Molecular Phylogeny Phylogeny is the inference of evolutionary











- Slides: 46

Molecular Phylogeny

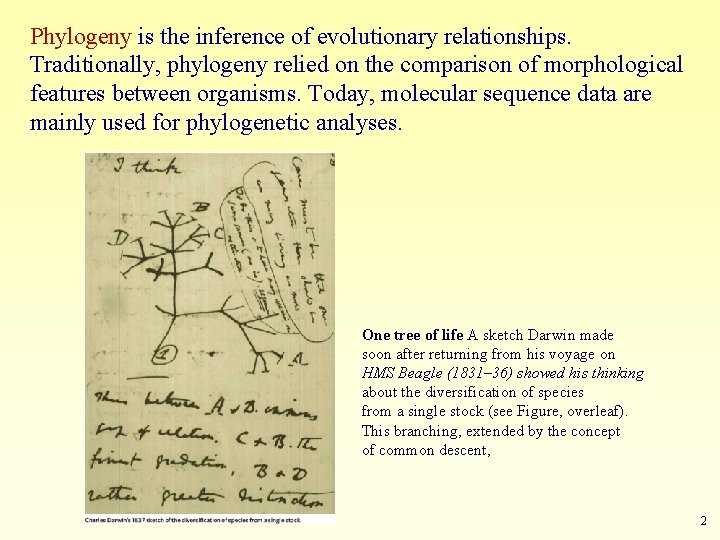
Phylogeny is the inference of evolutionary relationships. Traditionally, phylogeny relied on the comparison of morphological features between organisms. Today, molecular sequence data are mainly used for phylogenetic analyses. One tree of life A sketch Darwin made soon after returning from his voyage on HMS Beagle (1831– 36) showed his thinking about the diversification of species from a single stock (see Figure, overleaf). This branching, extended by the concept of common descent, 2

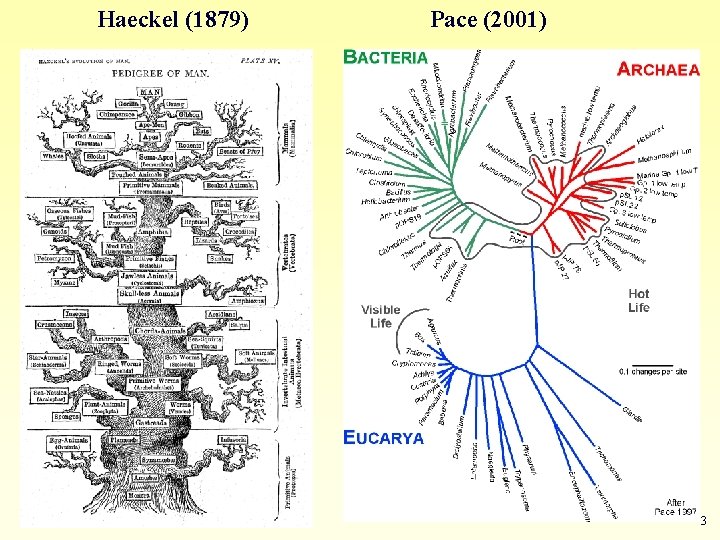
Haeckel (1879) Pace (2001) 3

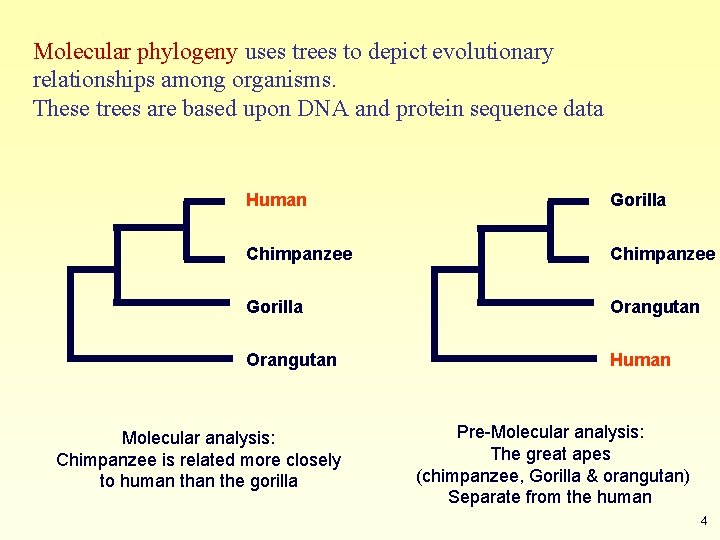
Molecular phylogeny uses trees to depict evolutionary relationships among organisms. These trees are based upon DNA and protein sequence data Human Gorilla Chimpanzee Gorilla Orangutan Human Molecular analysis: Chimpanzee is related more closely to human the gorilla Pre-Molecular analysis: The great apes (chimpanzee, Gorilla & orangutan) Separate from the human 4

What can we learn from phylogenetics tree? 5

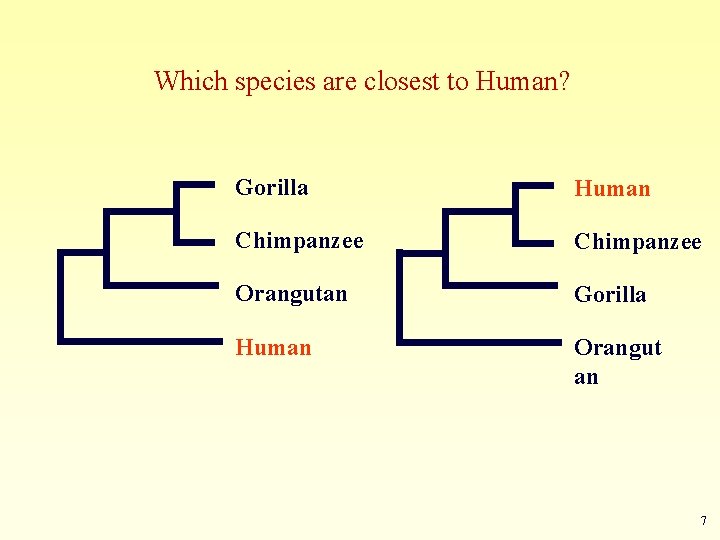
1. Determine the closest relatives of one organism in which we are interested • Was the extinct quagga more like a zebra or a horse?

Which species are closest to Human? Gorilla Human Chimpanzee Orangutan Gorilla Human Orangut an 7

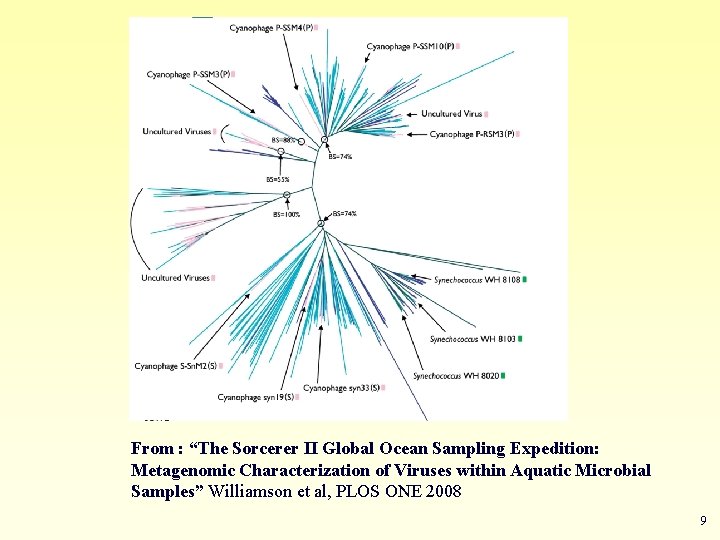
2. Help to find the relationship between the species and identify new species Example Metagenomics A new field in genomics aims the study the genomes recovered from environmental samples. A powerful tool to access the wealthy biodiversity of native environmental samples 8

From : “The Sorcerer II Global Ocean Sampling Expedition: Metagenomic Characterization of Viruses within Aquatic Microbial Samples” Williamson et al, PLOS ONE 2008 9

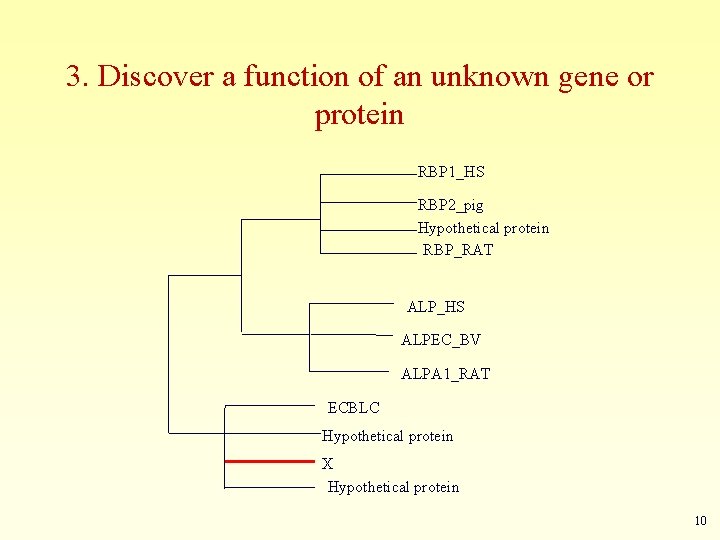
3. Discover a function of an unknown gene or protein RBP 1_HS RBP 2_pig Hypothetical protein RBP_RAT ALP_HS ALPEC_BV ALPA 1_RAT ECBLC Hypothetical protein X Hypothetical protein 10

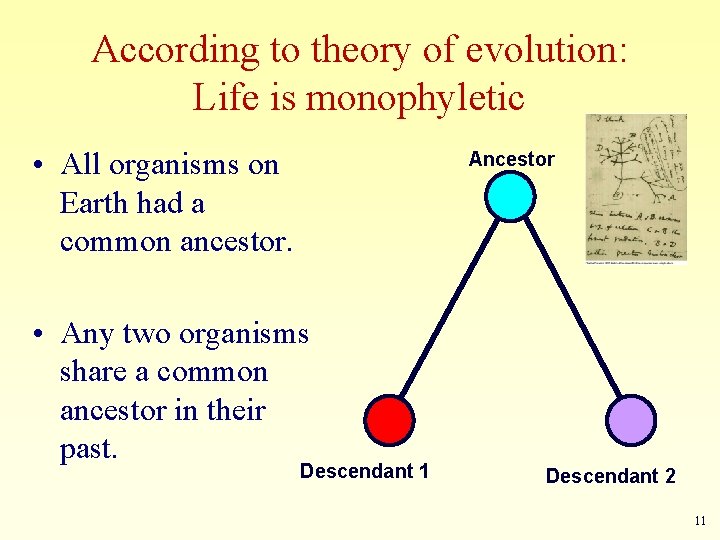
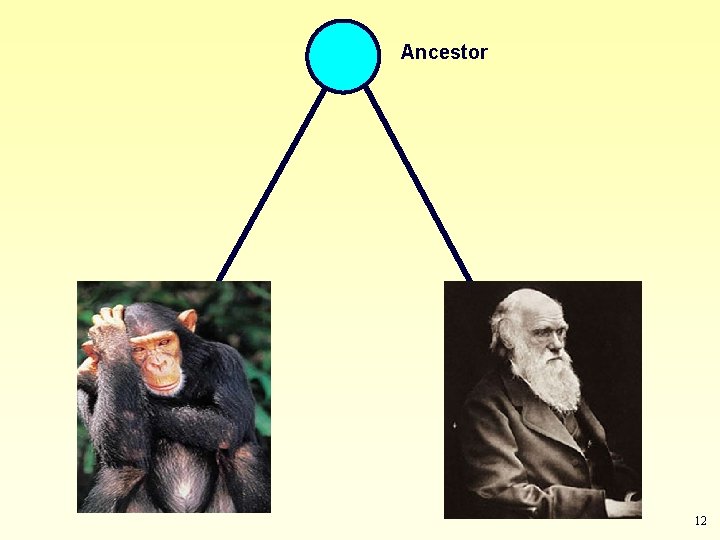
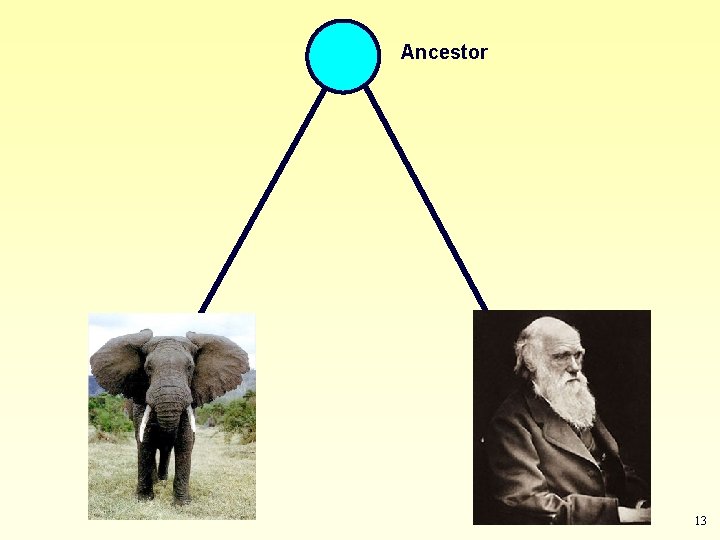
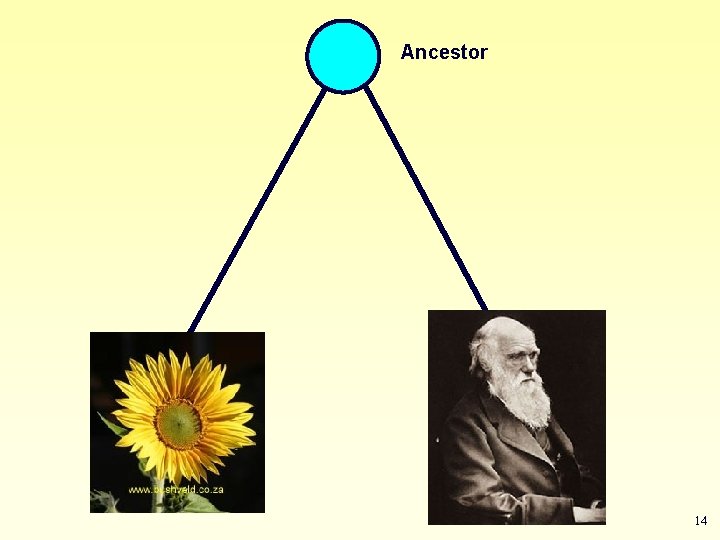
According to theory of evolution: Life is monophyletic • All organisms on Earth had a common ancestor. Ancestor • Any two organisms share a common ancestor in their past. Descendant 1 Descendant 2 11

Ancestor 12

Ancestor 13

Ancestor 14

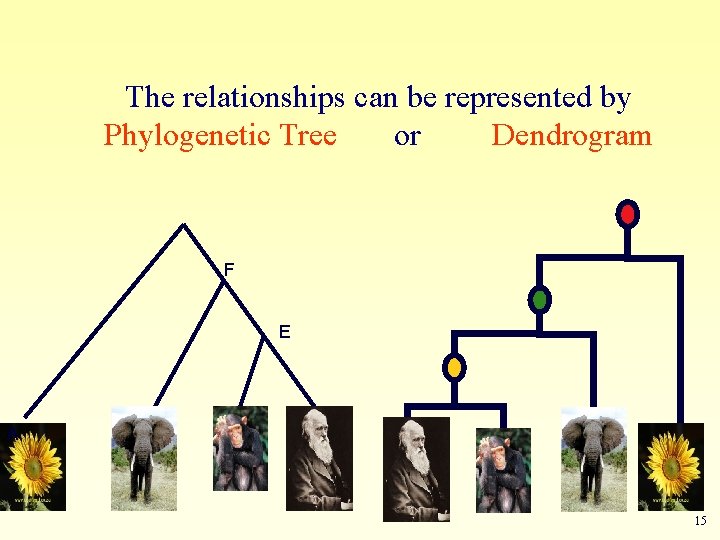
The relationships can be represented by Phylogenetic Tree or Dendrogram F E A B C D 15

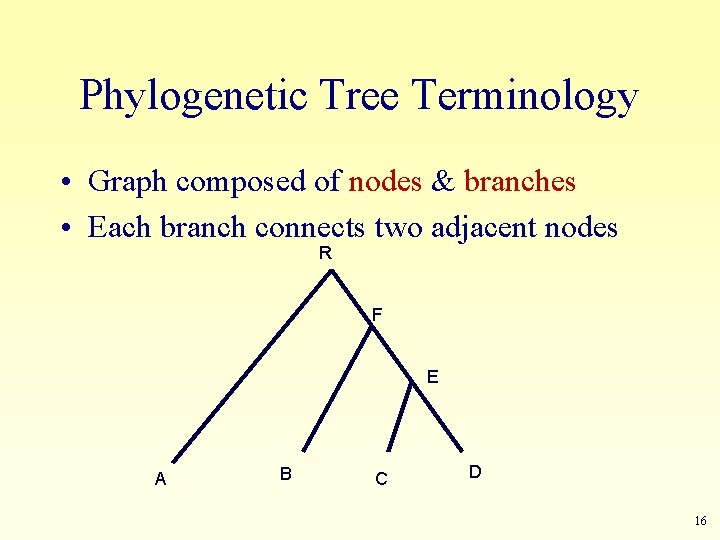
Phylogenetic Tree Terminology • Graph composed of nodes & branches • Each branch connects two adjacent nodes R F E A B C D 16

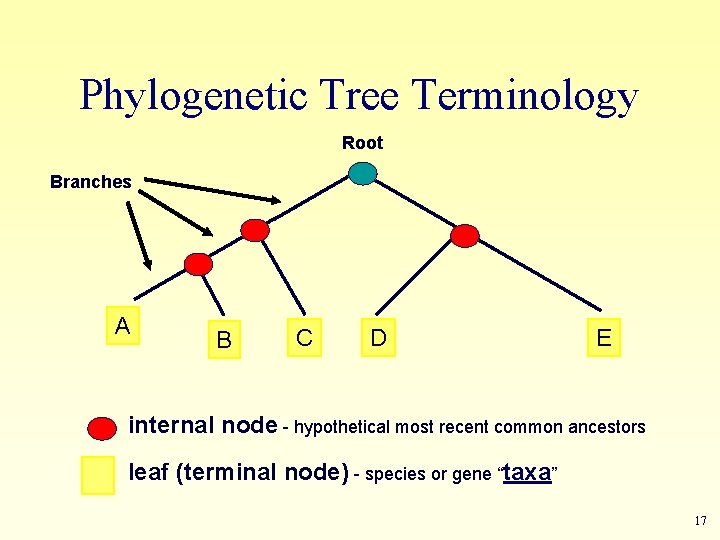
Phylogenetic Tree Terminology Root Branches A B C D E internal node - hypothetical most recent common ancestors leaf (terminal node) - species or gene “taxa” 17

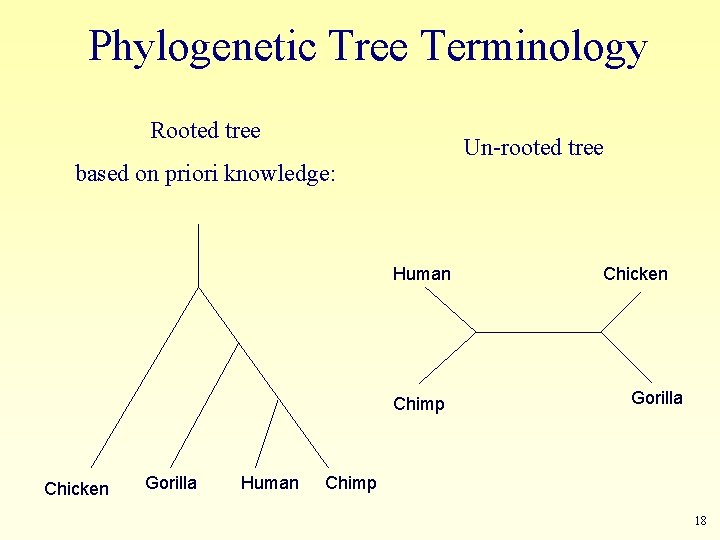
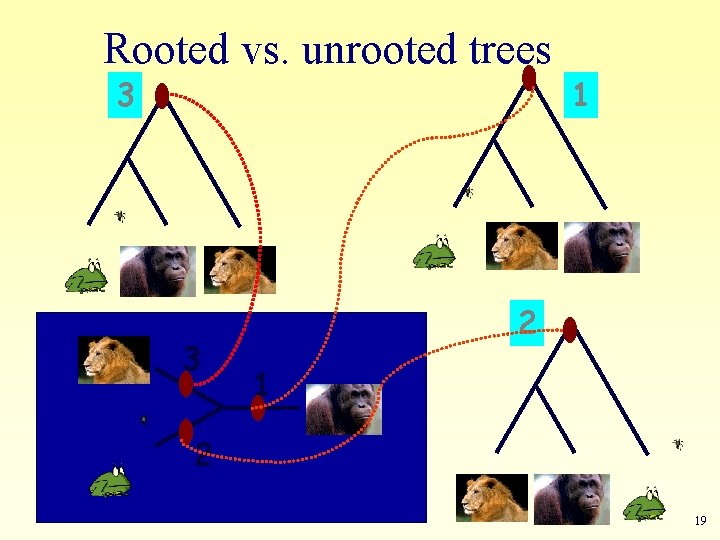
Phylogenetic Tree Terminology Rooted tree Un-rooted tree based on priori knowledge: Human Chimp Chicken Gorilla Human Chicken Gorilla Chimp 18

Rooted vs. unrooted trees 3 3 1 2 19

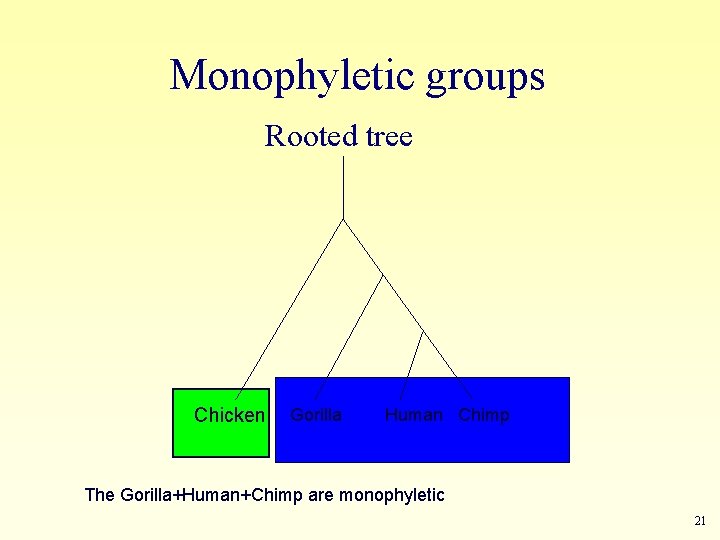
Monophyletic groups (clades): A group is monophyletic (clade) if it has a common ancestor and all the descendents of this ancestor are in the group. 20

Monophyletic groups Rooted tree Chicken Gorilla Human Chimp The Gorilla+Human+Chimp are monophyletic 21

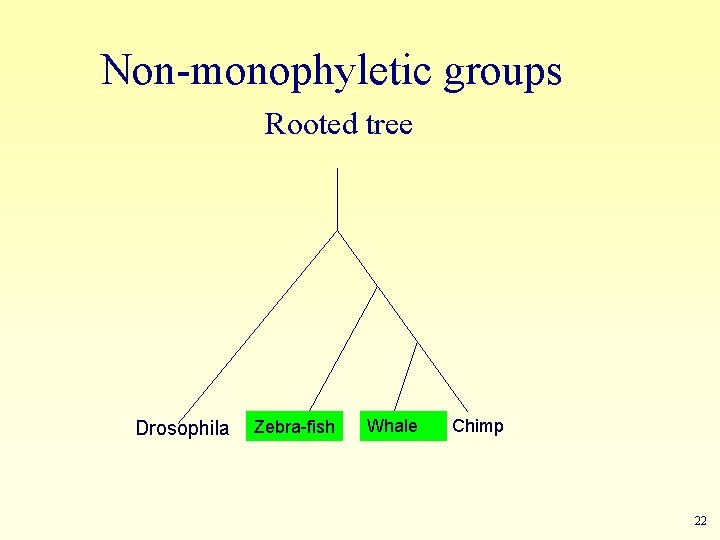
Non-monophyletic groups Rooted tree Drosophila Zebra-fish Whale Chimp 22

23

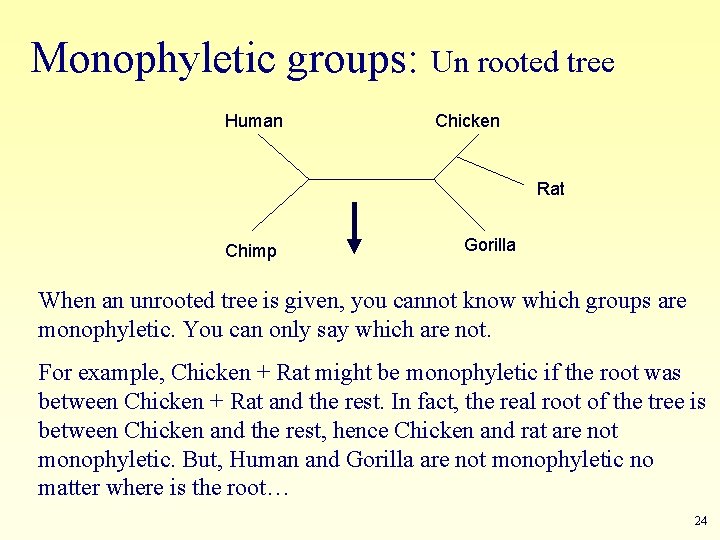
Monophyletic groups: Un rooted tree Human Chicken Rat Chimp Gorilla When an unrooted tree is given, you cannot know which groups are monophyletic. You can only say which are not. For example, Chicken + Rat might be monophyletic if the root was between Chicken + Rat and the rest. In fact, the real root of the tree is between Chicken and the rest, hence Chicken and rat are not monophyletic. But, Human and Gorilla are not monophyletic no matter where is the root… 24

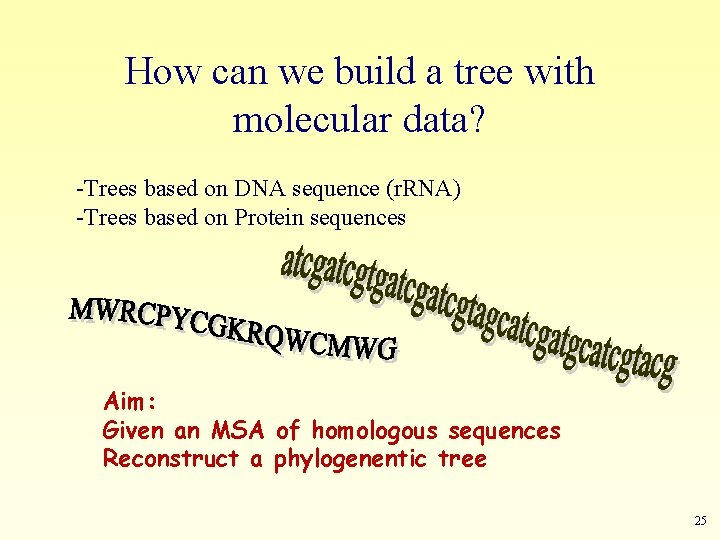
How can we build a tree with molecular data? -Trees based on DNA sequence (r. RNA) -Trees based on Protein sequences Aim: Given an MSA of homologous sequences Reconstruct a phylogenentic tree 25

Problems: • DNA and proteins from the same gene can produce different trees • Different genes may have different evolutionary history. • Different regions of the same gene can producec different trees 26

Methods 27

Approach 1 - Distance methods • Two steps : – Compute a distances between any two sequences from the MSA. – Find the tree that agrees most with the distance table. • Algorithms : -Neighbor joining Approach 2 - State methods • Algorithms: – Maximum parsimony (MP) – Maximum likelihood (ML) 28

Neighbor Joining (NJ) • Reconstructs unrooted tree • Calculates branch lengths Based on pairwise distance • In each stage, the two nearest nodes of the tree are chosen and defined as neighbors in our tree. This is done recursively until all of the nodes are paired together. 29

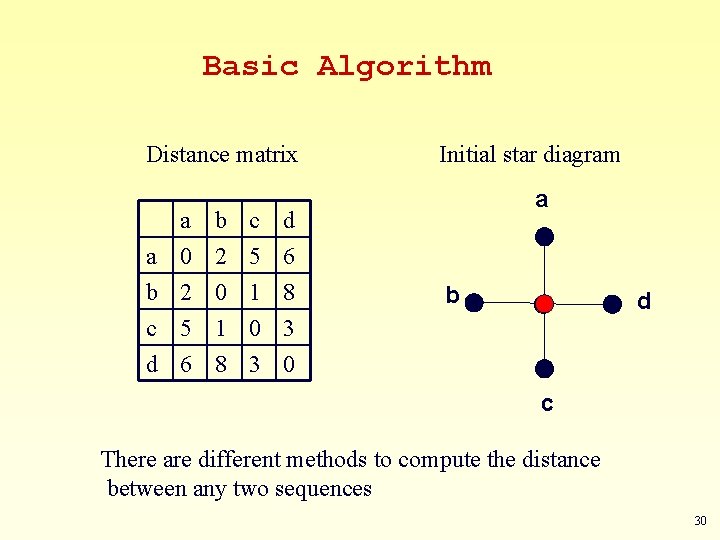
Basic Algorithm Distance matrix a a 0 b 2 c 5 b 2 0 1 c 5 1 0 d 6 8 3 Initial star diagram a b d d 6 8 3 0 c There are different methods to compute the distance between any two sequences 30

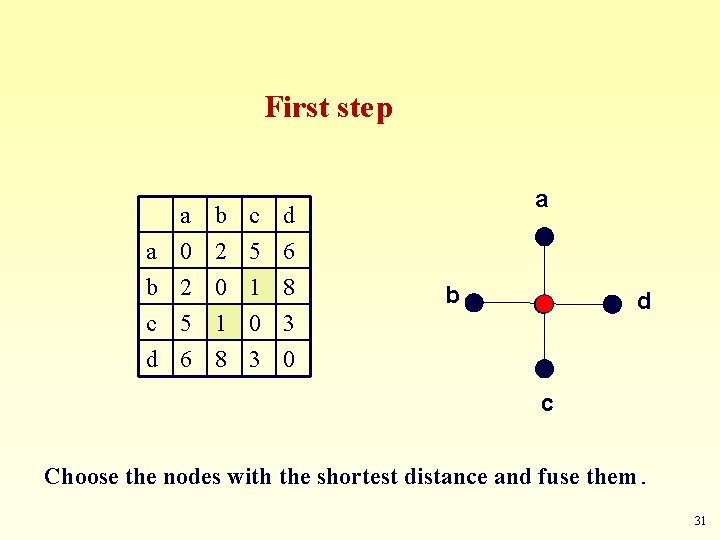
First step a a 0 b 2 c 5 b 2 0 1 c 5 1 0 d 6 8 3 a b d d 6 8 3 0 c Choose the nodes with the shortest distance and fuse them. 31

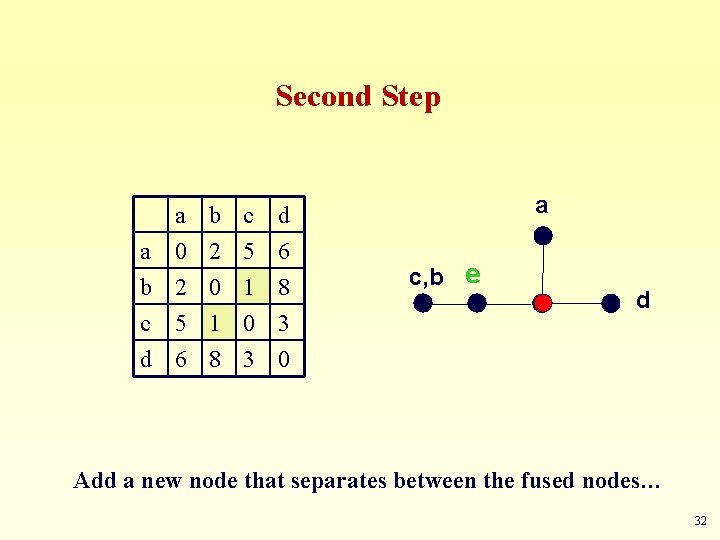
Second Step a a 0 b 2 c 5 b 2 0 1 c 5 1 0 d 6 8 3 a c, b e d d 6 8 3 0 Add a new node that separates between the fused nodes… 32

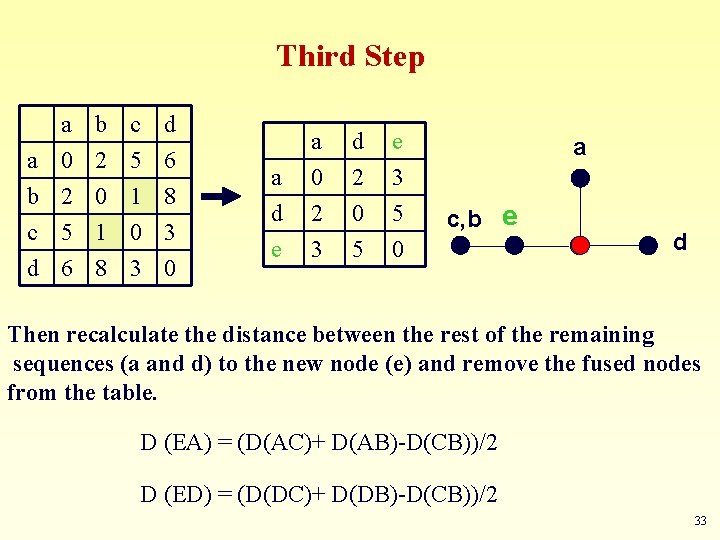
Third Step a a 0 b 2 c 5 b 2 0 1 c 5 1 0 d 6 8 3 0 a d e a 0 2 3 d 2 0 5 e 3 5 0 a c, b e d Then recalculate the distance between the rest of the remaining sequences (a and d) to the new node (e) and remove the fused nodes from the table. D (EA) = (D(AC)+ D(AB)-D(CB))/2 D (ED) = (D(DC)+ D(DB)-D(CB))/2 33

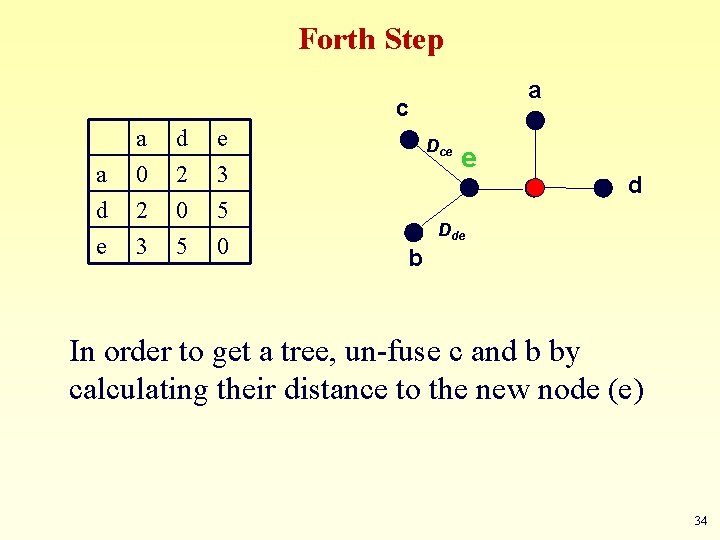
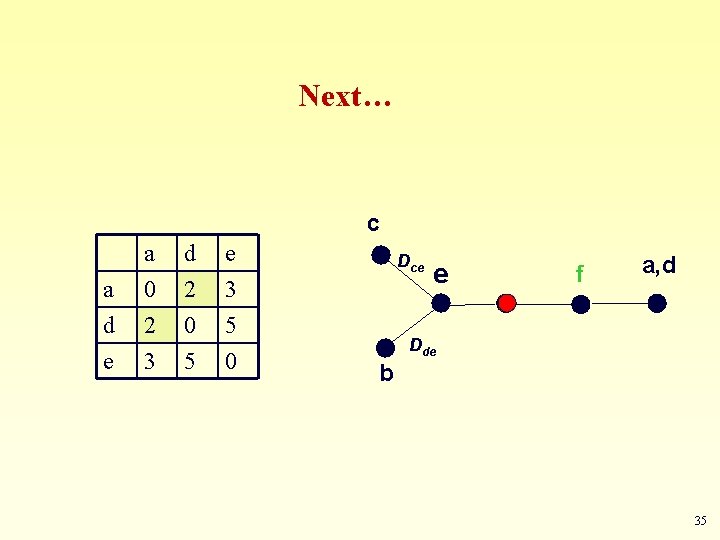
Forth Step a c a d e a 0 2 3 d 2 0 5 e 3 5 0 Dce e d Dde b In order to get a tree, un-fuse c and b by calculating their distance to the new node (e) 34

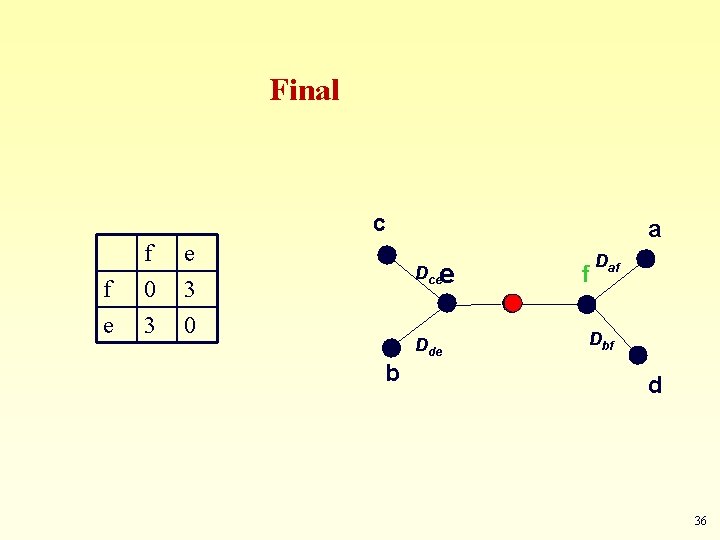
Next… c a d e a 0 2 3 d 2 0 5 e 3 5 0 Dce e f a, d Dde b 35

Final c f e f 0 3 e 3 0 a Dcee Dde b f Daf Dbf d 36

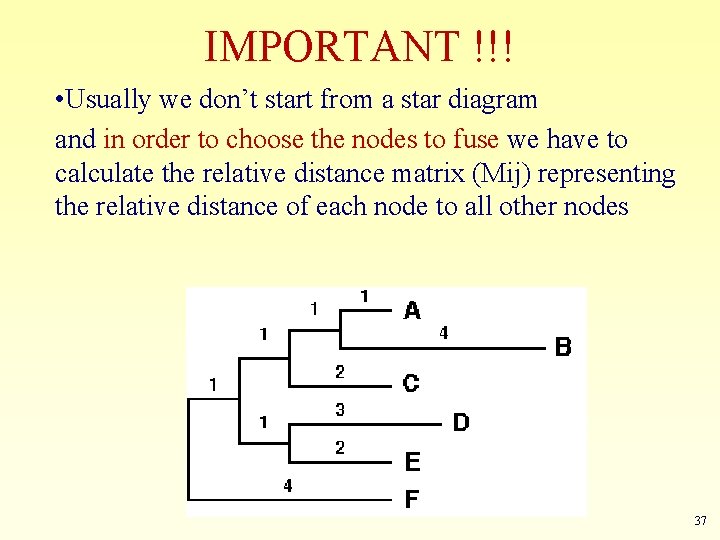
IMPORTANT !!! • Usually we don’t start from a star diagram and in order to choose the nodes to fuse we have to calculate the relative distance matrix (Mij) representing the relative distance of each node to all other nodes 37

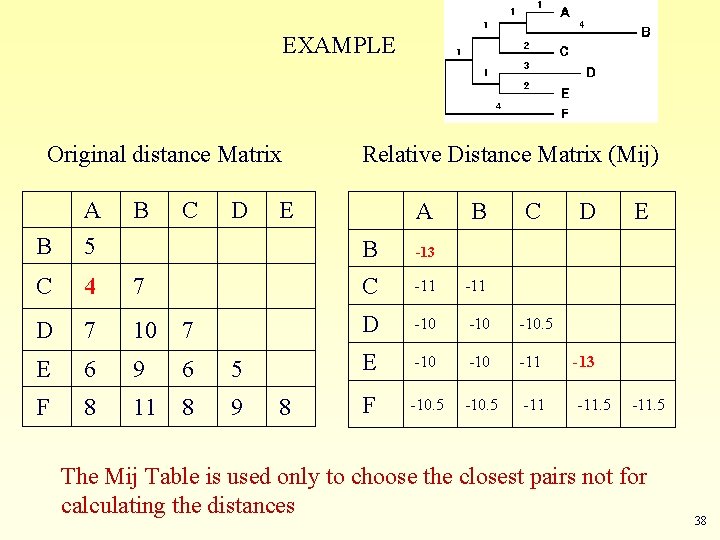
EXAMPLE Original distance Matrix Relative Distance Matrix (Mij) B A 5 B C D E A B C D E B -13 C 4 7 C -11 D 7 10 7 D -10 -10. 5 E 6 9 6 5 E -10 -11 -13 F 8 11 8 9 8 F -10. 5 -11. 5 The Mij Table is used only to choose the closest pairs not for calculating the distances 38

Advantages and disadvantages of the neighbor-joining method Advantages -It is fast and thus suited for large datasets -permits lineages with largely different branch lengths Disadvantages - sequence information is reduced - gives only one possible tree 39

More problems with phylogenetic trees • It is wrong to assume that branch length is proportional to speciation time (molecular clock). • It is wrong to produce a tree based on distance values of the whole alignment.

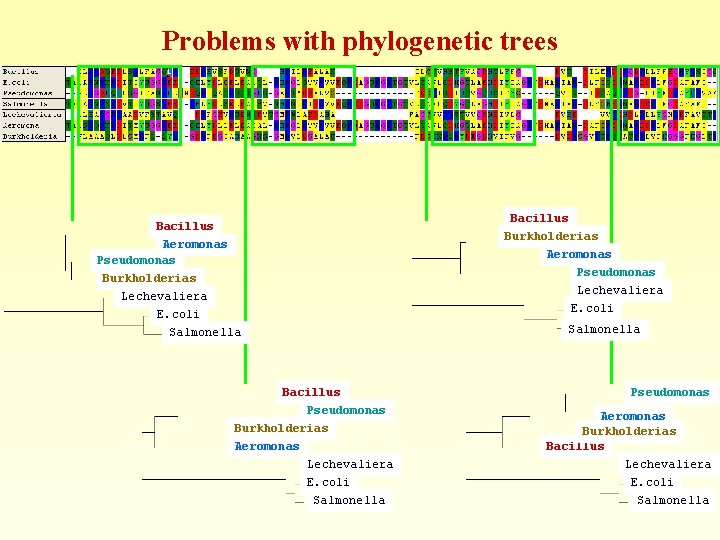
Problems with phylogenetic trees

Problems with phylogenetic trees Bacillus Aeromonas Pseudomonas Burkholderias Lechevaliera E. coli Salmonella Bacillus Pseudomonas Burkholderias Aeromonas Lechevaliera E. coli Salmonella Bacillus Burkholderias Aeromonas Pseudomonas Lechevaliera E. coli Salmonella Pseudomonas Aeromonas Burkholderias Bacillus Lechevaliera E. coli Salmonella

Problems with phylogenetic trees • It is wrong to assume that branch length is proportional to speciation time (molecular clock). • It is wrong to produce a tree based on distance values of the whole alignment : using different regions from a same alignment may produce different trees. • What to do? : use bootstrap

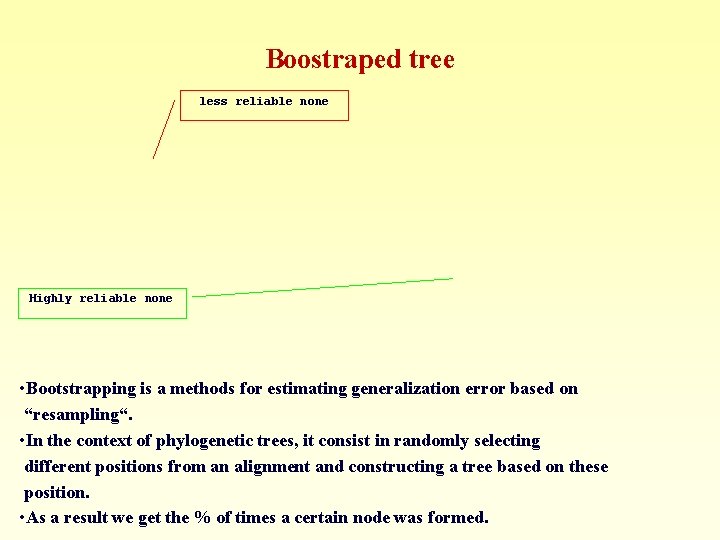
Boostraped tree less reliable none Highly reliable none • Bootstrapping is a methods for estimating generalization error based on “resampling“. • In the context of phylogenetic trees, it consist in randomly selecting different positions from an alignment and constructing a tree based on these position. • As a result we get the % of times a certain node was formed.

Tools for tree reconstruction • CLUSTALX (NJ method) • Phylip -PHYLogeny Inference Package – includes parsimony, distance matrix, and likelihood methods, including bootstrapping. • Phyml (maximum likelihood method) • More phylogeny programs 45

362 46
 Mega molecular
Mega molecular Molecular evolutionary genetics analysis
Molecular evolutionary genetics analysis Covalently bonded substances
Covalently bonded substances Ionic covalent metallic
Ionic covalent metallic Zinc oxide + nitric acid → zinc nitrate + water
Zinc oxide + nitric acid → zinc nitrate + water Closed circulatory system
Closed circulatory system Barnacle phylogeny
Barnacle phylogeny Craniata
Craniata Homologies
Homologies Animal kingdom cladogram
Animal kingdom cladogram Cluster omega
Cluster omega What is an outgroup in a cladogram
What is an outgroup in a cladogram Classification of platyhelminthes
Classification of platyhelminthes Phylogeny is the study of _____.
Phylogeny is the study of _____. Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Animal kingdom phylogenetic tree
Animal kingdom phylogenetic tree Anatomy of trout
Anatomy of trout Chapter 20 phylogeny and the tree of life
Chapter 20 phylogeny and the tree of life What is a mammal
What is a mammal Artiodactyla phylogeny
Artiodactyla phylogeny What is a sister group in phylogeny
What is a sister group in phylogeny Metazoan phylogeny
Metazoan phylogeny Taxa phylogenetic tree
Taxa phylogenetic tree Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Enantiornithes
Enantiornithes Symplesiomorphy
Symplesiomorphy Ontogeny recapitulates phylogeny
Ontogeny recapitulates phylogeny Protochordates are exclusively marine and include
Protochordates are exclusively marine and include Fish phylogeny
Fish phylogeny Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Aaabbbcccdd
Aaabbbcccdd Amphibian heart
Amphibian heart Rooted vs unrooted phylogenetic tree
Rooted vs unrooted phylogenetic tree Cat family tree
Cat family tree Monophyletic group
Monophyletic group Tỉ lệ cơ thể trẻ em
Tỉ lệ cơ thể trẻ em Lời thề hippocrates
Lời thề hippocrates đại từ thay thế
đại từ thay thế Vẽ hình chiếu đứng bằng cạnh của vật thể
Vẽ hình chiếu đứng bằng cạnh của vật thể Quá trình desamine hóa có thể tạo ra
Quá trình desamine hóa có thể tạo ra Các môn thể thao bắt đầu bằng tiếng bóng
Các môn thể thao bắt đầu bằng tiếng bóng Công thức tính thế năng
Công thức tính thế năng Thế nào là mạng điện lắp đặt kiểu nổi
Thế nào là mạng điện lắp đặt kiểu nổi Hát kết hợp bộ gõ cơ thể
Hát kết hợp bộ gõ cơ thể Khi nào hổ mẹ dạy hổ con săn mồi
Khi nào hổ mẹ dạy hổ con săn mồi Dot
Dot Thế nào là sự mỏi cơ
Thế nào là sự mỏi cơ