RNA Secondary Structure Prediction Dynamic Programming Approaches Sarah



- Slides: 16

RNA Secondary Structure Prediction Dynamic Programming Approaches Sarah Aerni http: //www. tbi. univie. ac. at/ 1

Outline l l RNA folding Dynamic programming for RNA secondary structure prediction 2

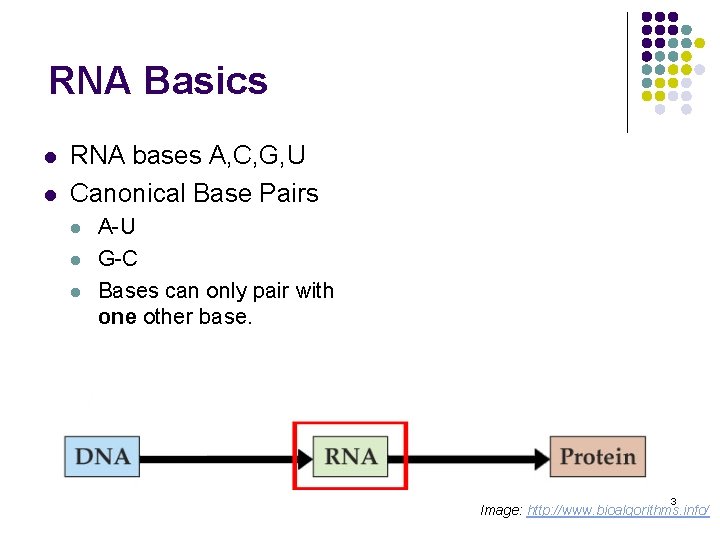
RNA Basics l l RNA bases A, C, G, U Canonical Base Pairs l l l A-U G-C Bases can only pair with one other base. 3 Image: http: //www. bioalgorithms. info/

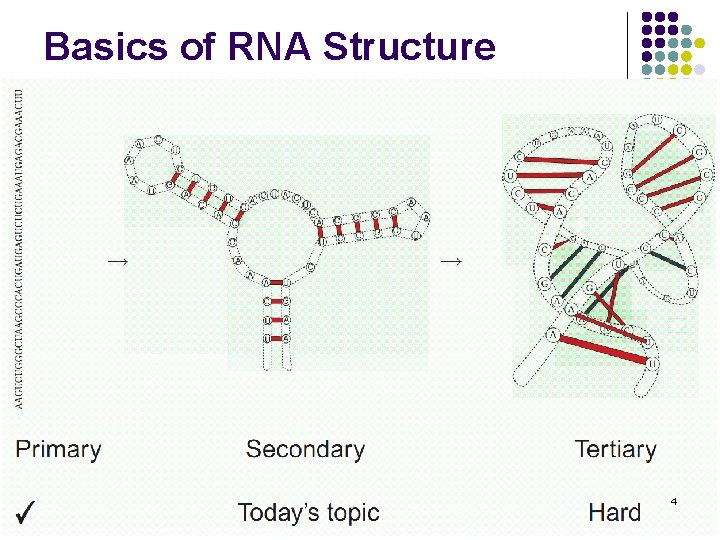
Basics of RNA Structure 4

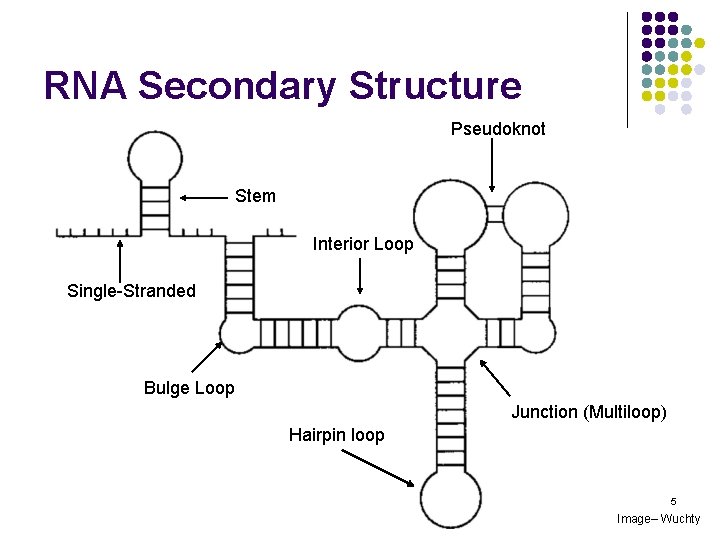
RNA Secondary Structure Pseudoknot Stem Interior Loop Single-Stranded Bulge Loop Junction (Multiloop) Hairpin loop 5 Image– Wuchty

Nesting 6

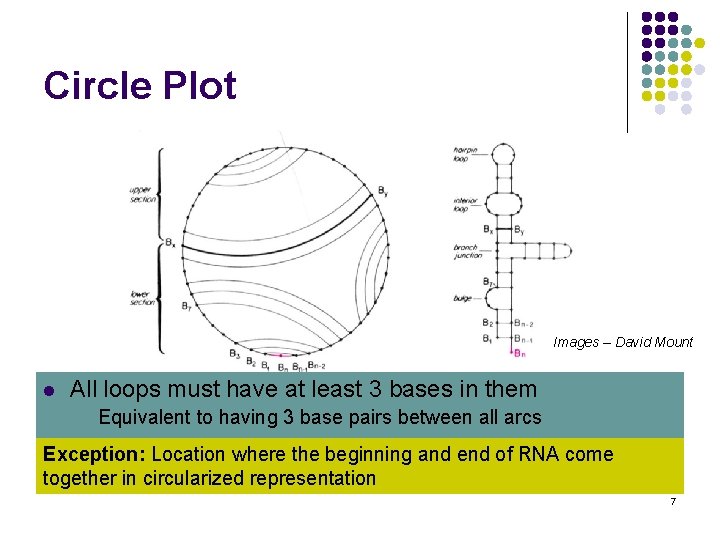
Circle Plot Images – David Mount l l Linear RNA strand folded back 3 on itself to All loops must have at least bases in create them secondary structure l Equivalent to having 3 base pairs between all arcs Circularized representation uses this requirement Exception: Location where the beginning and end of RNA come l Arcs represent base pairing together in circularized representation 7

Pseudoknots 8

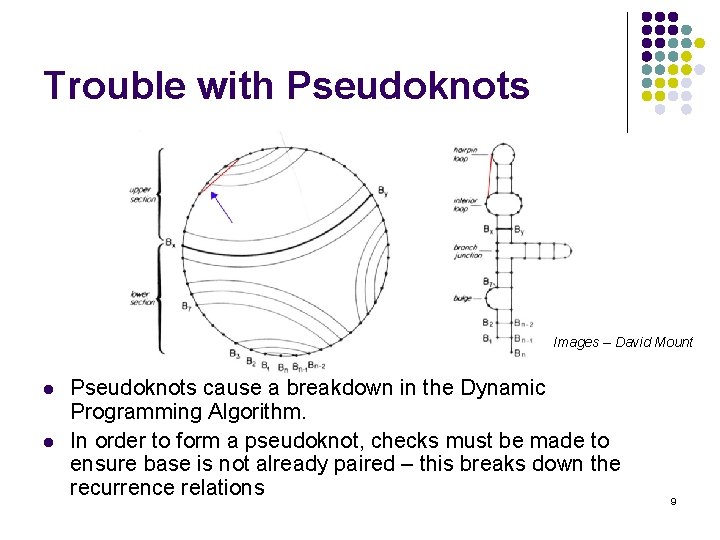
Trouble with Pseudoknots Images – David Mount l l Pseudoknots cause a breakdown in the Dynamic Programming Algorithm. In order to form a pseudoknot, checks must be made to ensure base is not already paired – this breaks down the recurrence relations 9

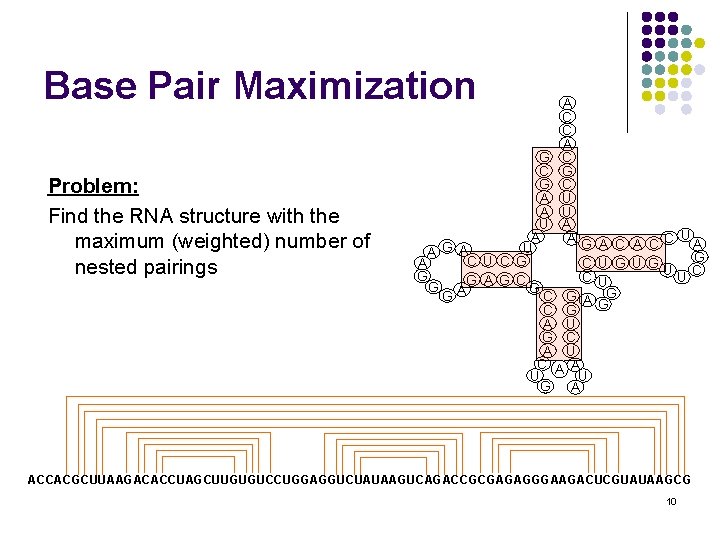
Base Pair Maximization Problem: Find the RNA structure with the maximum (weighted) number of nested pairings A C C A G C C G G C A U U A A AG G U A A C U C G C A G C G A G G C G A U G C A U A U G A U A CC A G U G UUC U G G ACCACGCUUAAGACACCUAGCUUGUGUCCUGGAGGUCUAUAAGUCAGACCGCGAGAGGGAAGACUCGUAUAAGCG 10

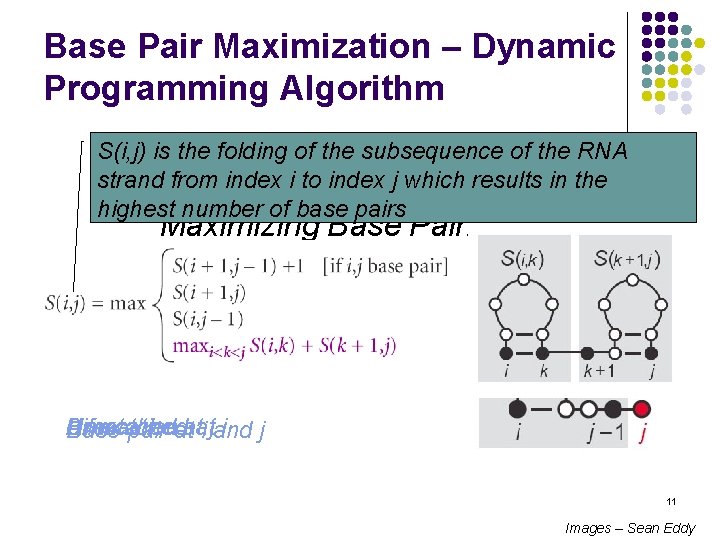
Base Pair Maximization – Dynamic Programming Algorithm S(i, j) is the folding of the subsequence of the RNA strand from. Simple index i to. Example: index j which results in the highest number of base pairs Maximizing Base Pairing Bifurcation Unmatched Umatched i j Base pair atatat i jand 11 Images – Sean Eddy

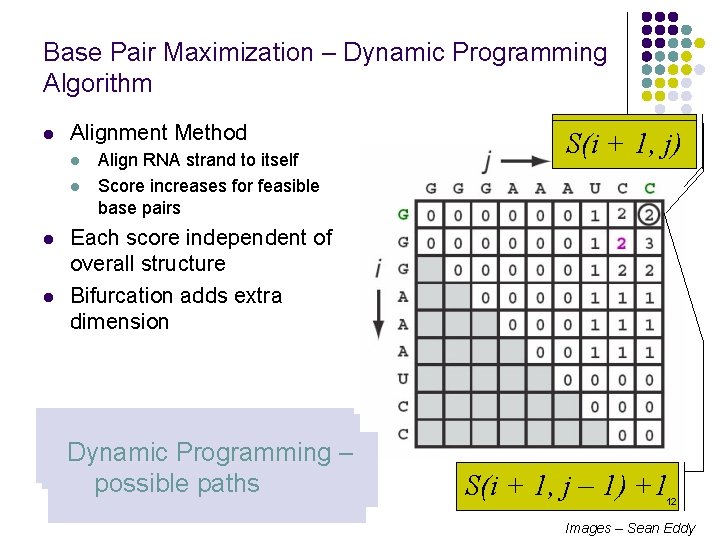
Base Pair Maximization – Dynamic Programming Algorithm l Alignment Method l l Align RNA strand to itself Score increases for feasible base pairs S(i, +j – 1, 1) S(i j) Each score independent of overall structure Bifurcation adds extra dimension Initialize first two diagonal Fill in squares sweeping Bases cannot pair, similar Bases can pair, similar Dynamic Programming – arrays to 0 diagonally to alignment to unmatched alignment possible paths S(i + 1, j – 1) +1 12 Images – Sean Eddy

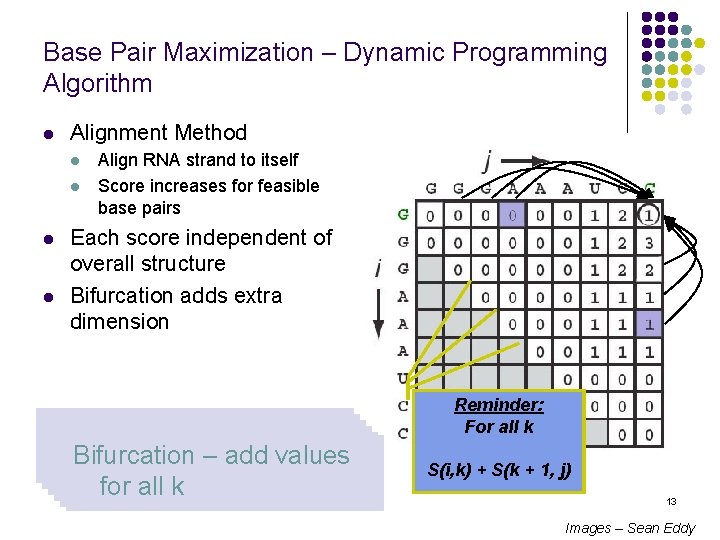
Base Pair Maximization – Dynamic Programming Algorithm l Alignment Method l l Align RNA strand to itself Score increases for feasible base pairs Each score independent of overall structure Bifurcation adds extra dimension Initialize first two diagonal Fill in squares sweeping Bases cannot pair, similar Bases can pair, similar Dynamic Bifurcation arrays to Programming 0 – add values– diagonally to matched alignment possible for all k paths k=0 Reminder: : Bifurcation max. For in this all kcase S(i, k) + S(k + 1, j) 13 Images – Sean Eddy

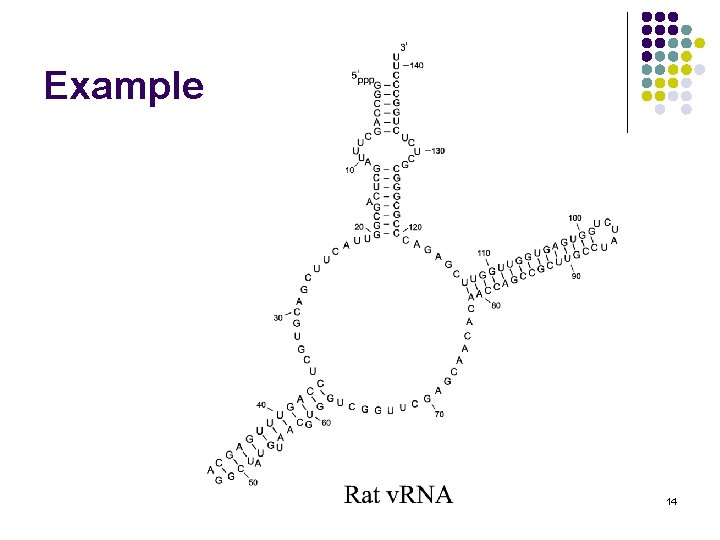
Example 14

Base Pair Maximization Drawbacks l Base pair maximization will not necessarily lead to the most stable structure l l May create structure with many interior loops or hairpins which are energetically unfavorable Not biologically reasonable 15

References l How Do RNA Folding Algorithms Work? . S. R. Eddy. Nature Biotechnology, 22: 1457 -1458, 2004. 16