Protein Tertiary Structure Prediction Dong Xu Computer Science









- Slides: 57

Protein Tertiary Structure Prediction Dong Xu Computer Science Department 271 C Life Sciences Center 1201 East Rollins Road University of Missouri-Columbia, MO 65211 -2060 E-mail: xudong@missouri. edu 573 -882 -7064 (O) http: //digbio. missouri. edu

Lecture Outline l Introduction to protein structure prediction l Concept of threading l Template l Scoring function l Alignment l Confidence Assessment l Mini-threading

Protein Structure Prediction Structure: Traditional experimental methods: X-Ray or NMR to solve structures; generate a few structures per day worldwide cannot keep pace for new protein sequences Strong demand for structure prediction: more than 30, 000 human genes; 10, 000 genomes will be sequenced in the next 10 years. Unsolved problem after efforts of two decades.

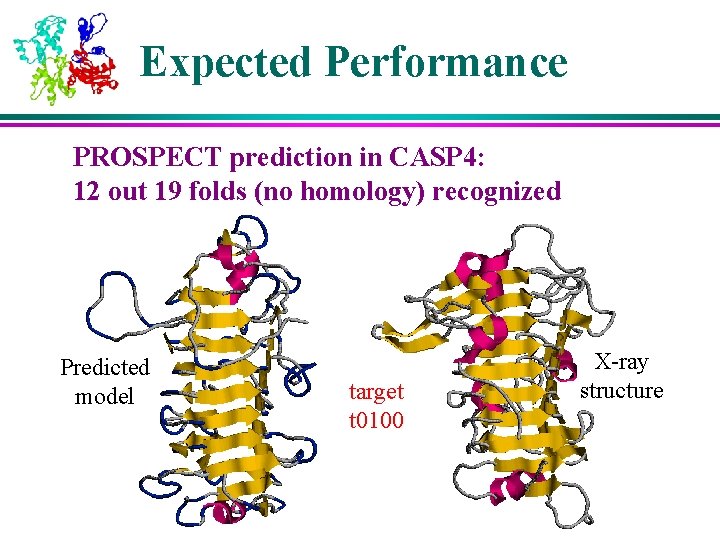
Expected Performance PROSPECT prediction in CASP 4: 12 out 19 folds (no homology) recognized Predicted model target t 0100 X-ray structure

Evaluating Structure Prediction 1. By eye 2. Number of amino acid predicted? 3. RMSD of predicted residues? 4. Match between contact maps? 5. Fold recognition? 6. Evolutionary or functional relationship? No universally agreed upon criteria.

Ab initio Structure Prediction Ø An energy function to describe the protein o bond energy o bond angle energy o dihedral angel energy o van der Waals energy o electrostatic energy Ø Minimize the function and obtain the structure. Not practical in general o Computationally too expensive o Accuracy is poor Ø

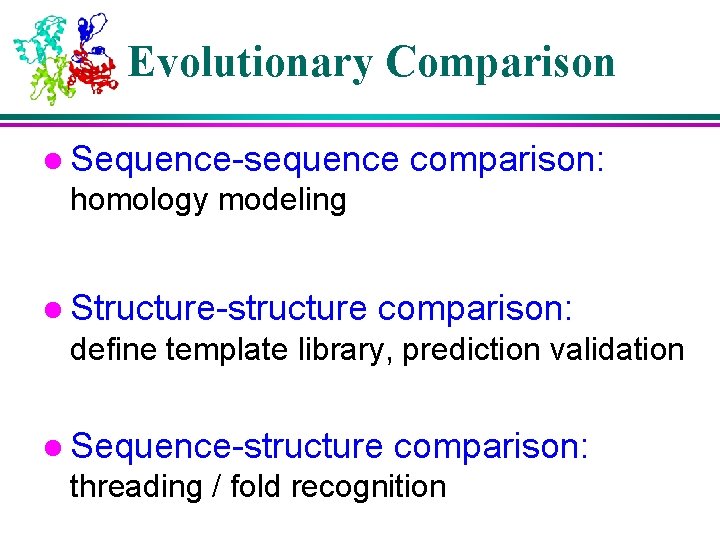
Template-Based Prediction Structure is better conserved than sequence Structure can adopt a wide range of mutations. Physical forces favor certain structures. Number of fold is limited. Currently ~700 Total: 1, 000 ~10, 000 TIM barrel

Evolutionary Comparison l Sequence-sequence comparison: homology modeling l Structure-structure comparison: define template library, prediction validation l Sequence-structure comparison: threading / fold recognition

Scope of the Problem Ø ~90% of new globular proteins share similar folds with known structures, implying the general applicability of comparative modeling methods for structure prediction Ø general applicability of template-based modeling methods for structure prediction (currently 60 -70% of new proteins, and this number is growing as more structures being solved) Ø NIH Structural Genomics Initiative plans to experimentally solve ~10, 000 “unique” structures and predict the rest using computational methods

Homology Modeling l Sequence is aligned with sequence of known structure, usually sharing sequence identity of 30% or more. l Superimpose sequence onto the template, replacing equivalent sidechain atoms where necessary. l Refine the model by minimizing an energy function

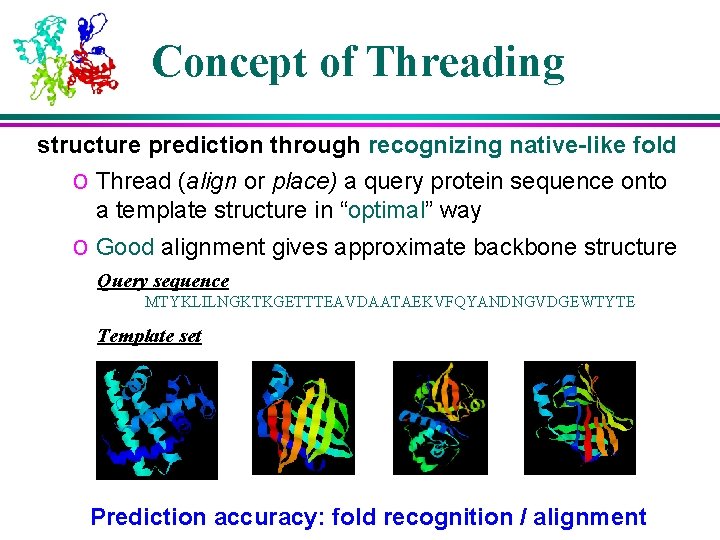
Concept of Threading structure prediction through recognizing native-like fold o Thread (align or place) a query protein sequence onto a template structure in “optimal” way o Good alignment gives approximate backbone structure Query sequence MTYKLILNGKTKGETTTEAVDAATAEKVFQYANDNGVDGEWTYTE Template set Prediction accuracy: fold recognition / alignment

Application of Threading l Predict structure l Identify distant homologues of protein families l Predict function of protein with low degree of sequence similarity with other proteins

4 Components of Threading Ø Template Ø Scoring library function Ø Alignment Ø Confidence assessment

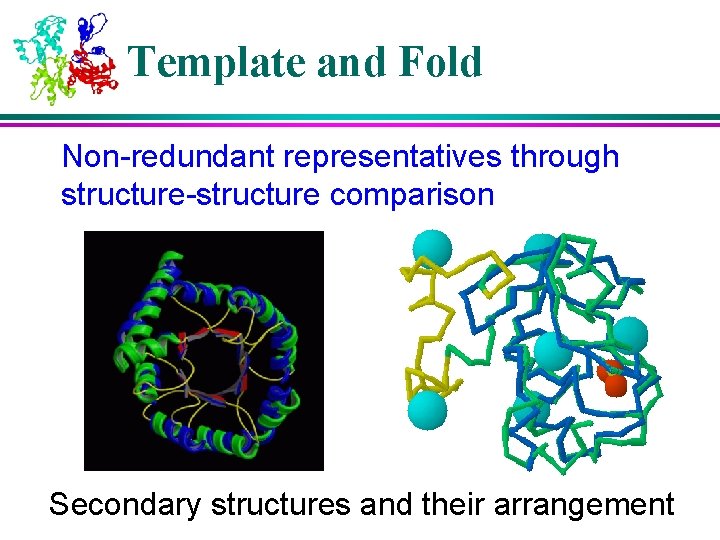
Template and Fold Non-redundant representatives through structure-structure comparison Secondary structures and their arrangement

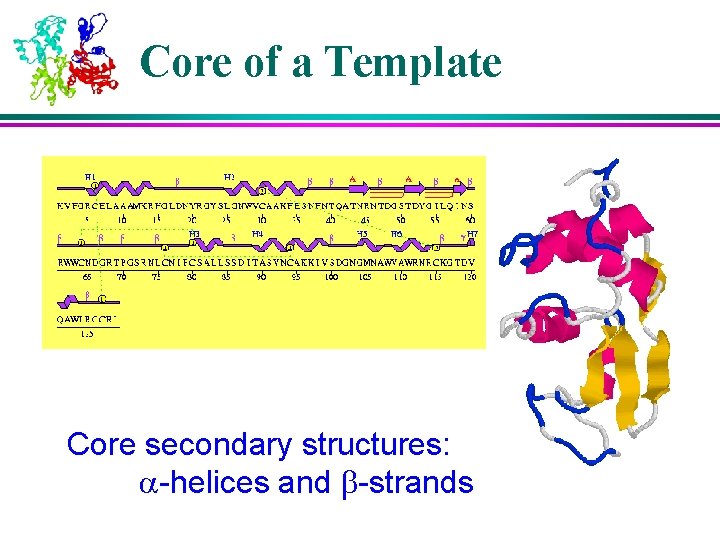
Core of a Template Core secondary structures: a-helices and b-strands

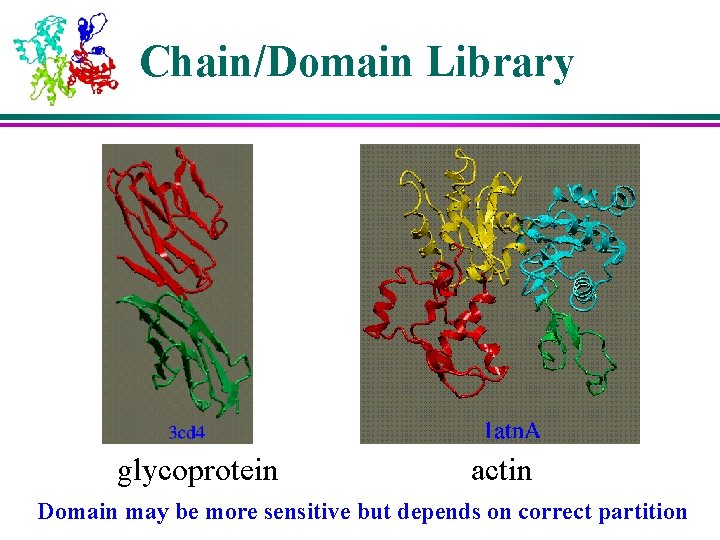
Chain/Domain Library glycoprotein actin Domain may be more sensitive but depends on correct partition

Structure Families SCOP: http: //scop. mrc-lmb. cam. ac. uk/scop/ (domains, good annotation) CATH: http: //www. biochem. ucl. ac. uk/bsm/cath/ CE: http: //cl. sdsc. edu/ce. html Dali Domain Dictionary: http: //columba. ebi. ac. uk: 8765/holm/ddd 2. cgi FSSP: http: //www 2. ebi. ac. uk/dali/fssp/ (chains, updated weekly) HOMSTRAD: http: //www-cryst. bioc. cam. ac. uk/~homstrad/ HSSP: http: //swift. embl-heidelberg. de/hssp/

Hierarchy of Templates Homologous family: evolutionarily related with a significant sequence identity -- 1827 in SCOP Superfamily: different families whose structural and functional features suggest common evolutionary origin --1073 in SCOP (good tradeoff for accuracy/computing) Fold: different superfamilies having same major secondary structures in same arrangement and with same topological connections (energetics favoring certain packing arrangements); -- 686 out of 39, 893 in SCOP Class: secondary structure composition.

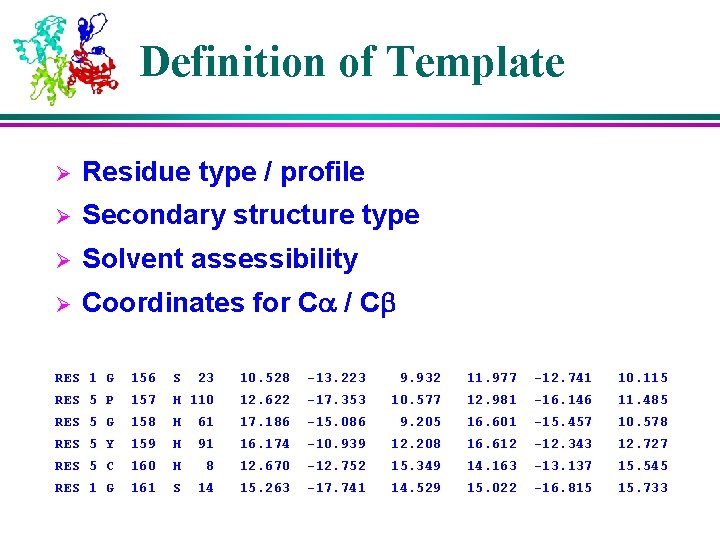
Definition of Template Ø Residue type / profile Ø Secondary structure type Ø Solvent assessibility Ø Coordinates for Ca / Cb RES 1 G 156 S 23 10. 528 -13. 223 9. 932 11. 977 -12. 741 10. 115 RES 5 P 157 H 110 12. 622 -17. 353 10. 577 12. 981 -16. 146 11. 485 RES 5 G 158 H 61 17. 186 -15. 086 9. 205 16. 601 -15. 457 10. 578 RES 5 Y 159 H 91 16. 174 -10. 939 12. 208 16. 612 -12. 343 12. 727 RES 5 C 160 H 8 12. 670 -12. 752 15. 349 14. 163 -13. 137 15. 545 RES 1 G 161 S 14 15. 263 -17. 741 14. 529 15. 022 -16. 815 15. 733

Scoring Function Ø Physical energy function: two sensitive o bond energy o van der Waals energy o electrostatic energy… Ø Knowledge-based scoring function (derived from known sequence/structure) Ø Two types of functions correlate each other

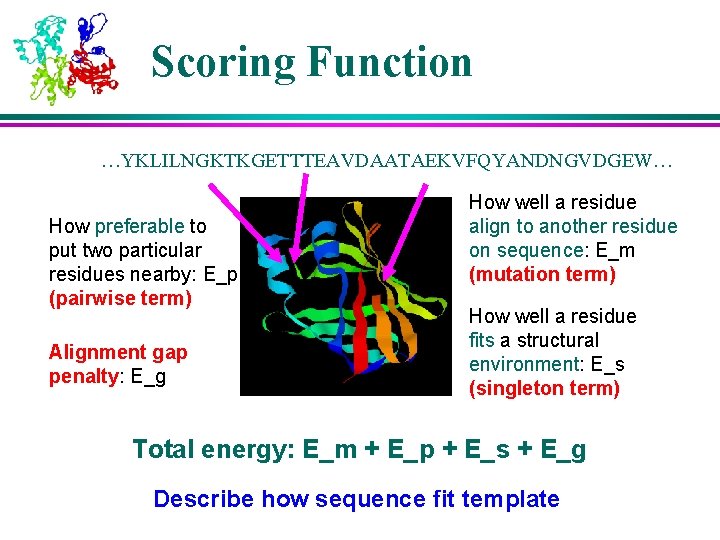
Scoring Function …YKLILNGKTKGETTTEAVDAATAEKVFQYANDNGVDGEW… How preferable to put two particular residues nearby: E_p (pairwise term) Alignment gap penalty: E_g How well a residue align to another residue on sequence: E_m (mutation term) How well a residue fits a structural environment: E_s (singleton term) Total energy: E_m + E_p + E_s + E_g Describe how sequence fit template

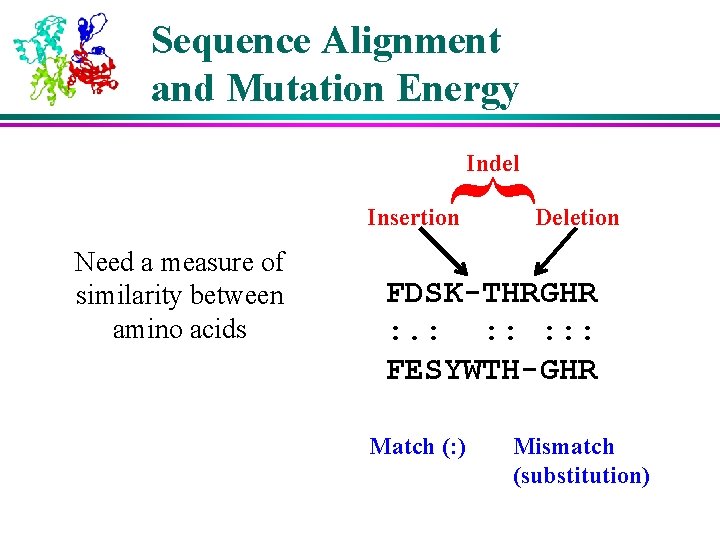
Sequence Alignment and Mutation Energy { Indel Insertion Need a measure of similarity between amino acids Deletion FDSK-THRGHR : . : : : FESYWTH-GHR Match (: ) Mismatch (substitution)

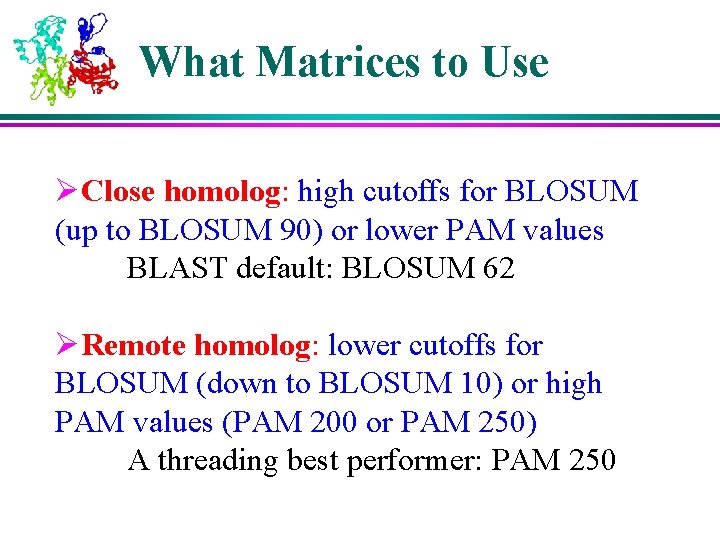
What Matrices to Use ØClose homolog: high cutoffs for BLOSUM (up to BLOSUM 90) or lower PAM values BLAST default: BLOSUM 62 ØRemote homolog: lower cutoffs for BLOSUM (down to BLOSUM 10) or high PAM values (PAM 200 or PAM 250) A threading best performer: PAM 250

Structure-based score l Structure provides additional (independent) information l Free energy (score) vs. distribution in thermal equilibrium (known protein structures) l Preference model of characteristics l Derive parameters for structure-based score using a non-redundant protein structure database (FSSP)

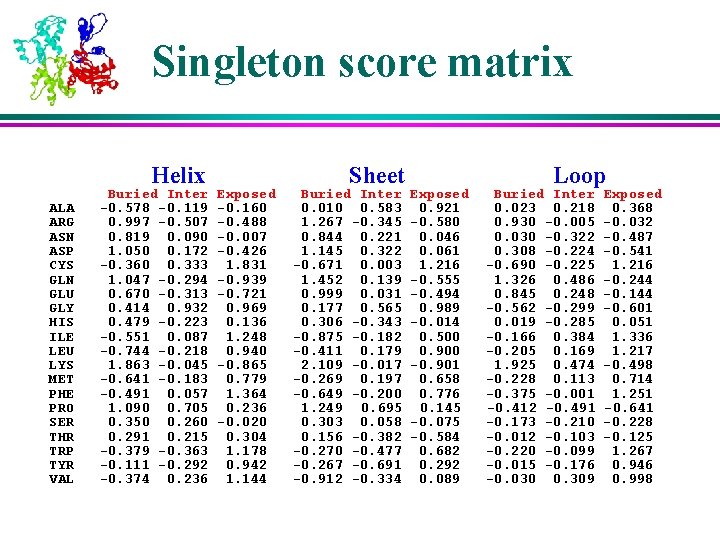
Singleton score l l A single residue’s preference in a specific structural environments. å secondary structure å solvent accessibility Compare actual occurrence against its “expected value” by chance

Singleton score matrix Helix ALA ARG ASN ASP CYS GLN GLU GLY HIS ILE LEU LYS MET PHE PRO SER THR TRP TYR VAL Buried Inter -0. 578 -0. 119 0. 997 -0. 507 0. 819 0. 090 1. 050 0. 172 -0. 360 0. 333 1. 047 -0. 294 0. 670 -0. 313 0. 414 0. 932 0. 479 -0. 223 -0. 551 0. 087 -0. 744 -0. 218 1. 863 -0. 045 -0. 641 -0. 183 -0. 491 0. 057 1. 090 0. 705 0. 350 0. 260 0. 291 0. 215 -0. 379 -0. 363 -0. 111 -0. 292 -0. 374 0. 236 Exposed -0. 160 -0. 488 -0. 007 -0. 426 1. 831 -0. 939 -0. 721 0. 969 0. 136 1. 248 0. 940 -0. 865 0. 779 1. 364 0. 236 -0. 020 0. 304 1. 178 0. 942 1. 144 Sheet Buried Inter 0. 010 0. 583 1. 267 -0. 345 0. 844 0. 221 1. 145 0. 322 -0. 671 0. 003 1. 452 0. 139 0. 999 0. 031 0. 177 0. 565 0. 306 -0. 343 -0. 875 -0. 182 -0. 411 0. 179 2. 109 -0. 017 -0. 269 0. 197 -0. 649 -0. 200 1. 249 0. 695 0. 303 0. 058 0. 156 -0. 382 -0. 270 -0. 477 -0. 267 -0. 691 -0. 912 -0. 334 Exposed 0. 921 -0. 580 0. 046 0. 061 1. 216 -0. 555 -0. 494 0. 989 -0. 014 0. 500 0. 900 -0. 901 0. 658 0. 776 0. 145 -0. 075 -0. 584 0. 682 0. 292 0. 089 Loop Buried Inter 0. 023 0. 218 0. 930 -0. 005 0. 030 -0. 322 0. 308 -0. 224 -0. 690 -0. 225 1. 326 0. 486 0. 845 0. 248 -0. 562 -0. 299 0. 019 -0. 285 -0. 166 0. 384 -0. 205 0. 169 1. 925 0. 474 -0. 228 0. 113 -0. 375 -0. 001 -0. 412 -0. 491 -0. 173 -0. 210 -0. 012 -0. 103 -0. 220 -0. 099 -0. 015 -0. 176 -0. 030 0. 309 Exposed 0. 368 -0. 032 -0. 487 -0. 541 1. 216 -0. 244 -0. 144 -0. 601 0. 051 1. 336 1. 217 -0. 498 0. 714 1. 251 -0. 641 -0. 228 -0. 125 1. 267 0. 946 0. 998

Side Chain Properties Neutral Hydrophobic Alanine Valine Leucine Isoleucine Proline Tryptophane Phenylalanine Methionine Acidic Aspartic Acid Glutamic Acid Neutral Polar Glycine Serine Threonine Tyrosine Cysteine Asparagine Glutamine Basic Lysine Arginine (Histidine)

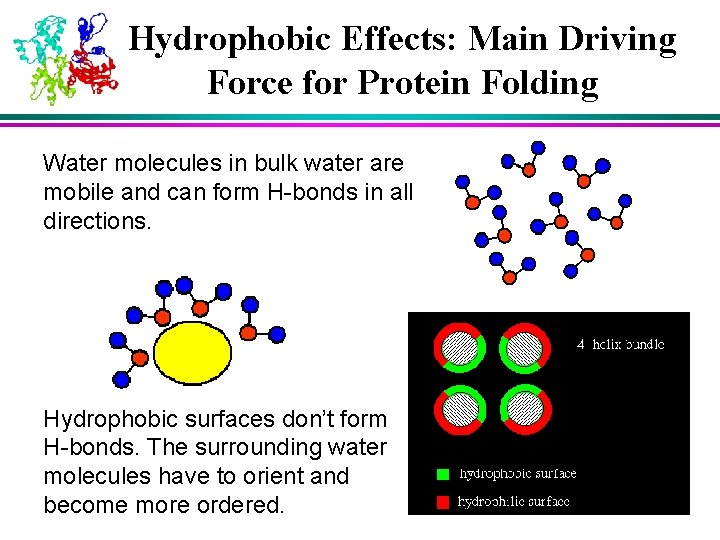
Hydrophobic Effects: Main Driving Force for Protein Folding Water molecules in bulk water are mobile and can form H-bonds in all directions. Hydrophobic surfaces don’t form H-bonds. The surrounding water molecules have to orient and become more ordered.

Using predicted secondary structure for singleton score l More reliable than single amino acid’s preference l Use probabilities of the three secondary structure states (a-helices, b-strand, and loop) l May have a risk of over-dependence on secondary structure prediction

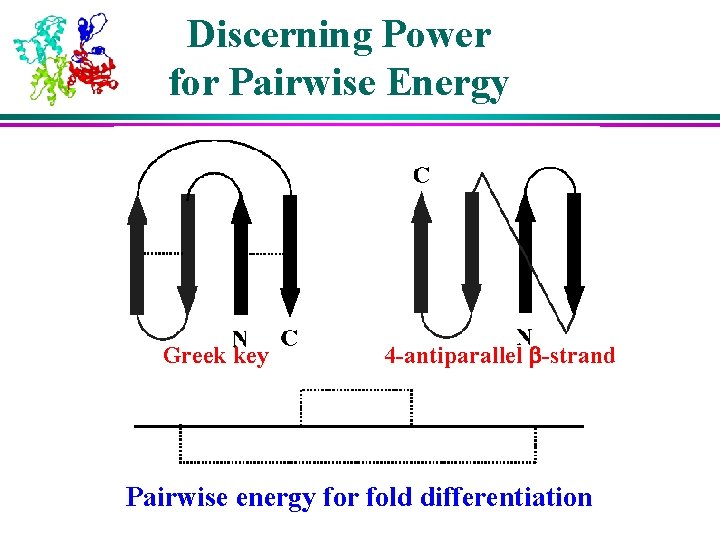
Discerning Power for Pairwise Energy Greek key 4 -antiparallel b-strand Pairwise energy for fold differentiation

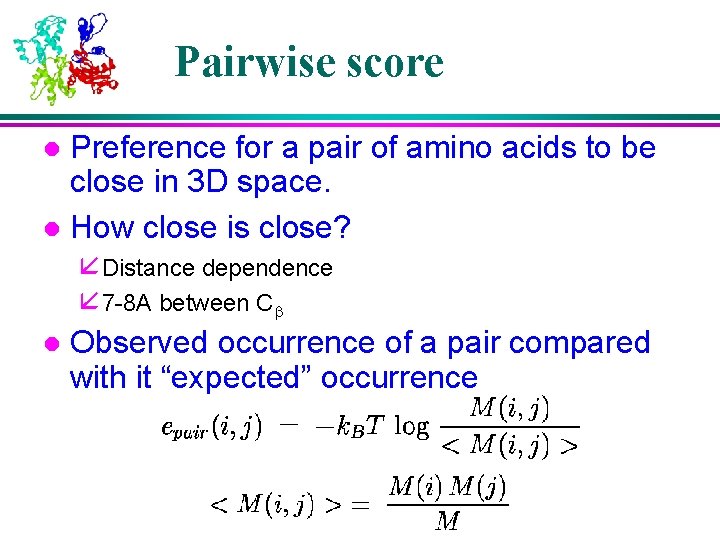
Pairwise score Preference for a pair of amino acids to be close in 3 D space. l How close is close? l å Distance dependence å 7 -8 A between Cb l Observed occurrence of a pair compared with it “expected” occurrence

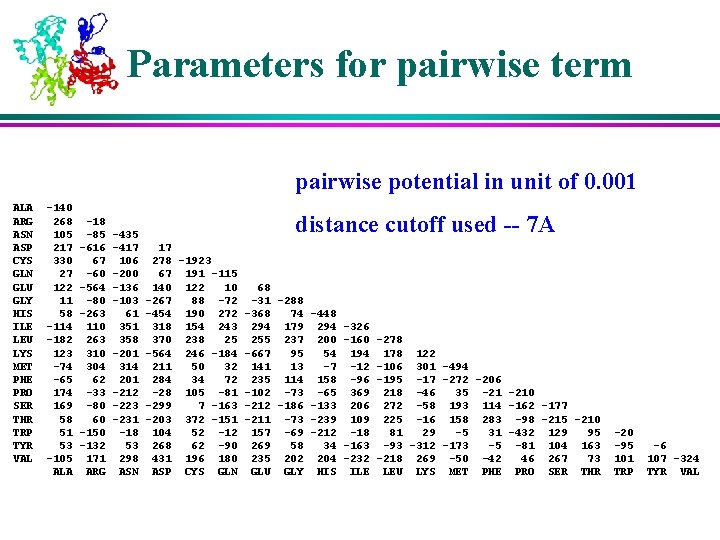
Parameters for pairwise term pairwise potential in unit of 0. 001 ALA ARG ASN ASP CYS GLN GLU GLY HIS ILE LEU LYS MET PHE PRO SER THR TRP TYR VAL -140 268 105 217 330 27 122 11 58 -114 -182 123 -74 -65 174 169 58 51 53 -105 ALA -18 -85 -616 67 -60 -564 -80 -263 110 263 310 304 62 -33 -80 60 -150 -132 171 ARG -435 -417 106 -200 -136 -103 61 358 -201 314 201 -212 -223 -231 -18 53 298 ASN distance cutoff used -- 7 A 17 278 -1923 67 191 -115 140 122 10 68 -267 88 -72 -31 -288 -454 190 272 -368 74 -448 318 154 243 294 179 294 -326 370 238 25 255 237 200 -160 -278 -564 246 -184 -667 95 54 194 178 122 211 50 32 141 13 -7 -12 -106 301 -494 284 34 72 235 114 158 -96 -195 -17 -272 -206 -28 105 -81 -102 -73 -65 369 218 -46 35 -210 -299 7 -163 -212 -186 -133 206 272 -58 193 114 -162 -177 -203 372 -151 -211 -73 -239 109 225 -16 158 283 -98 -215 -210 104 52 -12 157 -69 -212 -18 81 29 -5 31 -432 129 95 268 62 -90 269 58 34 -163 -93 -312 -173 -5 -81 104 163 431 196 180 235 202 204 -232 -218 269 -50 -42 46 267 73 ASP CYS GLN GLU GLY HIS ILE LEU LYS MET PHE PRO SER THR -20 -95 101 TRP -6 107 -324 TYR VAL

Optimizing Weights between different terms l Against threading performance l Place more weight on cores? l Different for different classes (superfamily vs. fold family) l Pure artificial scoring function based on threading performance

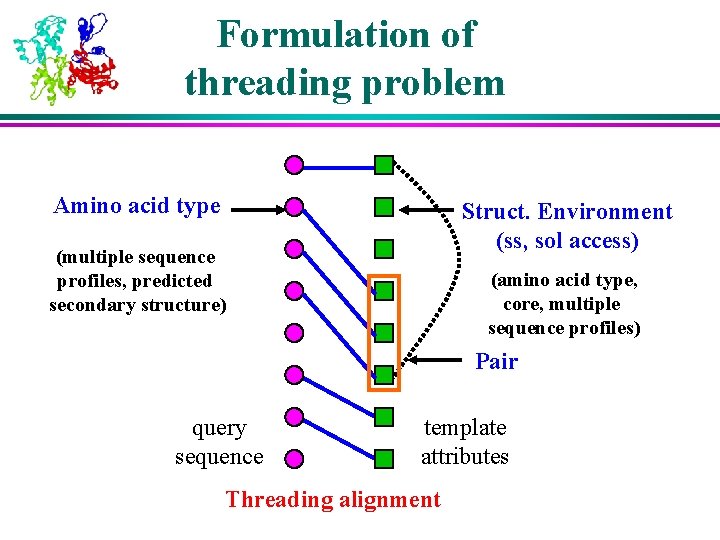
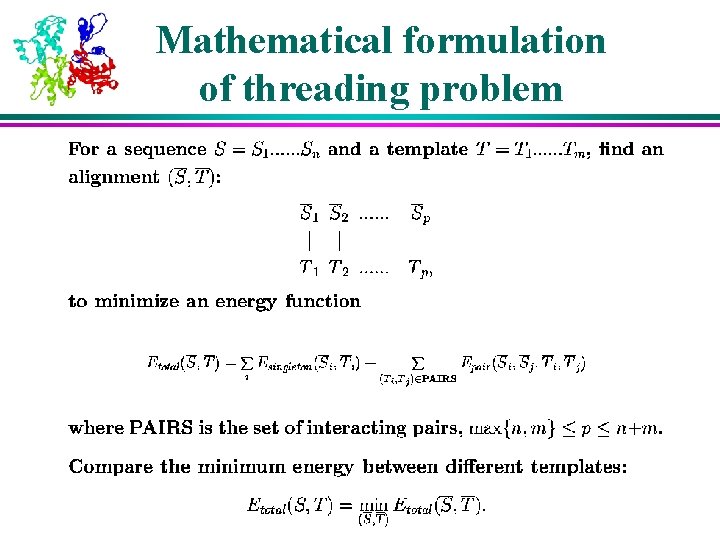
Formulation of threading problem Amino acid type Struct. Environment (ss, sol access) (multiple sequence profiles, predicted secondary structure) (amino acid type, core, multiple sequence profiles) Pair query sequence template attributes Threading alignment

Mathematical formulation of threading problem

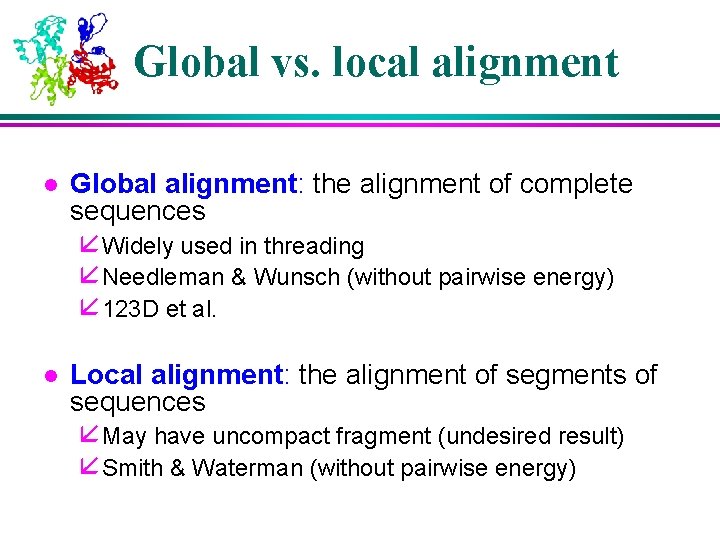
Global vs. local alignment l Global alignment: the alignment of complete sequences å Widely used in threading å Needleman & Wunsch (without pairwise energy) å 123 D et al. l Local alignment: the alignment of segments of sequences å May have uncompact fragment (undesired result) å Smith & Waterman (without pairwise energy)

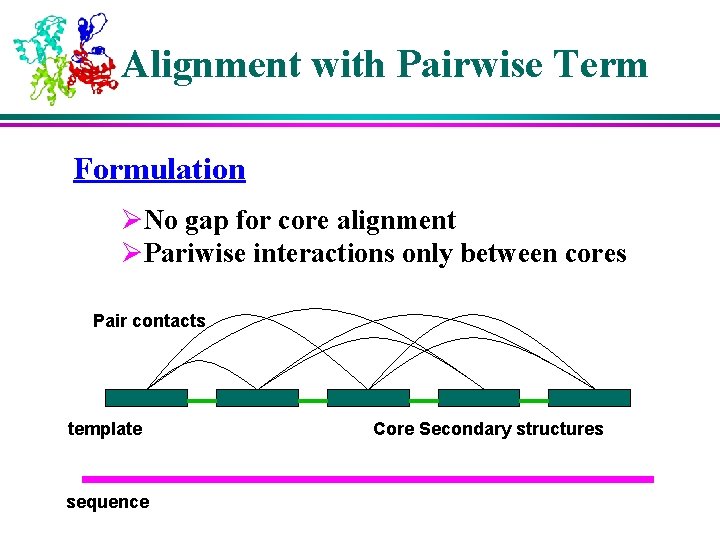
Alignment with Pairwise Term Formulation ØNo gap for core alignment ØPariwise interactions only between cores Pair contacts template sequence Core Secondary structures

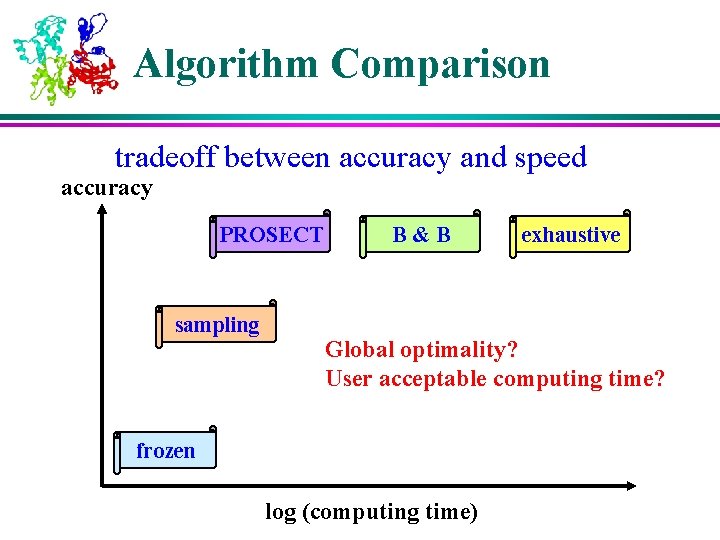
Algorithm Comparison tradeoff between accuracy and speed accuracy PROSECT sampling B&B exhaustive Global optimality? User acceptable computing time? frozen log (computing time)

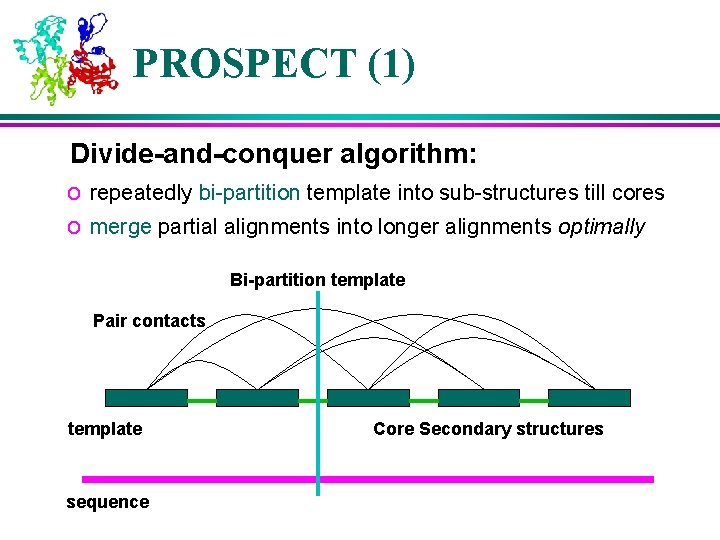
PROSPECT (1) Divide-and-conquer algorithm: o repeatedly bi-partition template into sub-structures till cores o merge partial alignments into longer alignments optimally Bi-partition template Pair contacts template sequence Core Secondary structures

PROSPECT (2) Partition a template to minimize computing time

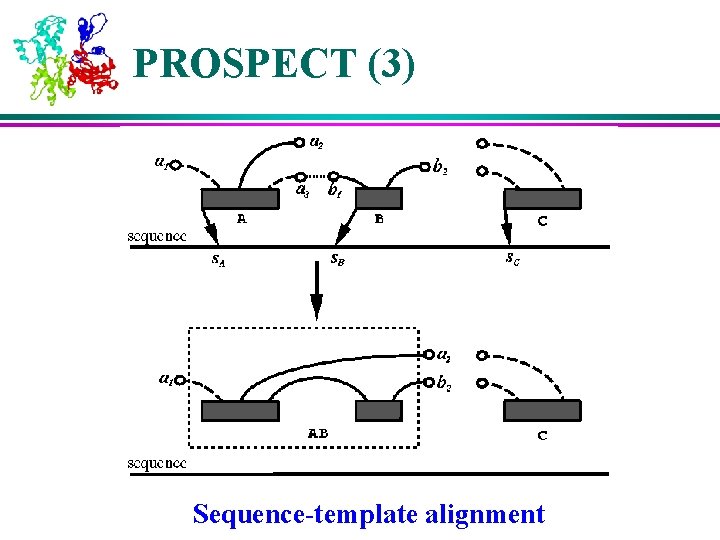
PROSPECT (3) Sequence-template alignment

PROSPECT (4) Computational complexity: mn + Mn. CNC m: length of template (~300) n: length of sequence (~300) M: number of cores in template (~20) N: maximum allowed gap for loop alignment (20) C: topological complexity (<6)

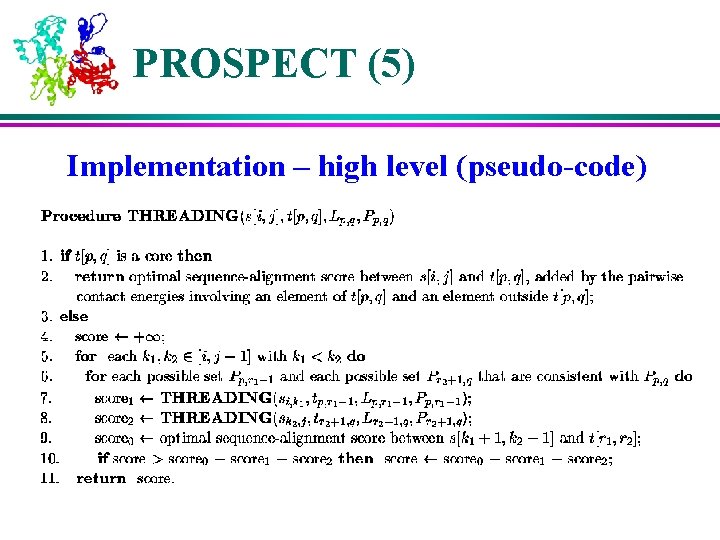
PROSPECT (5) Implementation – high level (pseudo-code)

Confidence Assessment of Threading Results l. A confidence score is need to normalized raw threading score l Z-score through random shuffling z-score = l Using score – ave_score standard_dev known correct pairs for training (neural networks / SVM)

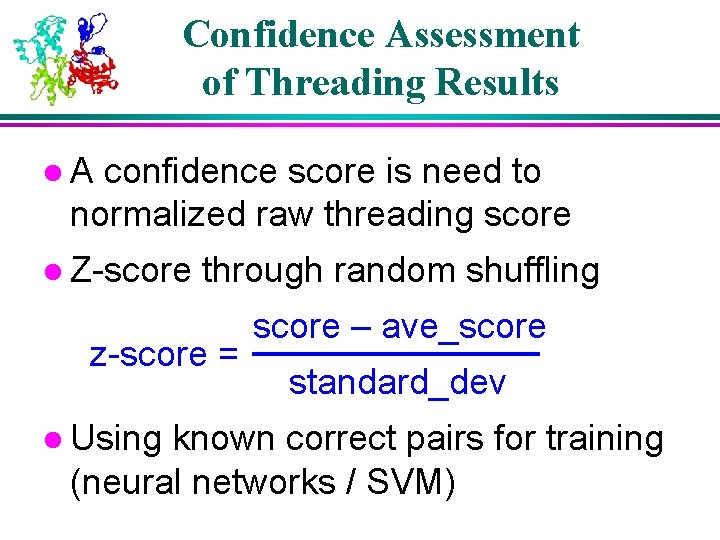
Threading Score Distribution

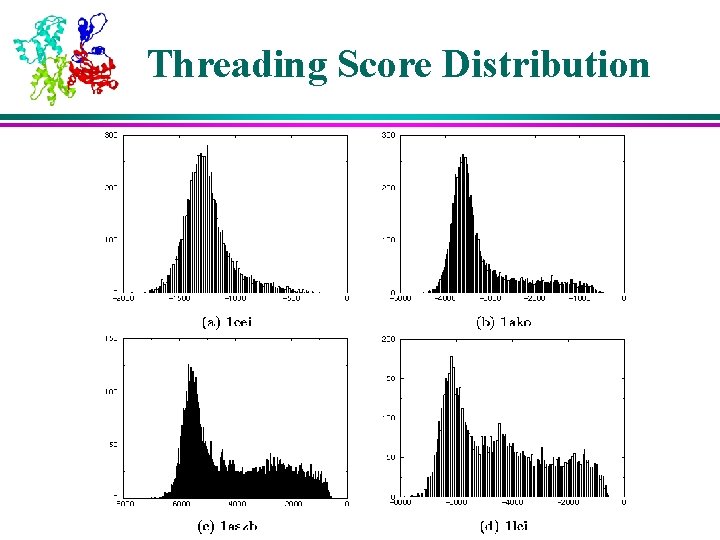
Neural Network Score Distribution

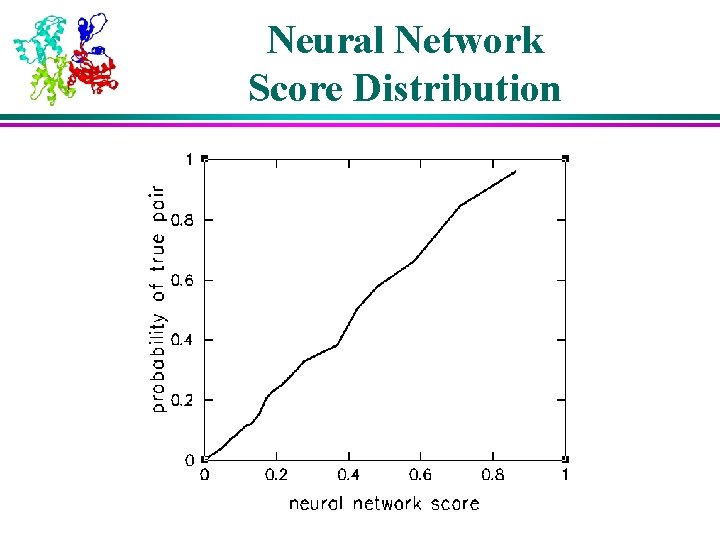
Performance of Confidence Assessment

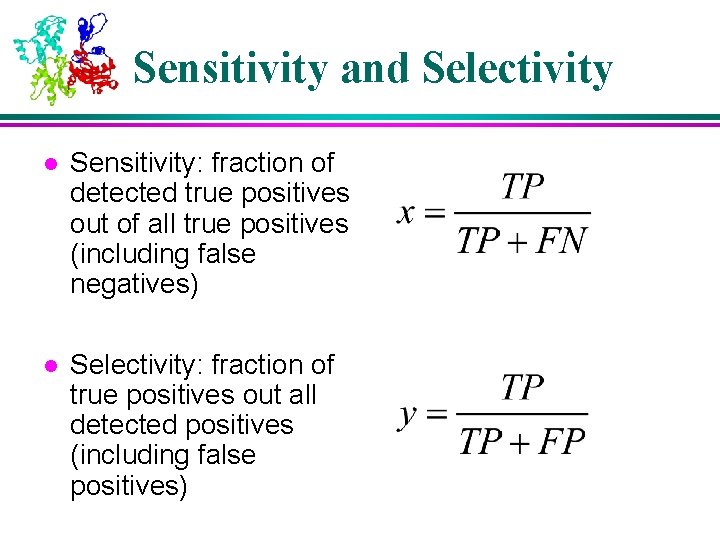
Sensitivity and Selectivity l Sensitivity: fraction of detected true positives out of all true positives (including false negatives) l Selectivity: fraction of true positives out all detected positives (including false positives)

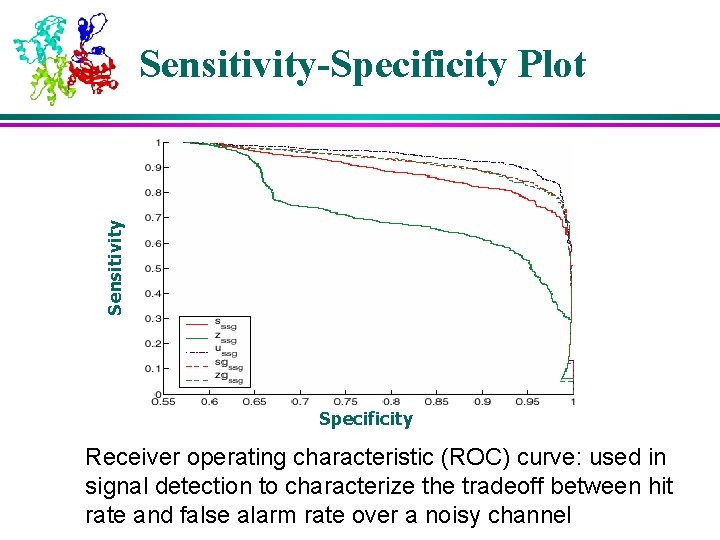
Sensitivity-Specificity Plot Specificity Receiver operating characteristic (ROC) curve: used in signal detection to characterize the tradeoff between hit rate and false alarm rate over a noisy channel

Rosetta Stone Approach Hieroglyphic Demotic Egyptian Greek

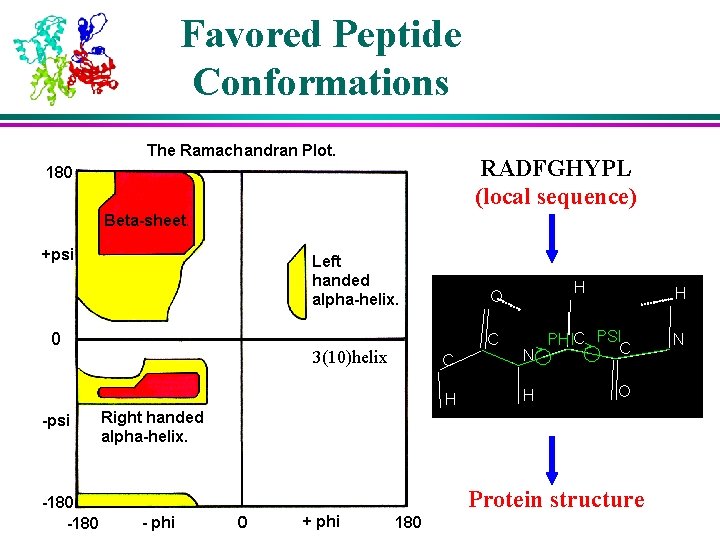
Favored Peptide Conformations RADFGHYPL (local sequence) 3(10)helix Protein structure

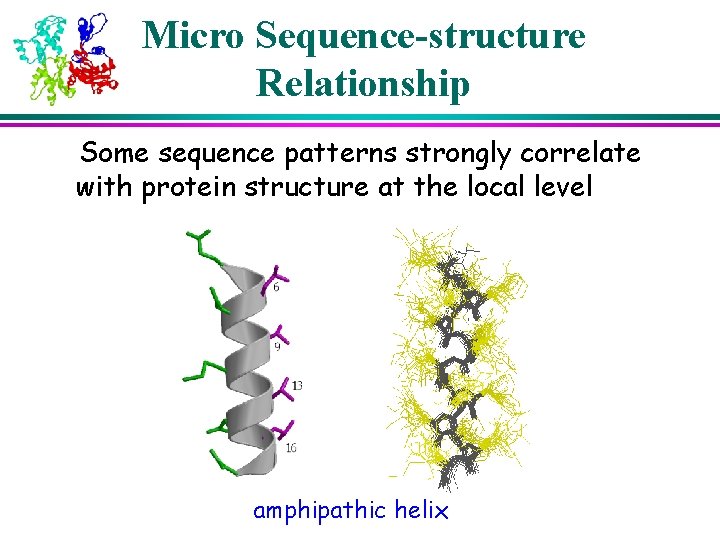
Micro Sequence-structure Relationship Some sequence patterns strongly correlate with protein structure at the local level amphipathic helix

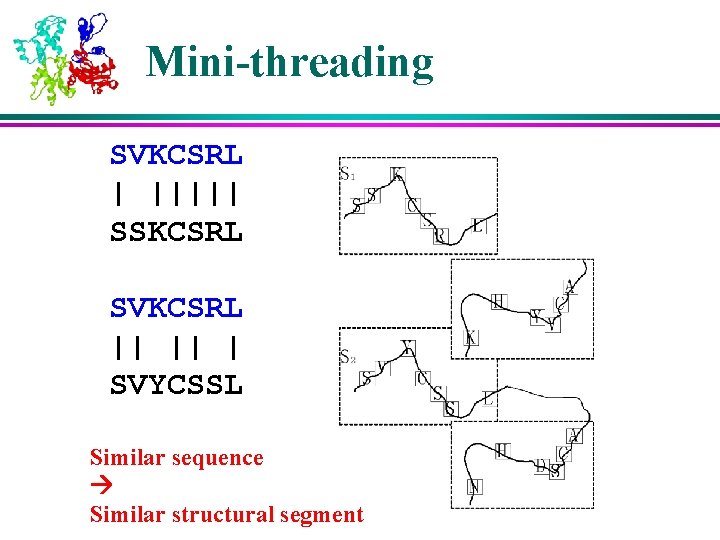
Mini-threading SVKCSRL | ||||| SSKCSRL SVKCSRL || || | SVYCSSL Similar sequence Similar structural segment

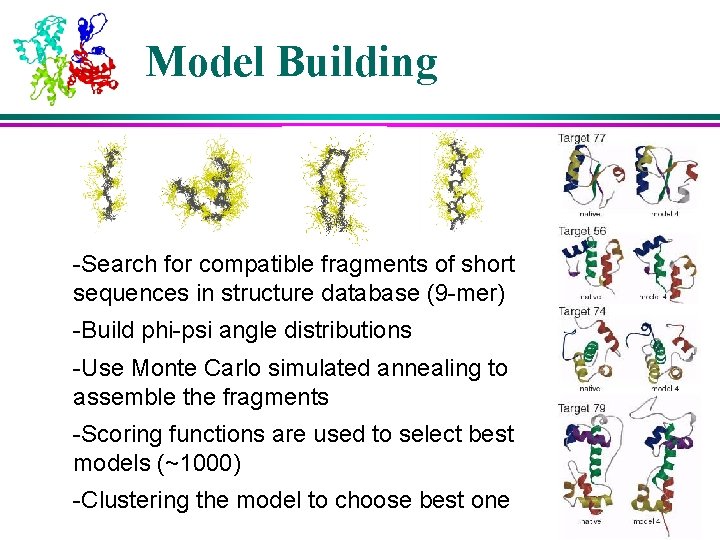
Model Building -Search for compatible fragments of short sequences in structure database (9 -mer) -Build phi-psi angle distributions -Use Monte Carlo simulated annealing to assemble the fragments -Scoring functions are used to select best models (~1000) -Clustering the model to choose best one

Reading Assignments l Suggested reading: å Chapter 18 in “Chapter 4 in “Current Topics in Computational Molecular Biology, edited by Tao Jiang, Ying Xu, and Michael Zhang. MIT Press. 2002. ” l Optional reading: å Ying Xu and Dong Xu. Protein threading using PROSPECT: Design and evaluation. Proteins: Structure, Function, and Genetics. 40: 343 -354. 2000.

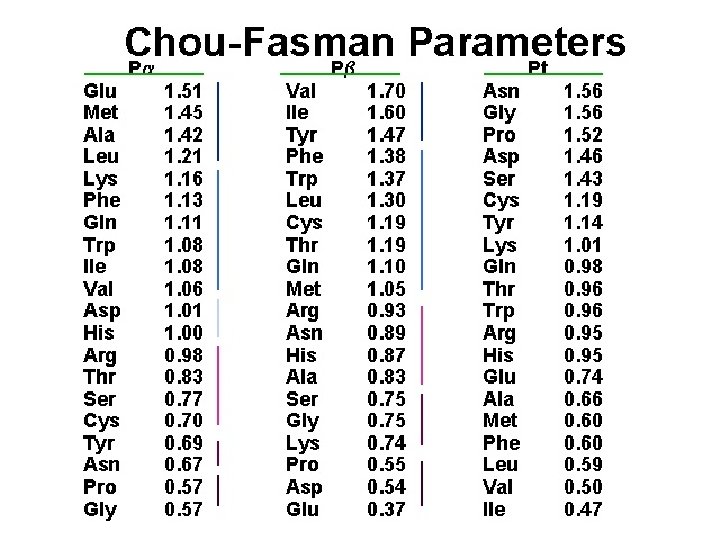
Project Assignment Develop a program that can perform a simple sequence-structure alignment: 1. Use global dynamic programming for alignment. 2. Use secondary structures for the template. 3. Use the score function of Chou-Fasman indices (no other factors to consider). For example, if Alanin (Ala, A) on the query sequence aligns to an a-hilex (H) on the template, add 1. 42 in the score. 4. Use – 3 for each opening gap and – 1 for each extension. For example, a gap of 3 is – 3 -1 -1=-5.
