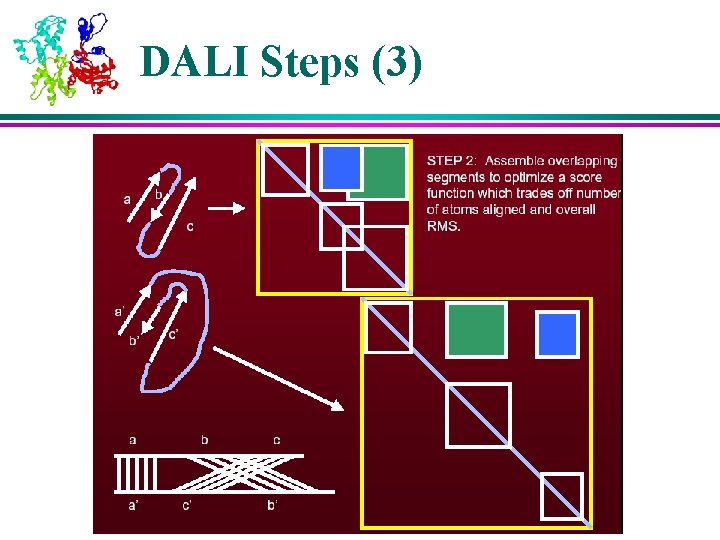
Structural Analysis Algorithms – DALI (1) l DALI is based on distance matrices – 2 D matrices containing all pairwise distances between points of a molecule l Distance matrices of two molecules are compared to find regions of similar patterns of distances, which indicate similarities in their 3 D structure l Key algorithm steps: 1. Divide distance matrices into overlapping sub-matrices of fixed size 2. Search through two matrices (of two molecules) to find similar patterns 3. Assemble matching pairs of sub-matrices in to larger sets to maximize their similarity score

Structural Analysis Algorithms – DALI (2) l l l Assembly of aligned sub-matrices is done using a Monte Carlo optimization is an iterative improvement by a random walk exploration of the search space, with occasional excursions in to non-optimal territory (i. e. occasionally, a move that reduces the overall score is carried out) The occasional non-optimal moves help avoid getting “trapped” in local optima of the score function, improving the chance of finding the global optimum

DALI Steps (1)

DALI Steps (2)

DALI Steps (3)

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

Fast Structural Similarity Search l Compare types and arrangements of secondary structures within two proteins l If elements similarly arranged, threedimensional structures are similar l LOCK, VAST and SARF are programs that use these fast methods

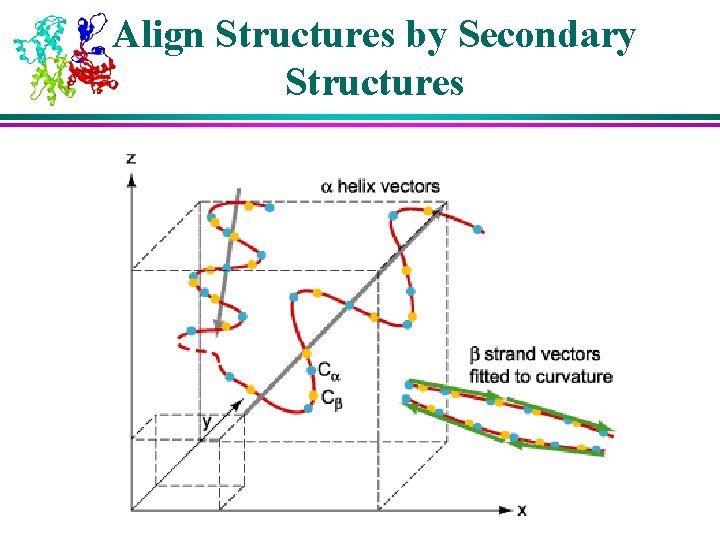
Align Structures by Secondary Structures

Structural Analysis Algorithms – LOCK l Both SSAP and DALI deal only with points (atoms) of the molecules l LOCK uses a hierarchical approach å Larger secondary structures such as helixes and strands are represented using vectors and dealt with first å Individual residues are dealt with afterwards å Assumes large secondary structures provide most stability and function to a protein, and are most likely to be preserved during evolution

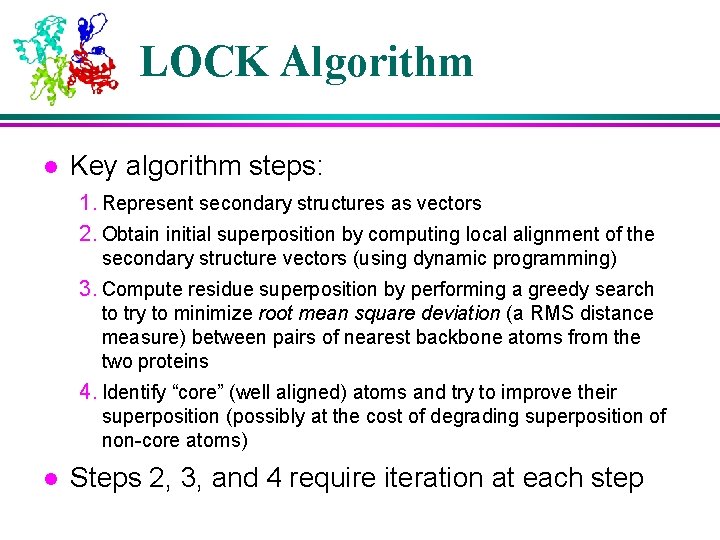
LOCK Algorithm l Key algorithm steps: 1. Represent secondary structures as vectors 2. Obtain initial superposition by computing local alignment of the secondary structure vectors (using dynamic programming) 3. Compute residue superposition by performing a greedy search to try to minimize root mean square deviation (a RMS distance measure) between pairs of nearest backbone atoms from the two proteins 4. Identify “core” (well aligned) atoms and try to improve their superposition (possibly at the cost of degrading superposition of non-core atoms) l Steps 2, 3, and 4 require iteration at each step

Protein. DBS Shyu, Chi, Scott, Xu. Nucleic Acid Research. 32, W 572 - CW 575, 2004

Comparison between different methods l CATH å Fully automated å SSAP l SCOP å Based on subjective interpretation of evolutionary history of proteins l FSSP å DALI l Agreement between CATH and SCOP may be at most 60%. å FSSP vs CATH 40% å FSSP vs SCOP 60%

Lecture Outline l Why structural alignment l Technical definition l SSAP l DALI l Fast search l Protein families

Structure Families (1) Homologous family: evolutionarily related with a significant sequence identity; Superfamily: different families whose structural and functional features suggest common evolutionary origin; Fold: different superfamilies having same major secondary structures in same arrangement and with same topological connections (energetics favoring certain packing arrangements); Class: secondary structure composition.

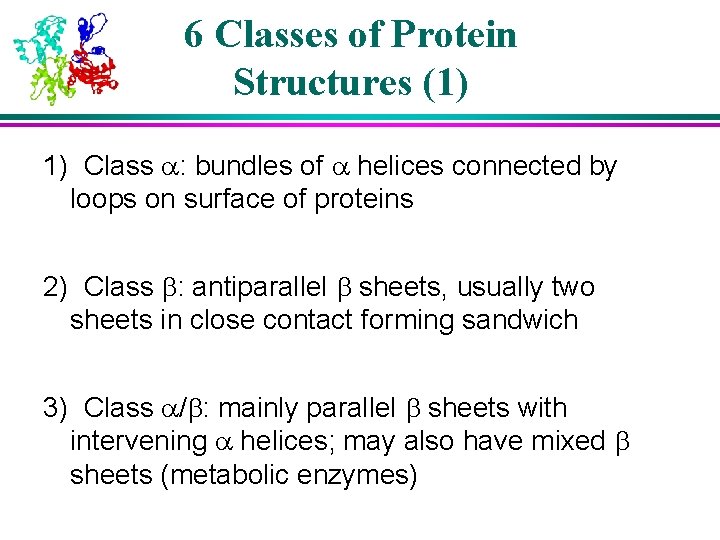
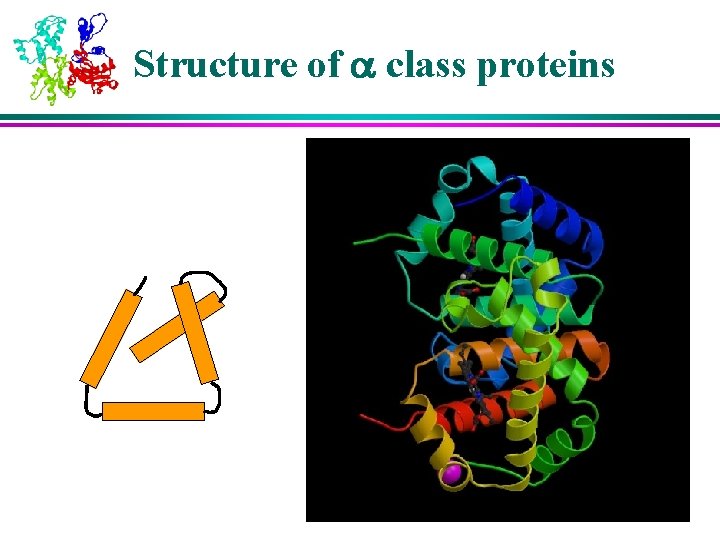
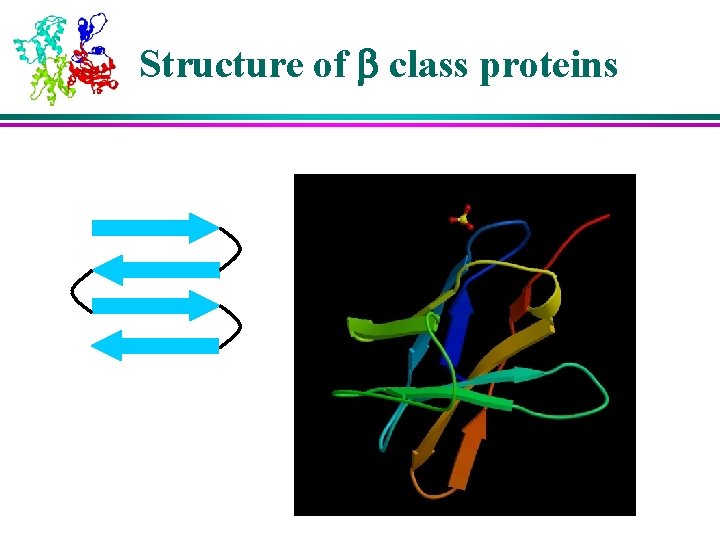
6 Classes of Protein Structures (1) 1) Class : bundles of helices connected by loops on surface of proteins 2) Class : antiparallel sheets, usually two sheets in close contact forming sandwich 3) Class / : mainly parallel sheets with intervening helices; may also have mixed sheets (metabolic enzymes)

6 Classes of Protein Structures (2) 4) Class + : mainly segregated helices and anti-parallel sheets 5) Multi-domain ( and ) proteins more than one of the above four domains 6) Membrane and cell-surface proteins and peptides excluding proteins of the immune system

Structure of a class proteins

Structure of b class proteins

Structure of a/b class proteins

Structure of a+b class proteins

20 most frequent common domains (folds)

Reading Assignments l Suggested reading: å Contemporary approaches to protein structure classification. Mark B. Swindells, et al. Bio. Essay. Volume 20, Issue 11, 1998, Pages: 884 -891 l Optional reading: å The structural alignment between two proteins: Is there a unique answer? Adam Godzik, Protein Science (1996), 5 13251338 å Protein Structure Similarities. Patrice Koehl, Current Opinions in Structural Biology (2001), 11 348 -353

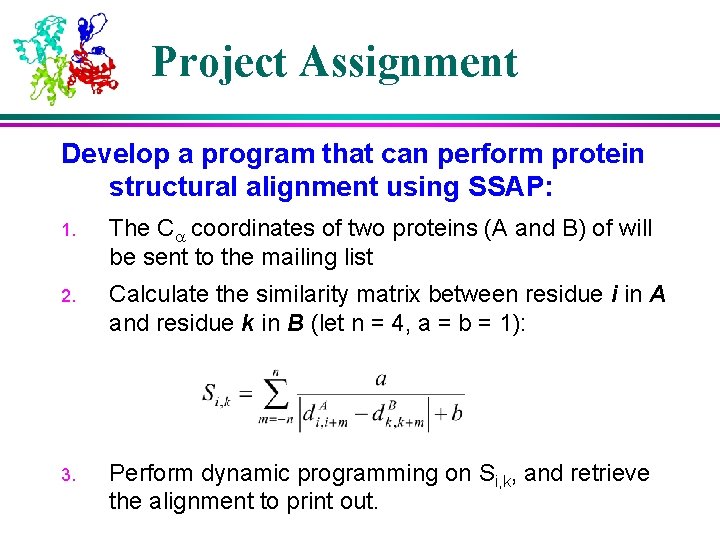
Project Assignment Develop a program that can perform protein structural alignment using SSAP: 1. The C coordinates of two proteins (A and B) of will be sent to the mailing list 2. Calculate the similarity matrix between residue i in A and residue k in B (let n = 4, a = b = 1): 3. Perform dynamic programming on Si, k, and retrieve the alignment to print out.

Project Phase III Report l l l Due on 11/17, send me through email Write on top of Phase II report. 7 -30 Pages As a draft of the final report Free style in writing (use 11 pt font or larger) Present key results å Software implementation å Benchmark (computing time) å Computational data å Interpret the meaning of the data