Protein structure determination prediction Tertiary protein structure protein

![Tertiary protein structure: protein folding Three main approaches: [1] experimental determination (X-ray crystallography, NMR) Tertiary protein structure: protein folding Three main approaches: [1] experimental determination (X-ray crystallography, NMR)](https://slidetodoc.com/presentation_image_h/21f5504e1ed297dd2115bebc7587a079/image-2.jpg)
![Experimental approaches to protein structure [1] X-ray crystallography -- Used to determine 80% of Experimental approaches to protein structure [1] X-ray crystallography -- Used to determine 80% of](https://slidetodoc.com/presentation_image_h/21f5504e1ed297dd2115bebc7587a079/image-3.jpg)







- Slides: 18

Protein structure determination & prediction
![Tertiary protein structure protein folding Three main approaches 1 experimental determination Xray crystallography NMR Tertiary protein structure: protein folding Three main approaches: [1] experimental determination (X-ray crystallography, NMR)](https://slidetodoc.com/presentation_image_h/21f5504e1ed297dd2115bebc7587a079/image-2.jpg)
Tertiary protein structure: protein folding Three main approaches: [1] experimental determination (X-ray crystallography, NMR) [2] Comparative modeling (based on homology) [3] Ab initio (de novo) prediction (Dr. Ingo Ruczinski at JHSPH)
![Experimental approaches to protein structure 1 Xray crystallography Used to determine 80 of Experimental approaches to protein structure [1] X-ray crystallography -- Used to determine 80% of](https://slidetodoc.com/presentation_image_h/21f5504e1ed297dd2115bebc7587a079/image-3.jpg)
Experimental approaches to protein structure [1] X-ray crystallography -- Used to determine 80% of structures -- Requires high protein concentration -- Requires crystals -- Able to trace amino acid side chains -- Earliest structure solved was myoglobin [2] NMR -- Magnetic field applied to proteins in solution -- Largest structures: 350 amino acids (40 k. D) -- Does not require crystallization

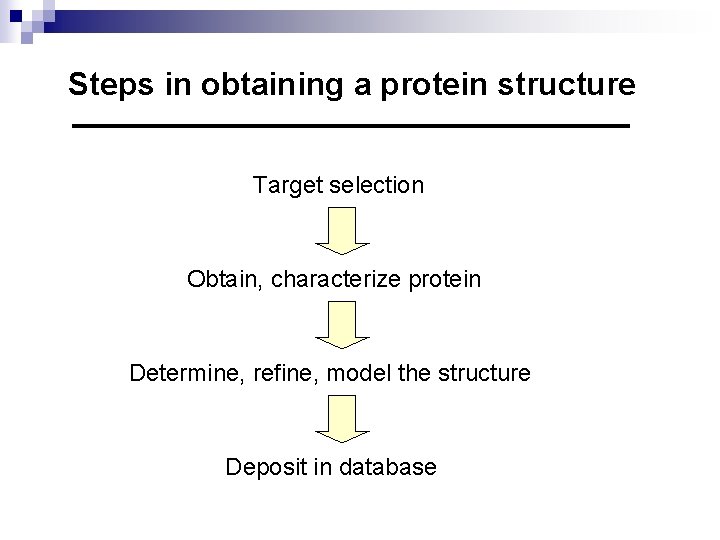
Steps in obtaining a protein structure Target selection Obtain, characterize protein Determine, refine, model the structure Deposit in database

X-ray crystallography http: //en. wikipedia. org/wiki/X-ray_diffraction Sperm Whale Myoglobin



Nuclear magnetic resonance spectroscopy http: //en. wikipedia. org/wiki/Nuclear_magnetic_resonance




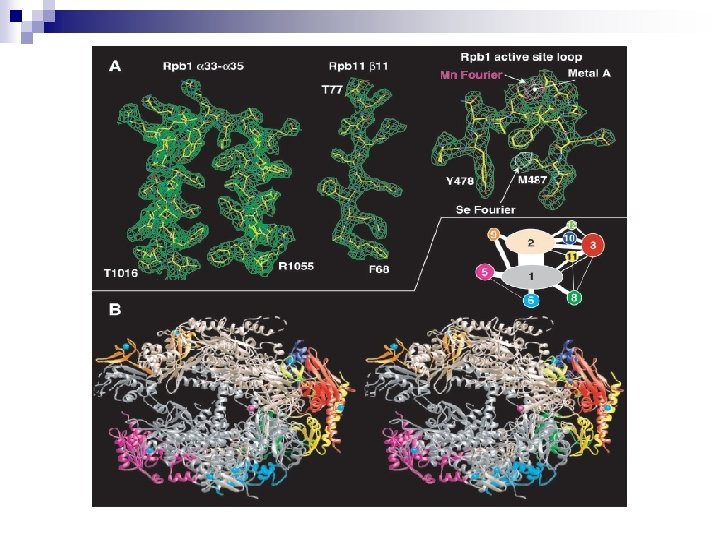
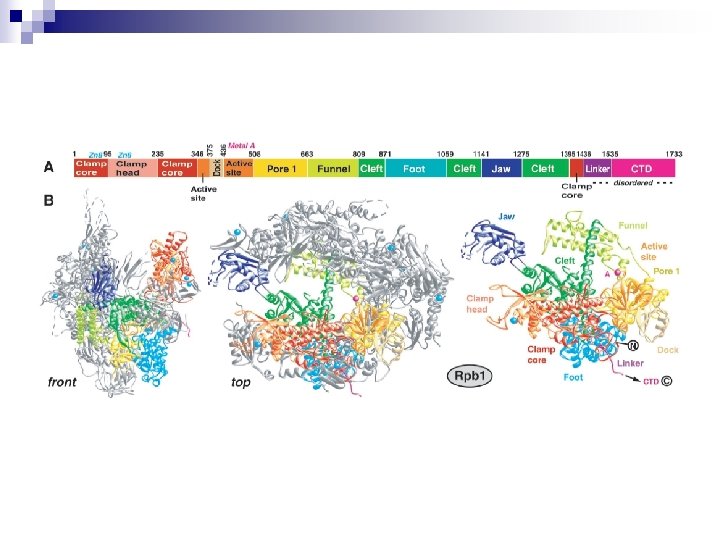
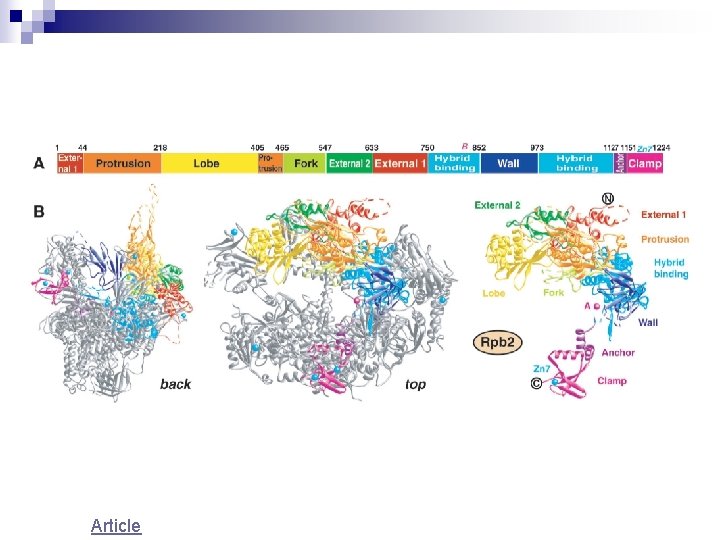
Article

Ab initio protein prediction n Starts with an attempt to derive secondary structure from the amino acid sequence Predicting the likelihood that a subsequence will fold into an alpha-helix, beta-sheet, or coil, using physicochemical parameters or HMMs and ANNs ¨ Able to accurately predict 3/4 of all local structures ¨

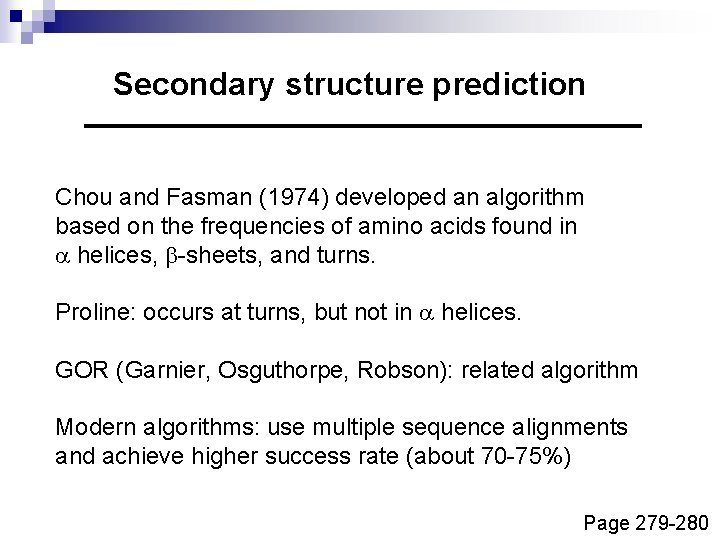
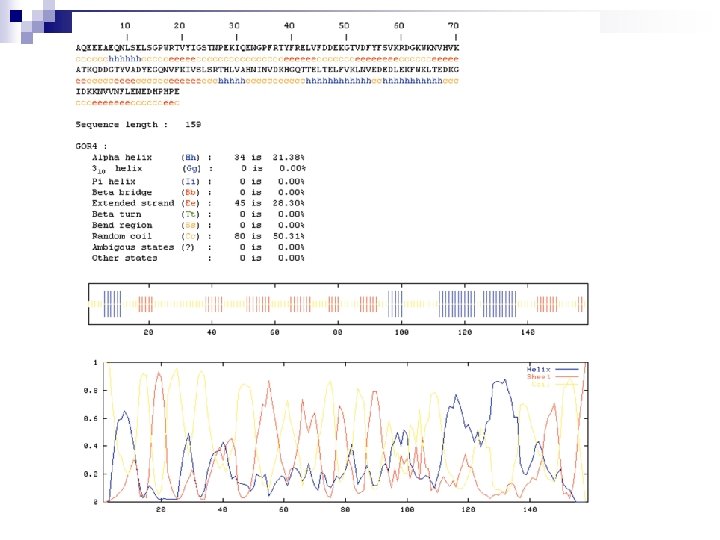
Secondary structure prediction Chou and Fasman (1974) developed an algorithm based on the frequencies of amino acids found in a helices, b-sheets, and turns. Proline: occurs at turns, but not in a helices. GOR (Garnier, Osguthorpe, Robson): related algorithm Modern algorithms: use multiple sequence alignments and achieve higher success rate (about 70 -75%) Page 279 -280



Fold recognition (structural profiles) Attempts to find the best fit of a raw polypeptide sequence onto a library of known protein folds n A prediction of the secondary structure of the unknown is made and compared with the secondary structure of each member of the library of folds n

Threading n Takes the fold recognition process a step further: ¨ Empirical-energy functions for residue pair interactions are used to mount the unknown onto the putative backbone in the best possible manner