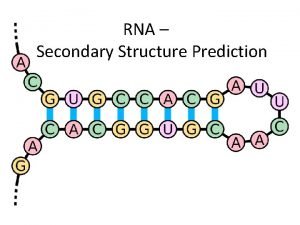
The role of prem RNA secondary structure in

- Slides: 11

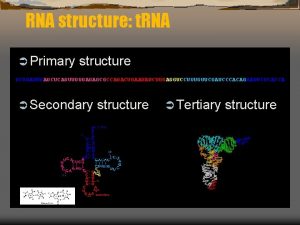
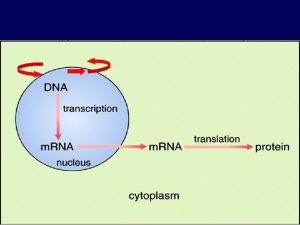
The role of pre-m. RNA secondary structure in gene splicing Sanja Rogic Computer Science Department, UBC pre-m. RNA structure and splicing Sanja Rogic

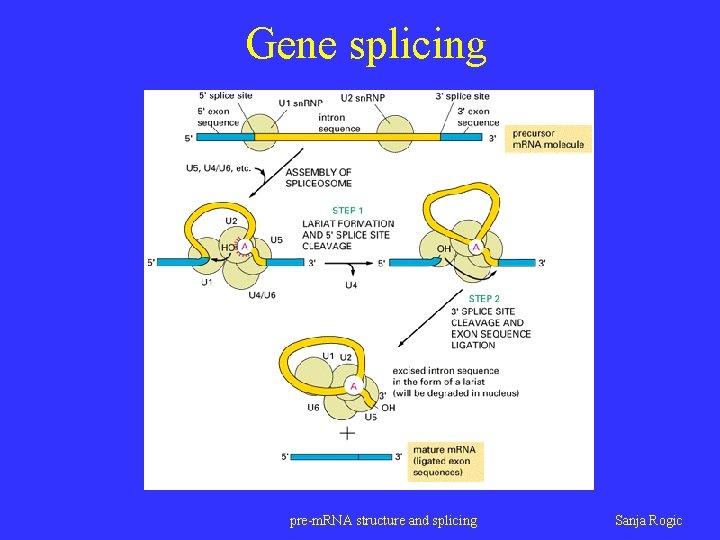
Gene splicing pre-m. RNA structure and splicing Sanja Rogic

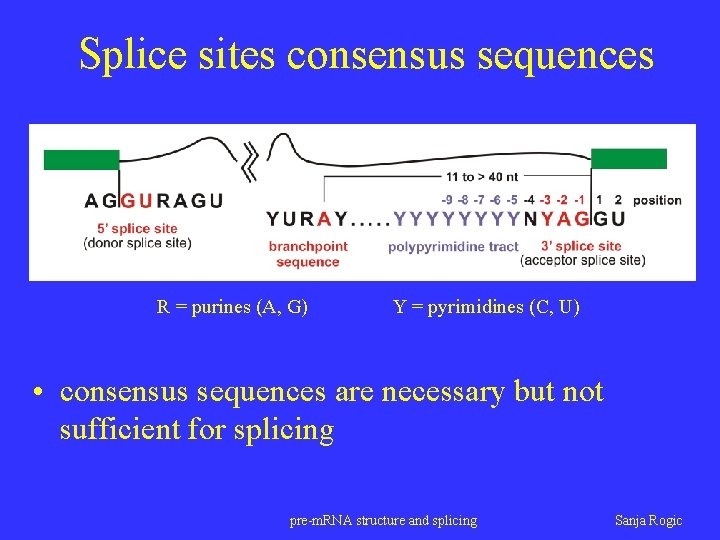
Splice sites consensus sequences R = purines (A, G) Y = pyrimidines (C, U) • consensus sequences are necessary but not sufficient for splicing pre-m. RNA structure and splicing Sanja Rogic

Motivation • splice site recognition by the spliceosome is still not well understood - new biological insights • low accuracy of computational splice site prediction is a major reason for limited accuracy of gene-finding programs - improving accuracy of gene-finding • pre-m. RNA is not linear but has secondary structure • biological studies suggest that pre-m. RNA secondary structure can affect splicing pre-m. RNA structure and splicing Sanja Rogic

Hypothesis and goal • secondary structure of pre-m. RNA plays a role in splicing Ø characterize secondary structure elements that are additional identifiers of intronic regions Dataset – S. cerevisiae introns • 215 introns with consistent annotation between three databases: AYID, YIDB, and CYGD pre-m. RNA structure and splicing Sanja Rogic

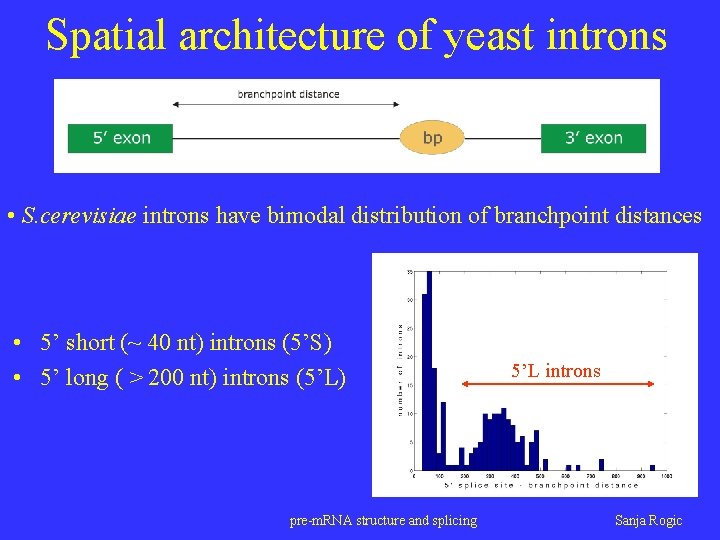
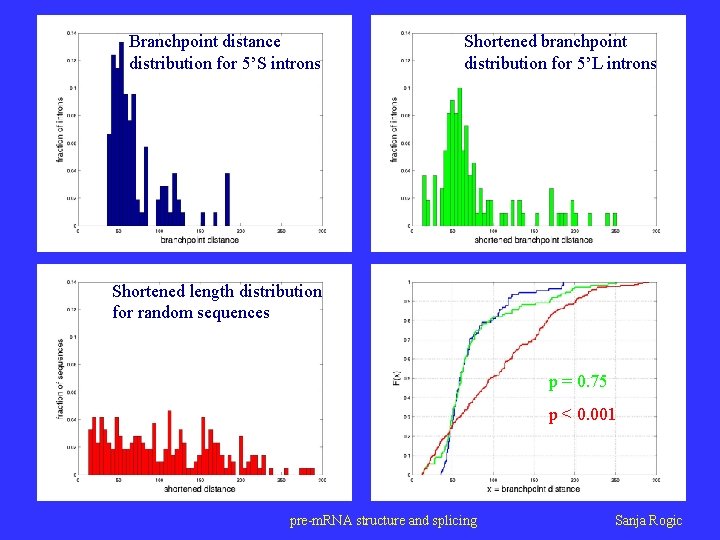
Spatial architecture of yeast introns • S. cerevisiae introns have bimodal distribution of branchpoint distances • 5’ short (~ 40 nt) introns (5’S) • 5’ long ( > 200 nt) introns (5’L) pre-m. RNA structure and splicing 5’L introns Sanja Rogic

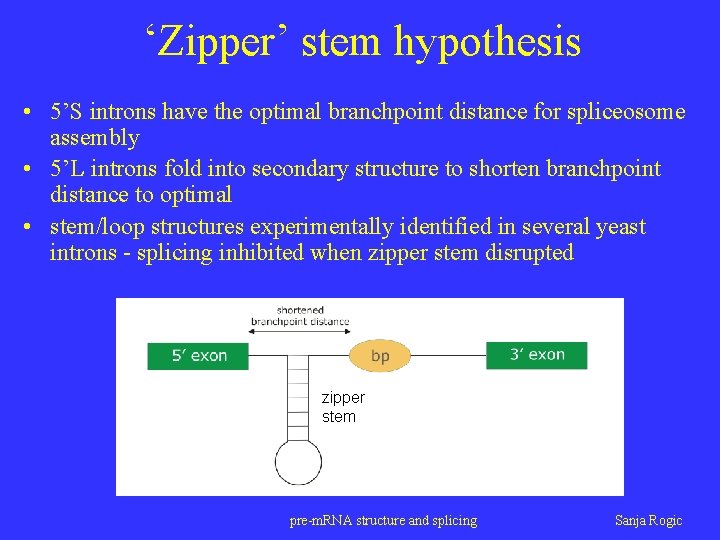
‘Zipper’ stem hypothesis • 5’S introns have the optimal branchpoint distance for spliceosome assembly • 5’L introns fold into secondary structure to shorten branchpoint distance to optimal • stem/loop structures experimentally identified in several yeast introns - splicing inhibited when zipper stem disrupted zipper stem pre-m. RNA structure and splicing Sanja Rogic

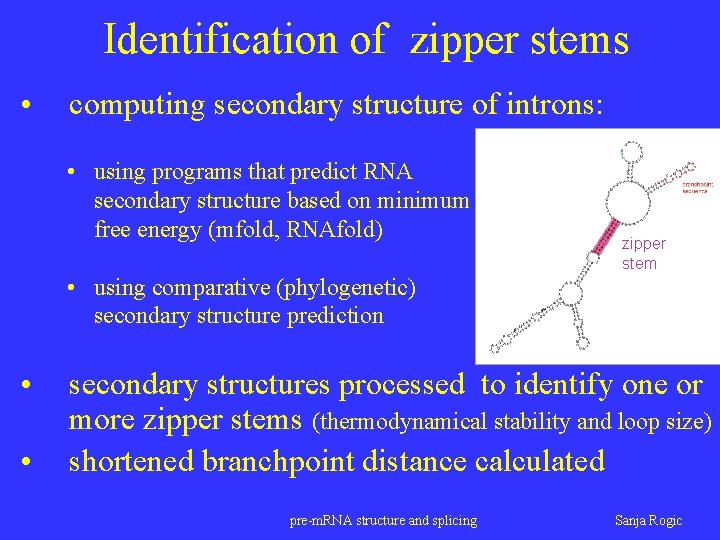
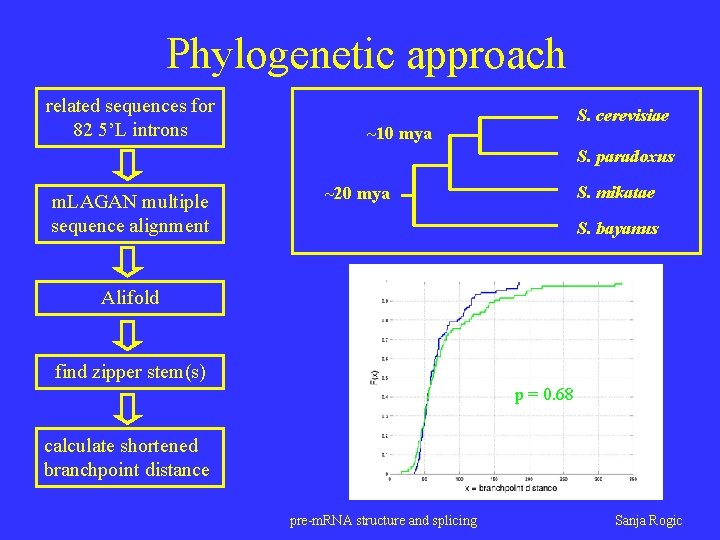
Identification of zipper stems • computing secondary structure of introns: • using programs that predict RNA secondary structure based on minimum free energy (mfold, RNAfold) zipper stem • using comparative (phylogenetic) secondary structure prediction • • secondary structures processed to identify one or more zipper stems (thermodynamical stability and loop size) shortened branchpoint distance calculated pre-m. RNA structure and splicing Sanja Rogic

Branchpoint distance distribution for 5’S introns Shortened branchpoint distribution for 5’L introns Shortened length distribution for random sequences p = 0. 75 p < 0. 001 pre-m. RNA structure and splicing Sanja Rogic

Phylogenetic approach related sequences for 82 5’L introns S. cerevisiae ~10 mya S. paradoxus m. LAGAN multiple sequence alignment S. mikatae ~20 mya S. bayanus Alifold find zipper stem(s) p = 0. 68 calculate shortened branchpoint distance pre-m. RNA structure and splicing Sanja Rogic

Acknowledgements • thesis supervisors: Alan Mackworth, CS Department, UBC Holger Hoos, CS Department, UBC Francis Ouellette, UBi. C • colleagues in Beta lab (Bioinformatics and Empirical & Theoretical Algorithmics) pre-m. RNA structure and splicing Sanja Rogic
 Rna secondary structure dynamic programming
Rna secondary structure dynamic programming Rna secondary structure
Rna secondary structure Rna secondary structure dynamic programming
Rna secondary structure dynamic programming Rna secondary structure prediction
Rna secondary structure prediction Rna secondary structure prediction
Rna secondary structure prediction Prem earth
Prem earth Raja rani vs prem adib
Raja rani vs prem adib Voltage secure email
Voltage secure email Prem
Prem Who gives two rupees to ghisho in the shroud
Who gives two rupees to ghisho in the shroud Mulk raj anand education
Mulk raj anand education On prem
On prem