Phylogeny reconstruction How do we reconstruct the tree

















- Slides: 60

Phylogeny reconstruction How do we reconstruct the tree of life? Outline: Terminology Methods distance parsimony maximum likelihood bootstrapping Problems homoplasy hybridisation Dr. Sean Graham, UBC.

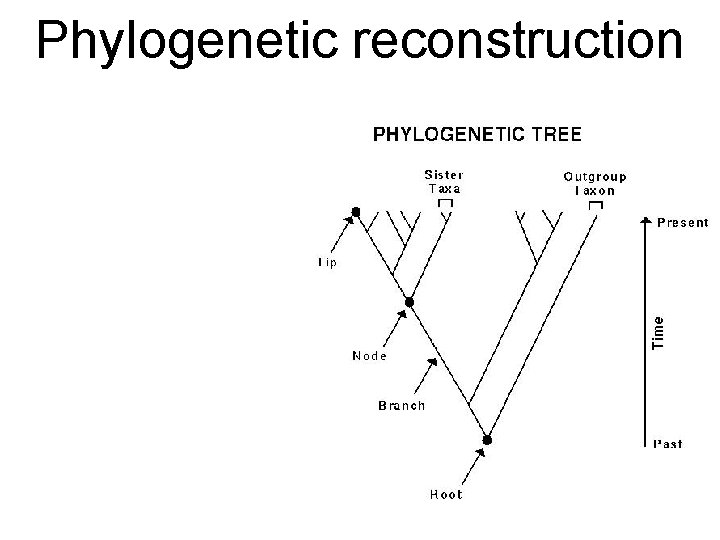
Phylogenetic reconstruction

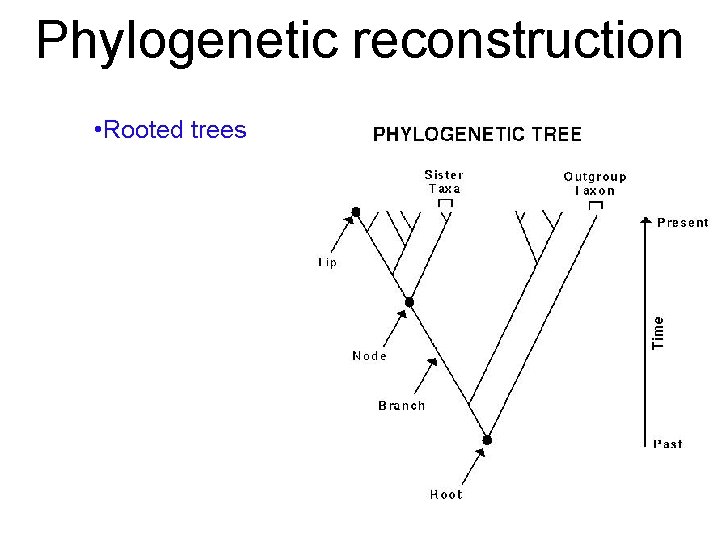
Phylogenetic reconstruction • Rooted trees

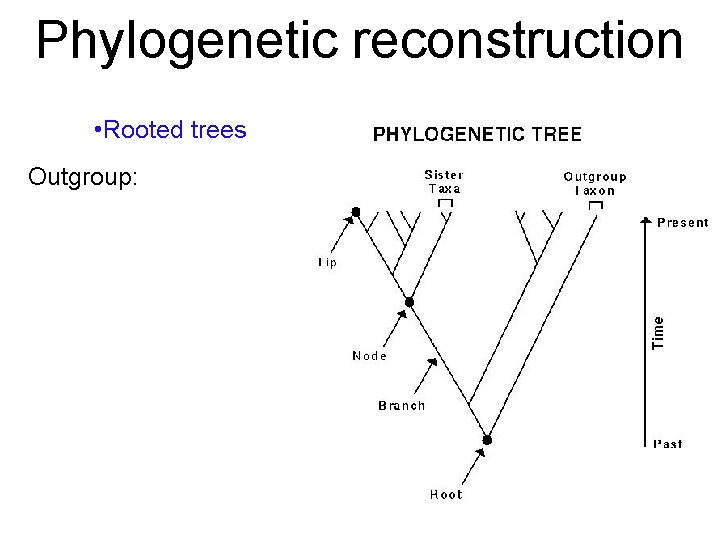
Phylogenetic reconstruction • Rooted trees Outgroup:

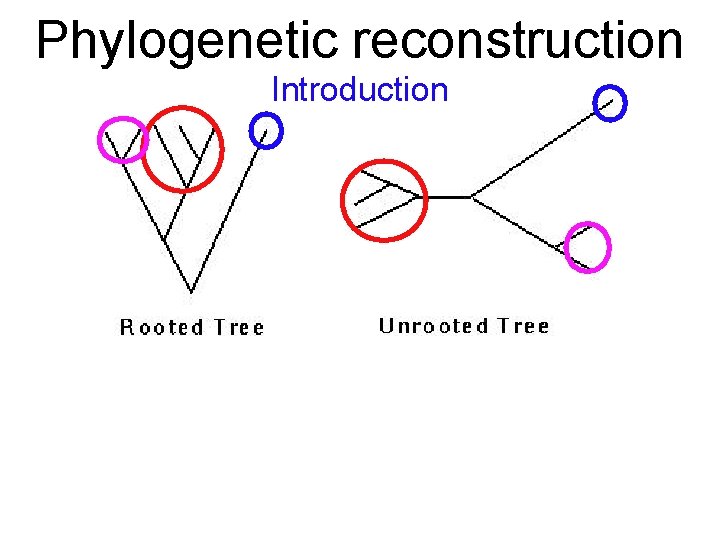
Phylogenetic reconstruction Introduction

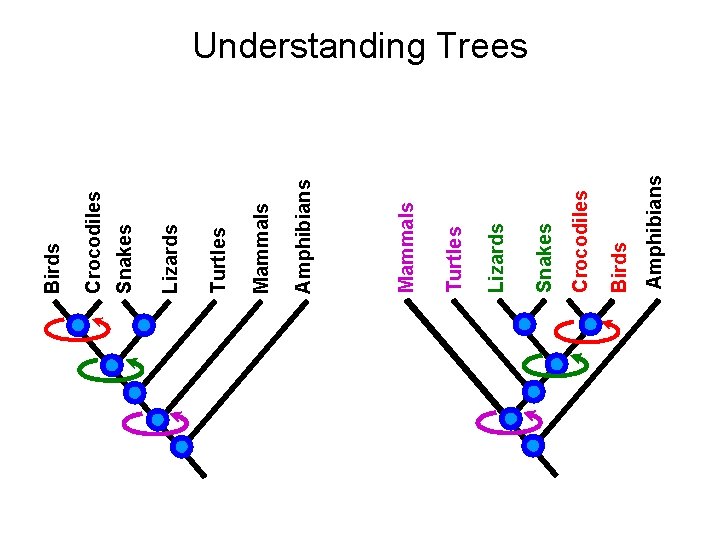
Amphibians Birds Crocodiles Snakes Lizards Turtles Mammals Amphibians Mammals Turtles Lizards Snakes Crocodiles Birds Understanding Trees

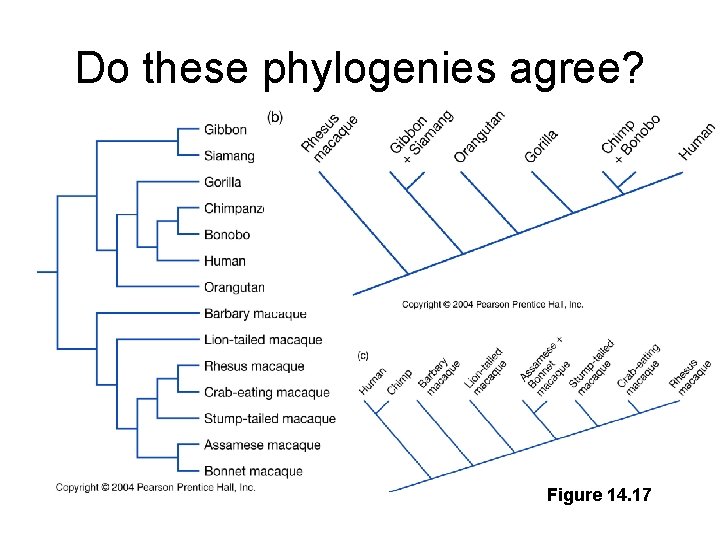
Do these phylogenies agree? Figure 14. 17

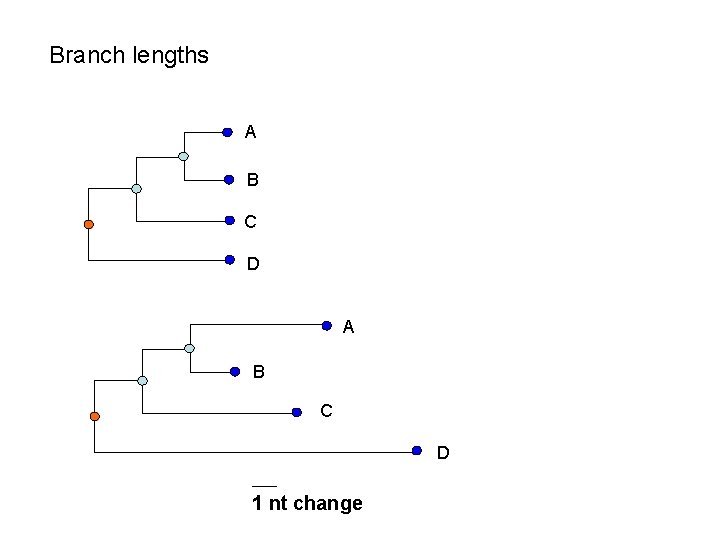
Branch lengths A B C D 1 nt change

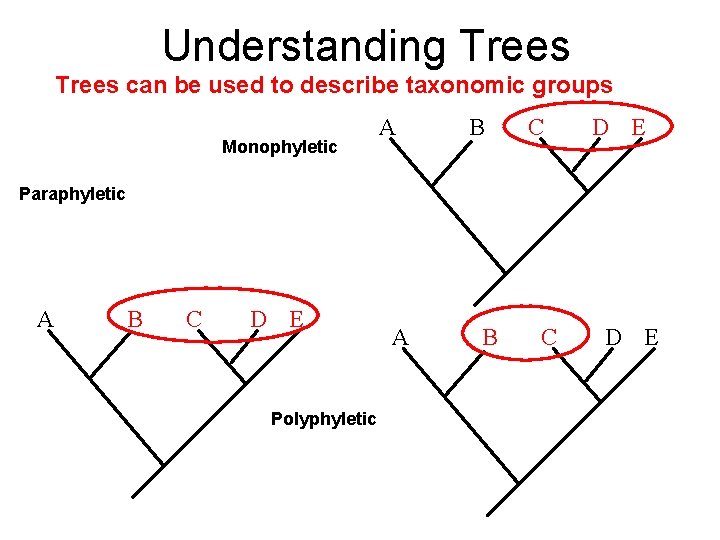
Understanding Trees can be used to describe taxonomic groups Monophyletic A B C D E Paraphyletic A B C D E Polyphyletic A B C D E

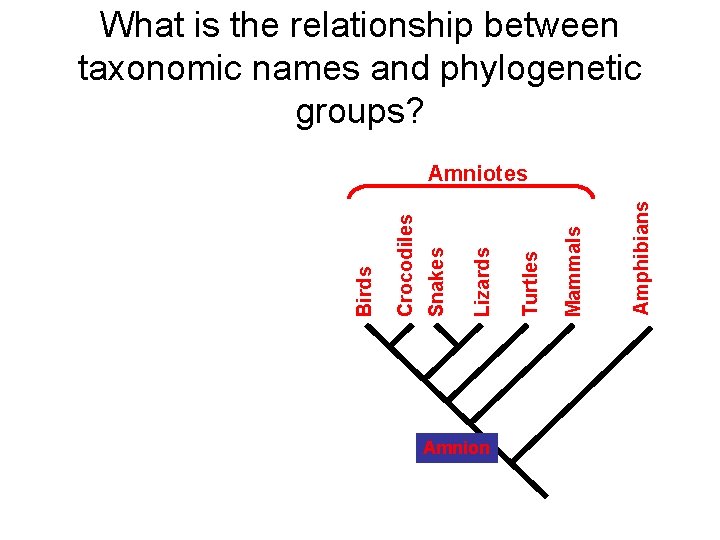
What is the relationship between taxonomic names and phylogenetic groups? Amnion Amphibians Mammals Turtles Lizards Snakes Crocodiles Birds Amniotes

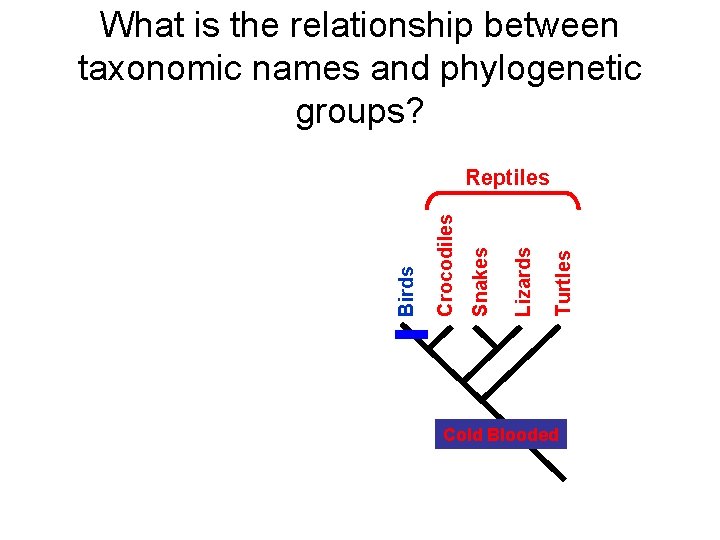
What is the relationship between taxonomic names and phylogenetic groups? Turtles Lizards Snakes Crocodiles Birds Reptiles Cold Blooded

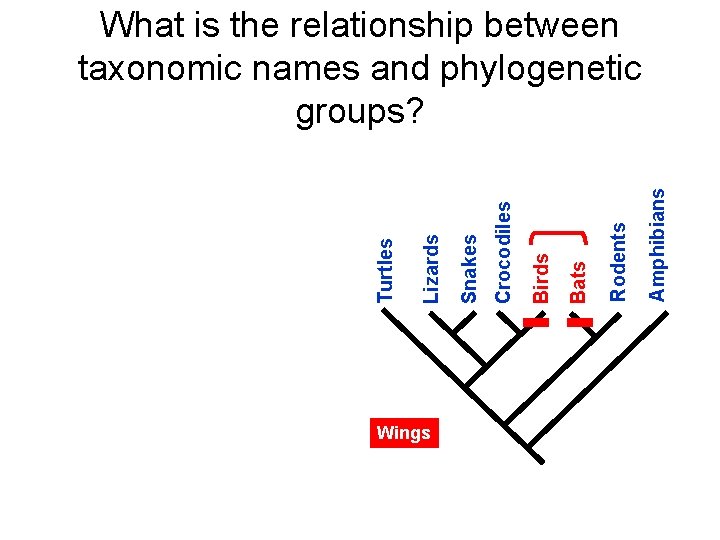
Wings Amphibians Rodents Bats Birds Crocodiles Snakes Lizards Turtles What is the relationship between taxonomic names and phylogenetic groups?

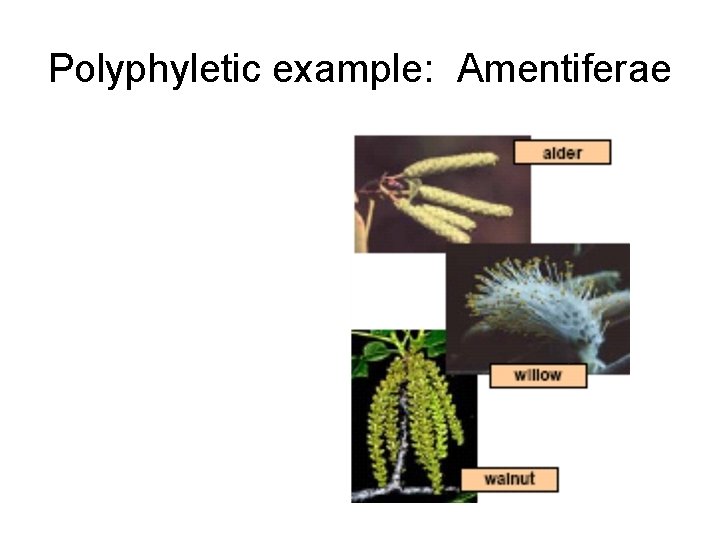
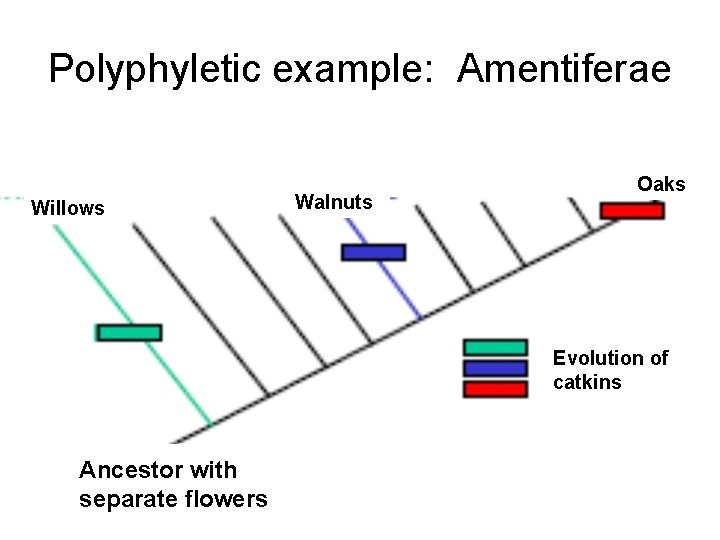
Polyphyletic example: Amentiferae

Polyphyletic example: Amentiferae Willows Walnuts Oaks Evolution of catkins Ancestor with separate flowers

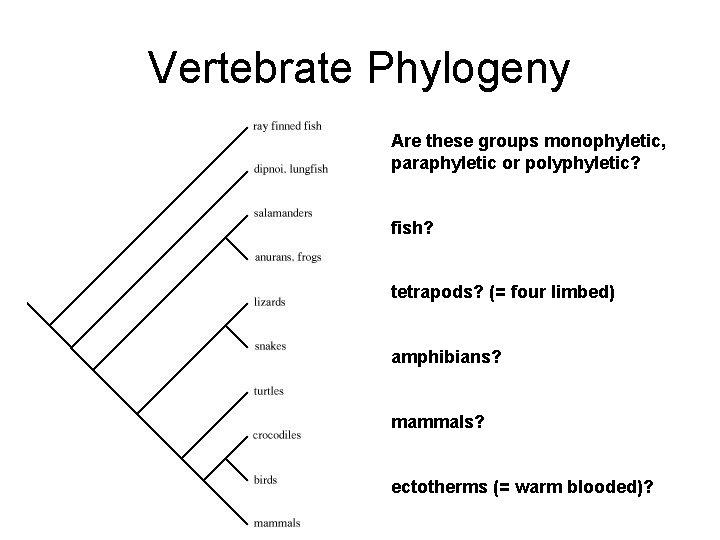
Vertebrate Phylogeny Are these groups monophyletic, paraphyletic or polyphyletic? fish? tetrapods? (= four limbed) amphibians? mammals? ectotherms (= warm blooded)?

Constructing Trees Methods: distance (UPGMA, Neighbor joining) parsimony maximum likelihood (Bayesian)

Distance Methods (phenetics)

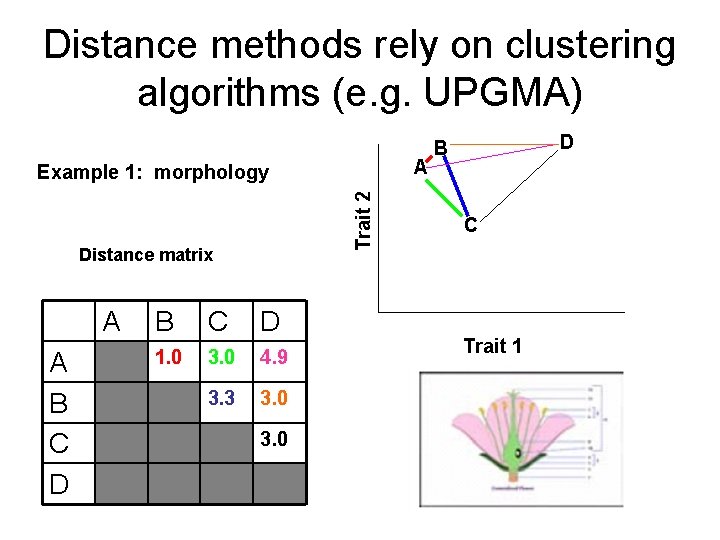
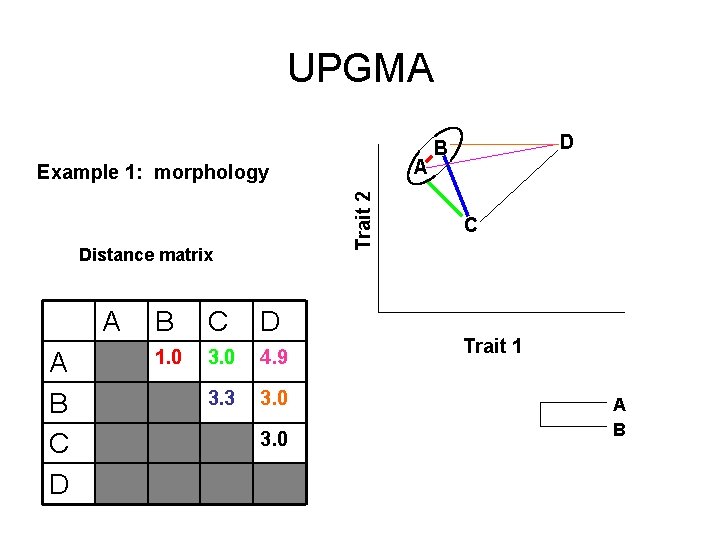
Distance methods rely on clustering algorithms (e. g. UPGMA) A Trait 2 Example 1: morphology Distance matrix A A B C D 1. 0 3. 0 4. 9 3. 3 3. 0 D B C Trait 1

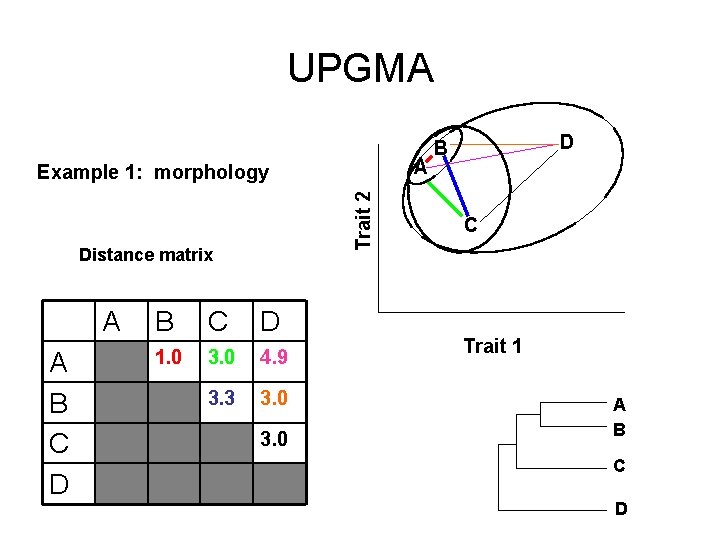
UPGMA A Trait 2 Example 1: morphology Distance matrix A A B C D 1. 0 3. 0 4. 9 3. 3 3. 0 D B C Trait 1 A B

UPGMA A Trait 2 Example 1: morphology Distance matrix A A B C D 1. 0 3. 0 4. 9 3. 3 3. 0 D B C Trait 1 A B C D

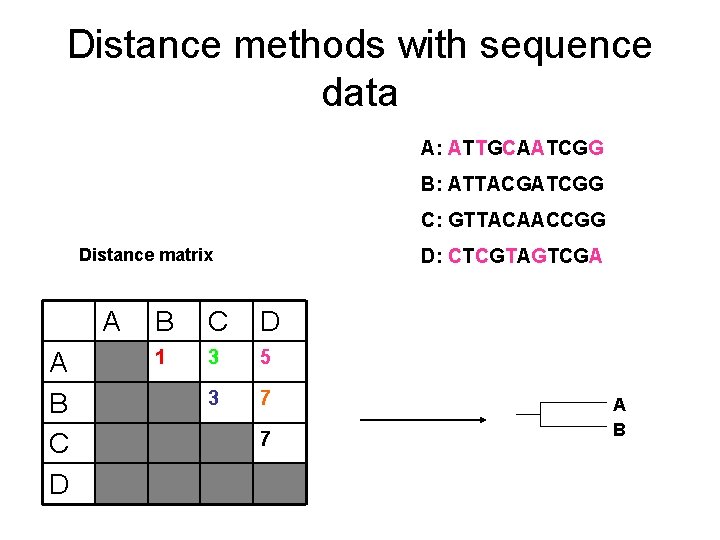
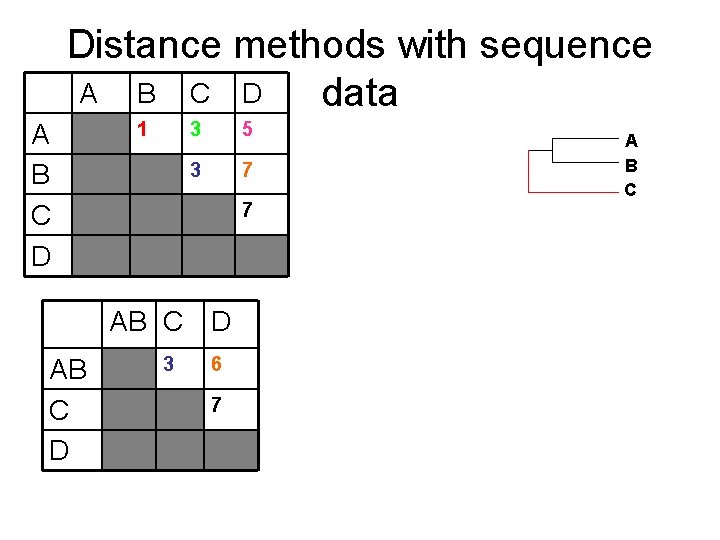
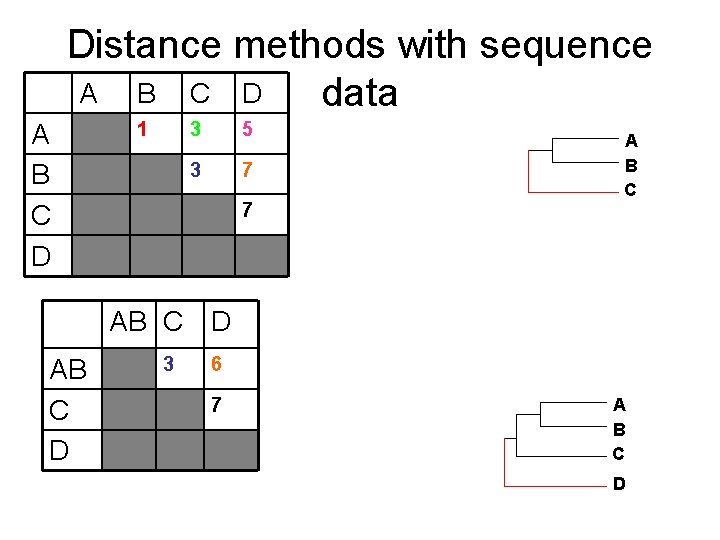
Distance methods with sequence data A: ATTGCAATCGG B: ATTACGATCGG C: GTTACAACCGG Distance matrix A A B C D D: CTCGTAGTCGA B C D 1 3 5 3 7 7 A B

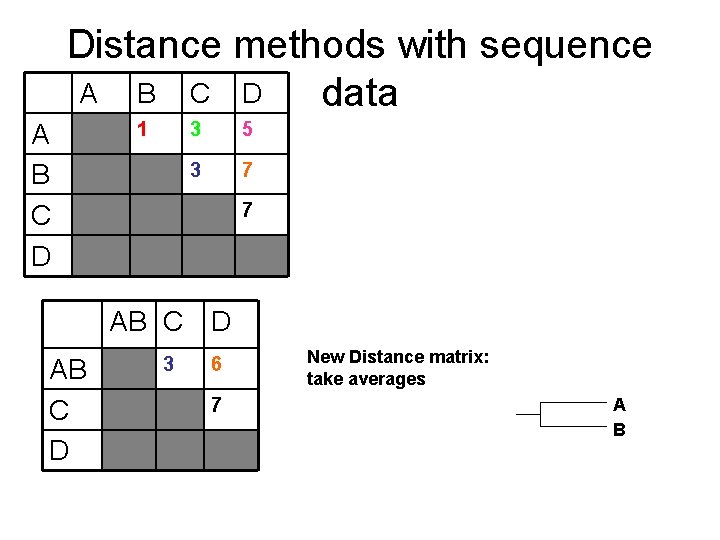
Distance methods with sequence A B C D data A B C D 1 3 5 3 7 7 AB C D 3 6 7 New Distance matrix: take averages A B

Distance methods with sequence A B C D data A B C D 1 3 5 3 7 7 A B C AB C D 3 6 7 A B C D

Distance methods with sequence A B C D data A B C D 1 3 5 3 7 7 A B C AB C D 3 6 7 A B C D

Assumptions of distance methods

Strengths and weaknesses of distance methods

II. Parsimony Methods (Cladistics) Hennig (German entomologist) wrote in 1966 Translated into English in 1976: very influential

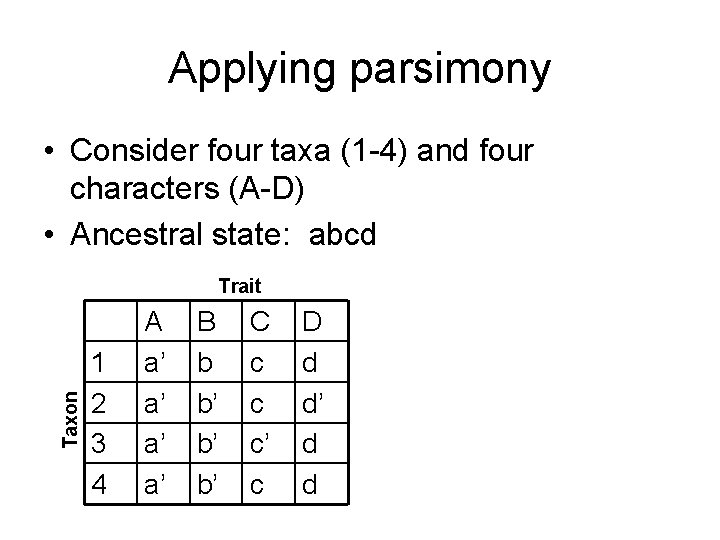
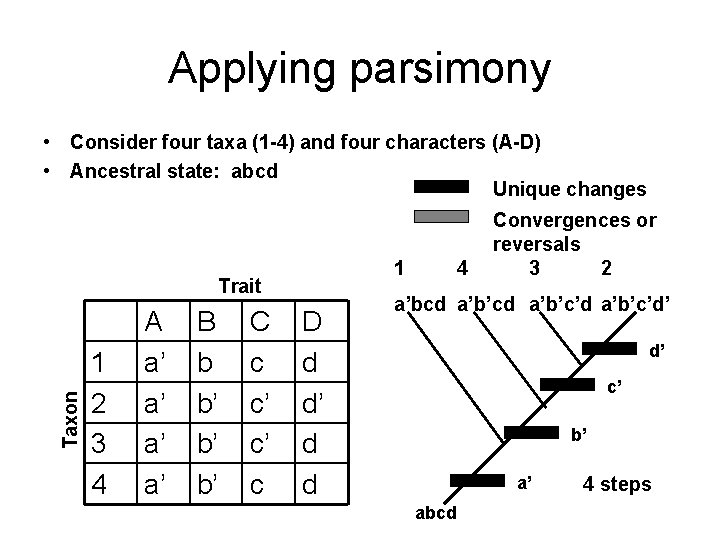
Applying parsimony • Consider four taxa (1 -4) and four characters (A-D) • Ancestral state: abcd Taxon Trait 1 2 3 4 A a’ a’ B b b’ b’ b’ C c c c’ c D d d’ d d

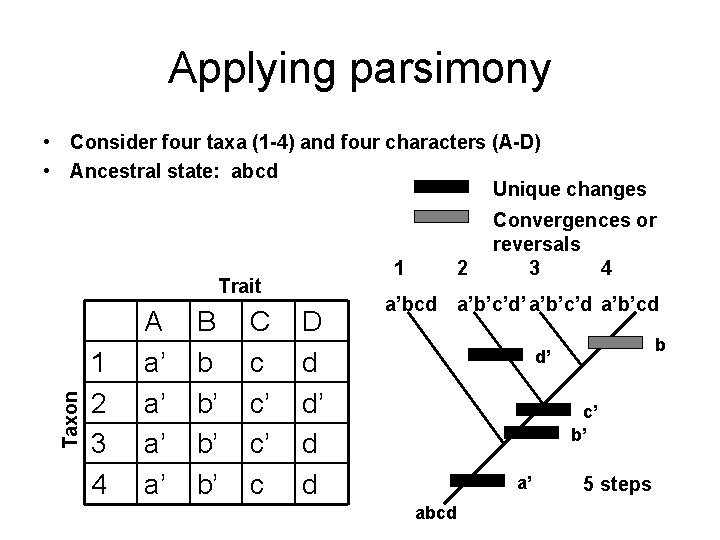
Applying parsimony • Consider four taxa (1 -4) and four characters (A-D) • Ancestral state: abcd Unique changes 1 Taxon Trait 1 2 3 4 A a’ a’ B b b’ b’ b’ C c c’ c’ c D d d’ d d 2 Convergences or reversals 3 4 a’bcd a’b’c’d’ a’b’c’d a’b’cd b d’ c’ b’ a’ abcd 5 steps

Applying parsimony • Consider four taxa (1 -4) and four characters (A-D) • Ancestral state: abcd Unique changes 1 Taxon Trait 1 2 3 4 A a’ a’ B b b’ b’ b’ C c c’ c’ c D d d’ d d 4 Convergences or reversals 3 2 a’bcd a’b’c’d’ d’ c’ b’ a’ abcd 4 steps

Strengths and weaknesses of parsimony Strengths Weaknesses.

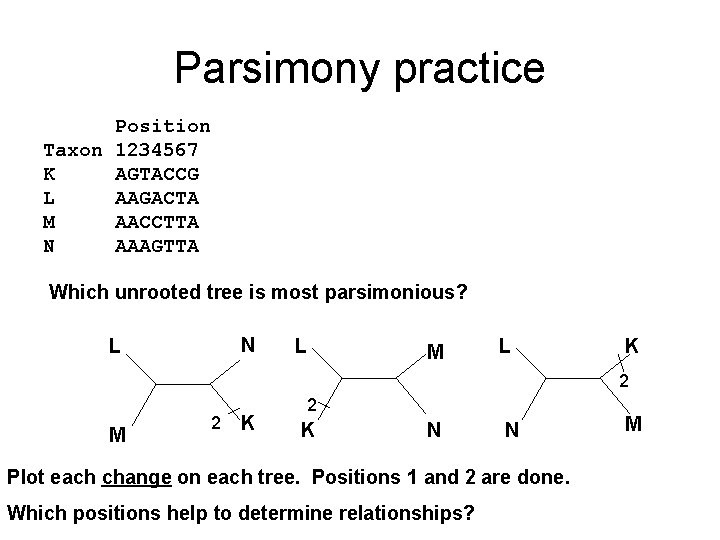
Parsimony practice Position Taxon 1234567 K AGTACCG L AAGACTA M AACCTTA N AAAGTTA Which unrooted tree is most parsimonious? N L L M L K 2 M 2 K N N Plot each change on each tree. Positions 1 and 2 are done. Which positions help to determine relationships? M

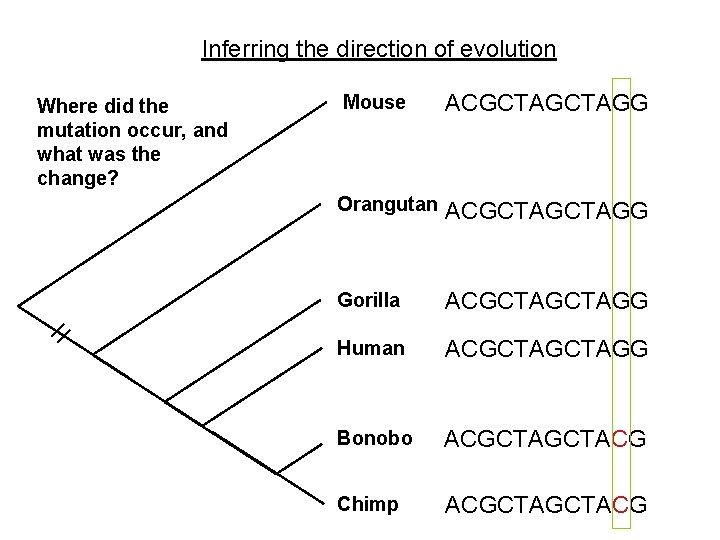
Inferring the direction of evolution Where did the mutation occur, and what was the change? Mouse ACGCTAGG Orangutan ACGCTAGG Gorilla ACGCTAGG Human ACGCTAGG Bonobo ACGCTACG Chimp ACGCTACG

III. Maximum likelihood (and Bayesian)

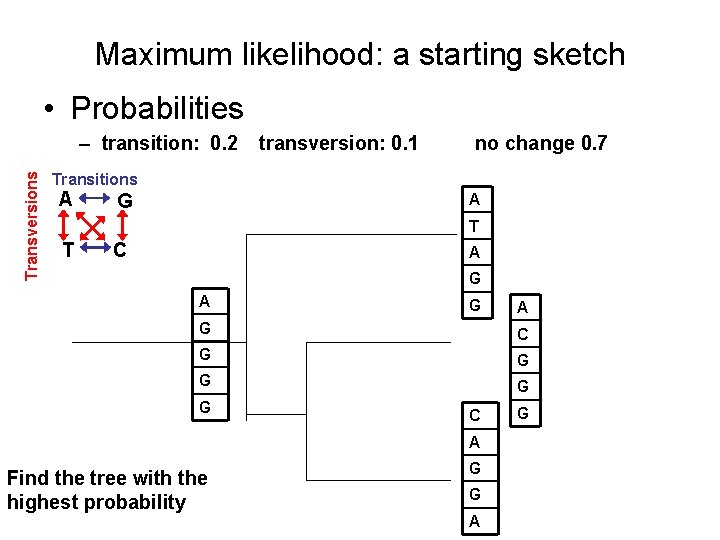
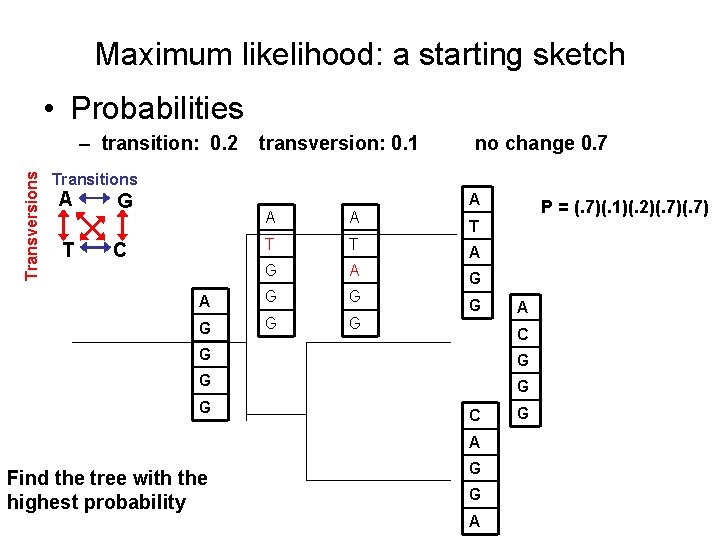
Maximum likelihood: a starting sketch • Probabilities Transversions – transition: 0. 2 transversion: 0. 1 no change 0. 7 Transitions A G A T T C A G A G C G G G C A Find the tree with the highest probability G G A G

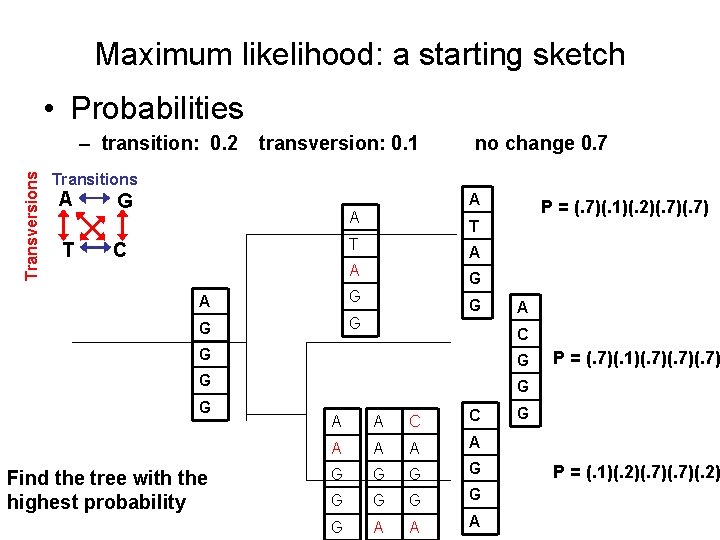
Maximum likelihood: a starting sketch • Probabilities Transversions – transition: 0. 2 transversion: 0. 1 no change 0. 7 Transitions A T G A A A T T G A A G G G C P = (. 7)(. 1)(. 2)(. 7) T A G G A C G G G C A Find the tree with the highest probability G G A G

Maximum likelihood: a starting sketch • Probabilities Transversions – transition: 0. 2 transversion: 0. 1 no change 0. 7 Transitions A T G A A T T C A A A G G G P = (. 7)(. 1)(. 2)(. 7) G G A C G G G Find the tree with the highest probability A A C C A A G G G G G A A A P = (. 7)(. 1)(. 7)(. 7) G P = (. 1)(. 2)(. 7)(. 2)

Assessment of Maximum Likelihood (also Bayesian) • Strengths • Weaknesses

Characters to use in phylogeny • Morphology • DNA sequence

Challenges of using DNA data Alignment can be very challenging! Taxon 1 AATGCGC Taxon 2 AATCGCT Taxon 1 Taxon 2 AATGCGC

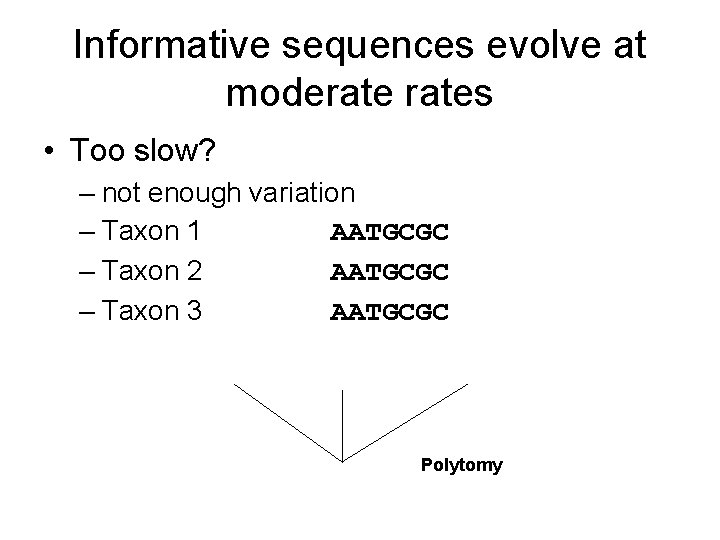
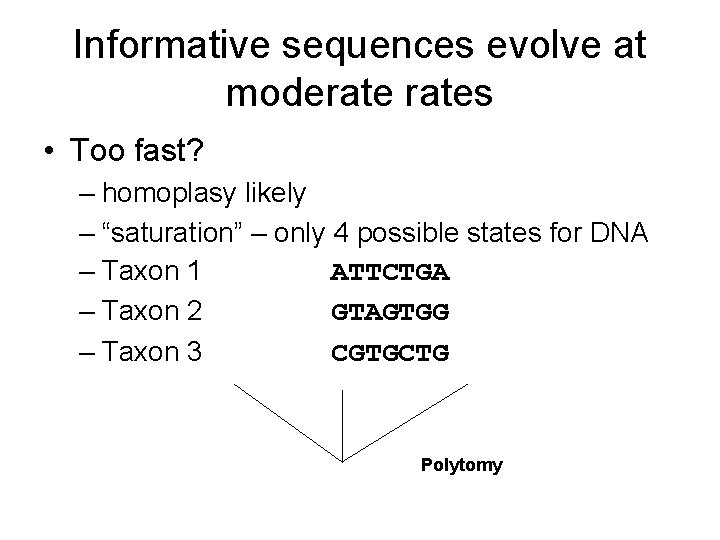
Informative sequences evolve at moderates • Too slow? – not enough variation – Taxon 1 AATGCGC – Taxon 2 AATGCGC – Taxon 3 AATGCGC Polytomy

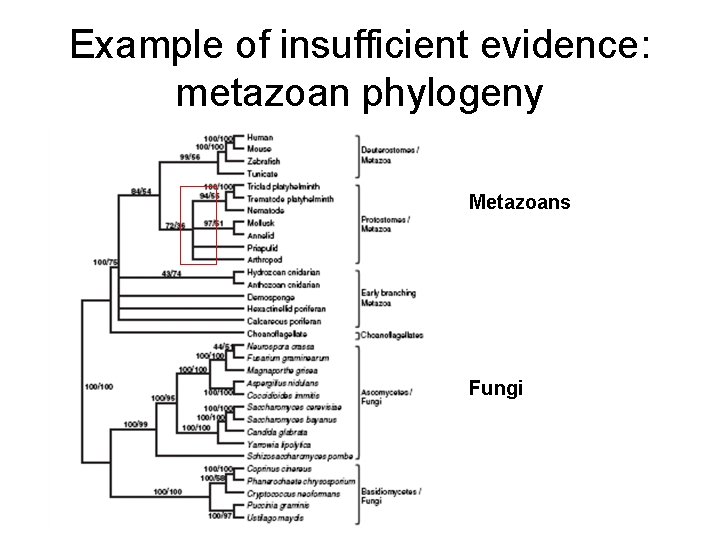
Example of insufficient evidence: metazoan phylogeny Metazoans Fungi

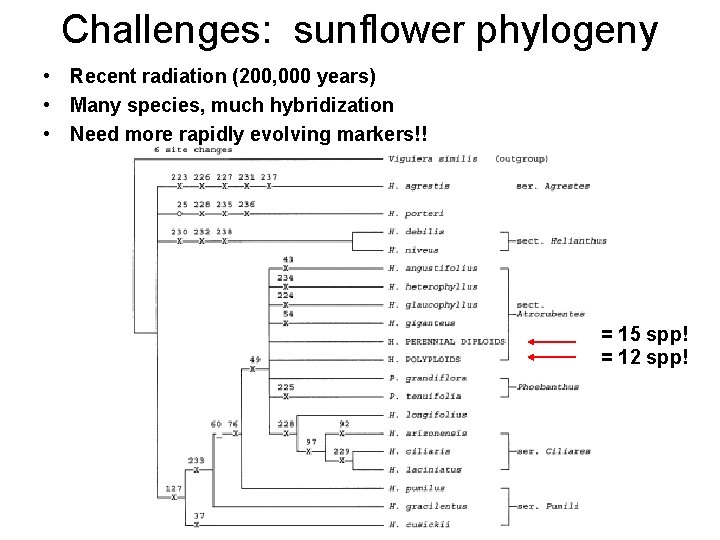
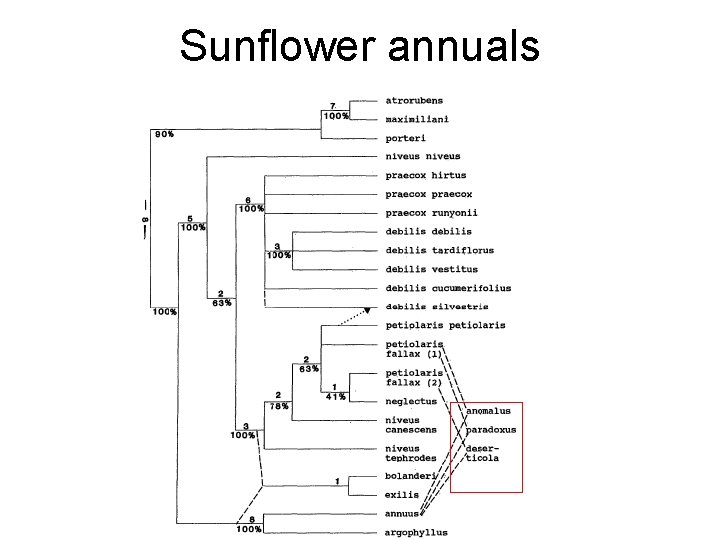
Challenges: sunflower phylogeny • Recent radiation (200, 000 years) • Many species, much hybridization • Need more rapidly evolving markers!! = 15 spp! = 12 spp!

Informative sequences evolve at moderates • Too fast? – homoplasy likely – “saturation” – only 4 possible states for DNA – Taxon 1 ATTCTGA – Taxon 2 GTAGTGG – Taxon 3 CGTGCTG Polytomy

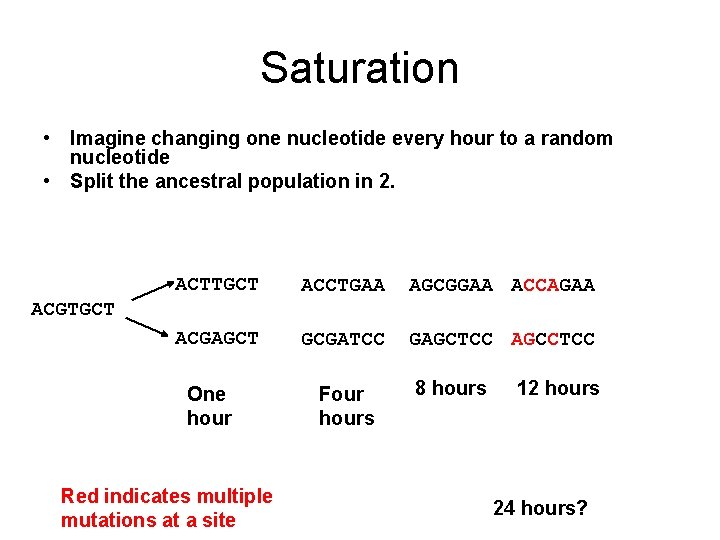
Saturation • Imagine changing one nucleotide every hour to a random nucleotide • Split the ancestral population in 2. ACTTGCT ACCTGAA AGCGGAA ACCAGAA ACGAGCT GCGATCC GAGCTCC AGCCTCC ACGTGCT One hour Red indicates multiple mutations at a site Four hours 8 hours 12 hours 24 hours?

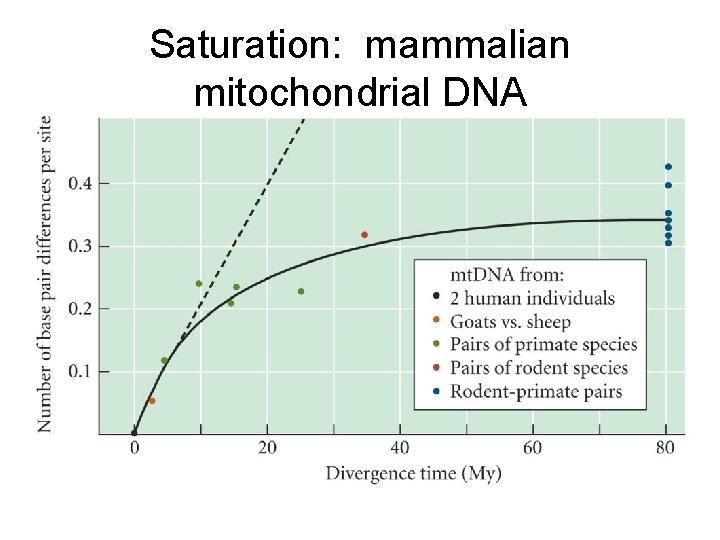
Saturation: mammalian mitochondrial DNA

Forces of evolution and phylogeny reconstruction How does each force affect the ability to reconstruct phylogeny? mutation? drift? selection? non-random mating? migration?

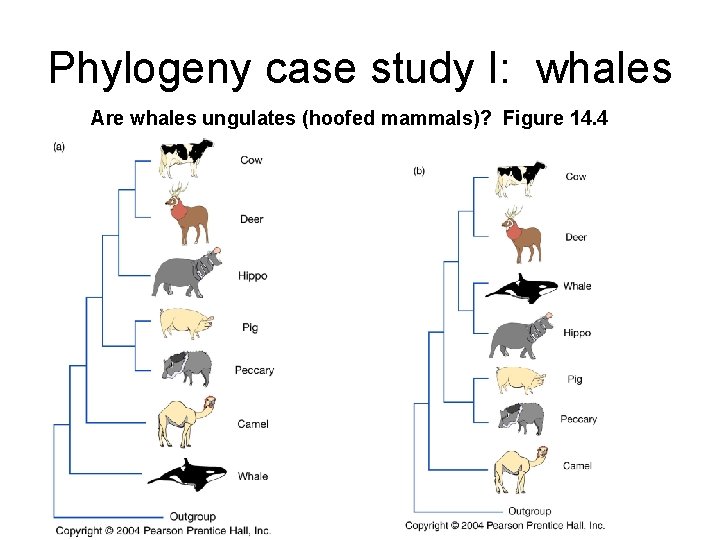
Phylogeny case study I: whales Are whales ungulates (hoofed mammals)? Figure 14. 4

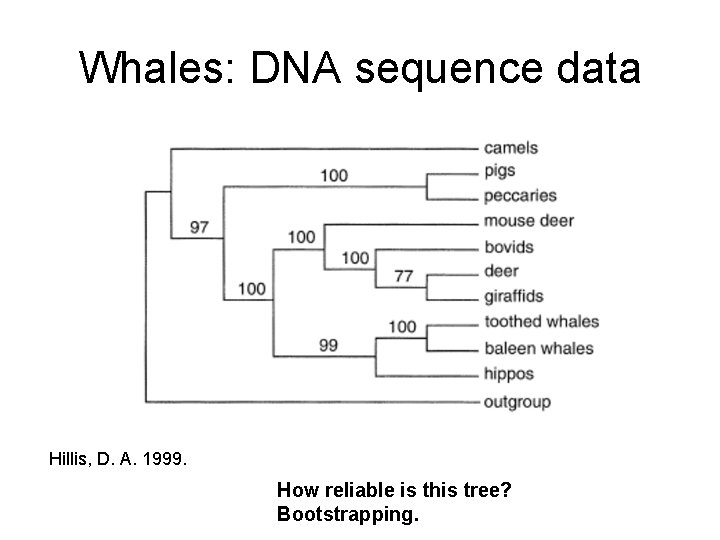
Whales: DNA sequence data Hillis, D. A. 1999. How reliable is this tree? Bootstrapping.

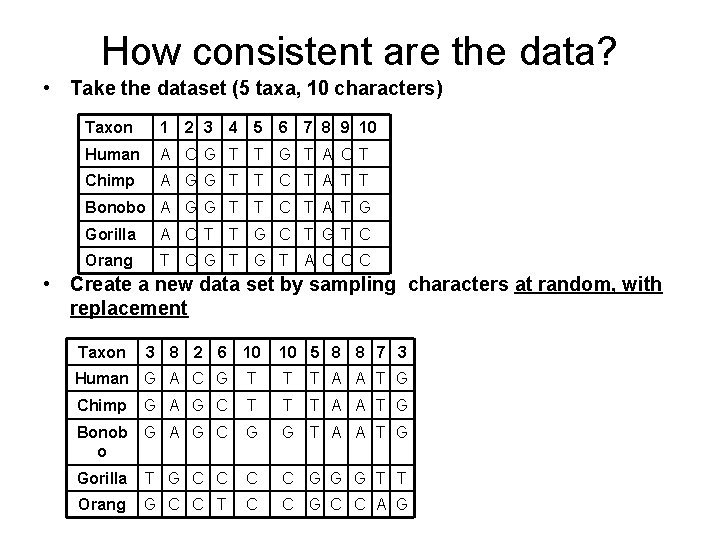
How consistent are the data? • Take the dataset (5 taxa, 10 characters) Taxon 1 2 3 4 5 6 7 8 9 10 Human A C G T T G T A C T Chimp A GG T T C T A T T Bonobo A G G T T C T A T G Gorilla A C T T G C T GT C Orang T C G T A C C C • Create a new data set by sampling characters at random, with replacement Taxon 3 8 2 6 10 10 5 8 8 7 3 Human G A C G T T T A A T G Chimp G A G C T T T A A T G Bonob o G A G C G G T A A T G Gorilla T G C C G G G T T Orang G C C T C C G C C A G

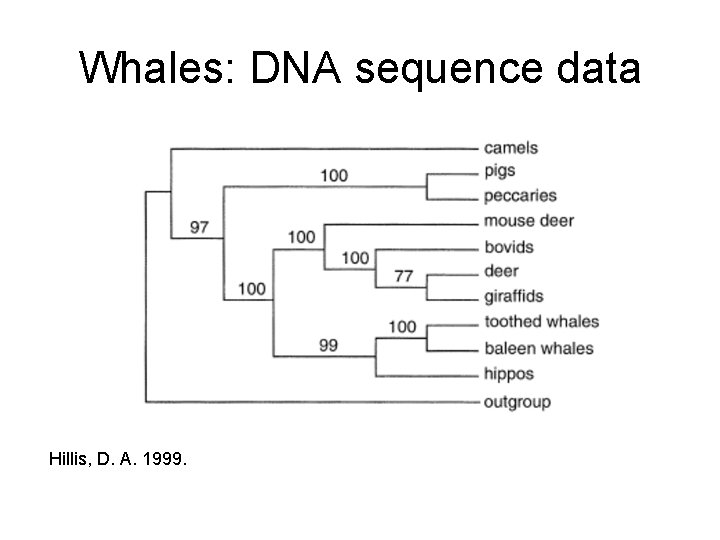
Whales: DNA sequence data Hillis, D. A. 1999.

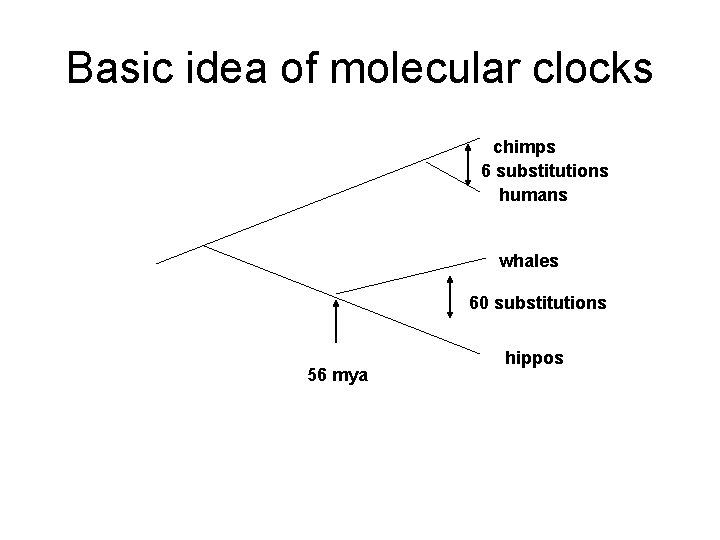
Molecular clocks

Basic idea of molecular clocks chimps 6 substitutions humans whales 60 substitutions 56 mya hippos

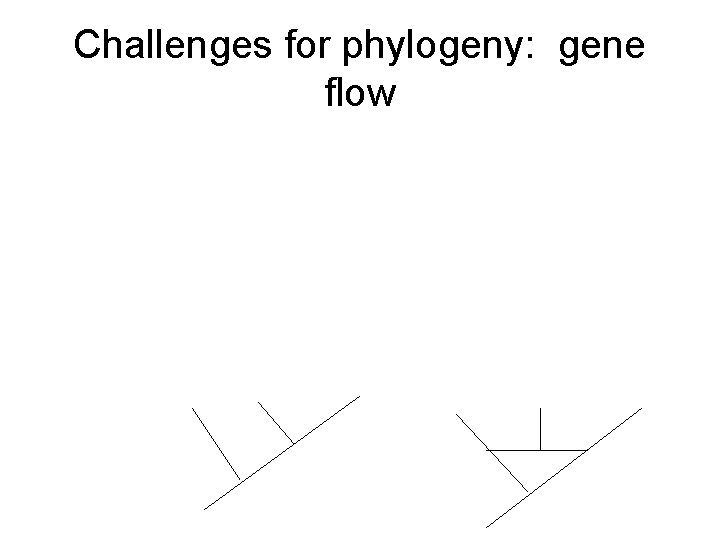
Challenges for phylogeny: gene flow

Sunflower annuals

Different genes may have different histories!

Phylogeny summary

Phylogeny study questions 1) Explain in words the difference between monophyletic, paraphyletic, and polyphyletic taxa. Draw a hypothetical phylogeny representing each type. Give an actual example of a commonly recognized paraphyletic taxon in both animals and in plants. 2) How can a reconstructed phylogeny be used to determine if a similar character in two taxa is due to homoplasy? 3) Whales are classified as cetaceans, not artiodactyl ungulates. This makes artiodactyls paraphyletic – why? What is the evidence that whales belong in the artiodactyls? 4) Phenetics (distance methods) and cladistics (parsimony) differ in the ways they recognize and use similarities among taxa to form phylogenetic groupings. What types of similarity does each school recognize, and how useful is each type of similarity considered to be for identifying groups?

Phylogeny study questions 5) What is “bootstrapping” in the context of phylogenetic analysis, and why is this procedure performed? 6) Why are maximum likelihood methods increasing in popularity for reconstructing phylogenies? In your answer, include a short description of how this method identifies the best phylogeny. 7) For what kinds of data can maximum likelihood methods of phylogeny construction be used? Why is this so? What types of data are typically not used, and why? 8) Would animal mitochondrial DNA provide a reasonable molecular tool for evaluating deep phylogenetic relationships between animal phyla? What about ribosomal DNA? Justify your answers. 9) Integrative question: Draw a pair of axes with “Time since divergence” on the x axis and “percent of sites that are the same” on the y axis. Draw a graph that shows the basic pattern for third codon sites: is your graph linear? Explain why or why not.

Phylogeny study questions 10) You are studying a group of species that lives in two very different environments. You build two phylogenies: one is based on a locus that is probably under divergent selection in the two environments, while the other phylogeny is based on a neutral locus. Which phylogeny would be more likely to represent the species history? why? 11) For a number of years, Anolis lizards are found in similar microhabitats on many separate islands in the Carribean are very similar to each other (for example, large lizards that feed on the ground, smaller lizards that feed on tree trunks, and very small lizards that feed at the tops of branches). Two different, historical explanations have been proposed to explain this pattern: each morph has evolved repeatedly on each island, or each morph has evolved just once, then dipsersed. Sketch a phylogeny that would support each hypothesis. 12) Integrative question: the Cameroon lake cichlid phylogeny, showing that the lake species were monophyletic, was based on mitochondrial DNA. Explain why this might not reflect the species history. How could you be more certain about the phylogeny? 13) Explain why allopolyploid taxa pose problems for phylogenies.