Chapter 26 Phylogeny and the Tree of Life



- Slides: 27

Chapter 26 Phylogeny and the Tree of Life

What you need to know: • The taxonomic categories and how they indicate relatedness. • How systematics is used to develop phylogenetic trees. • The three domains of life including their similarities and their differences.

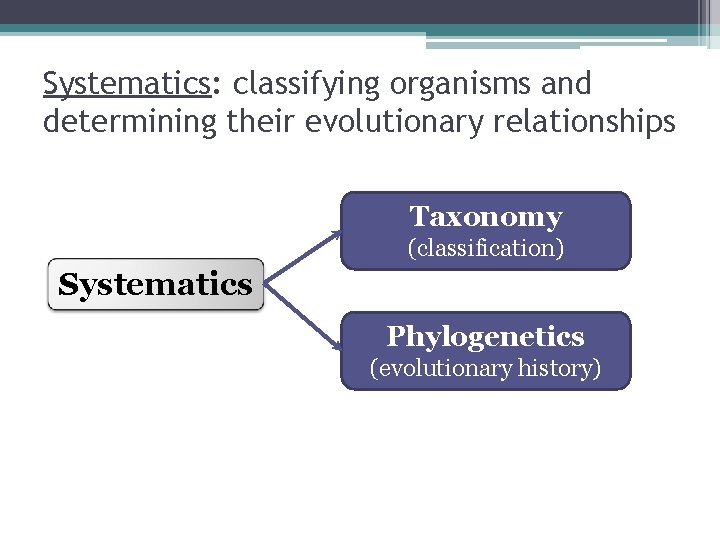
Systematics: classifying organisms and determining their evolutionary relationships Taxonomy (classification) Systematics Phylogenetics (evolutionary history)

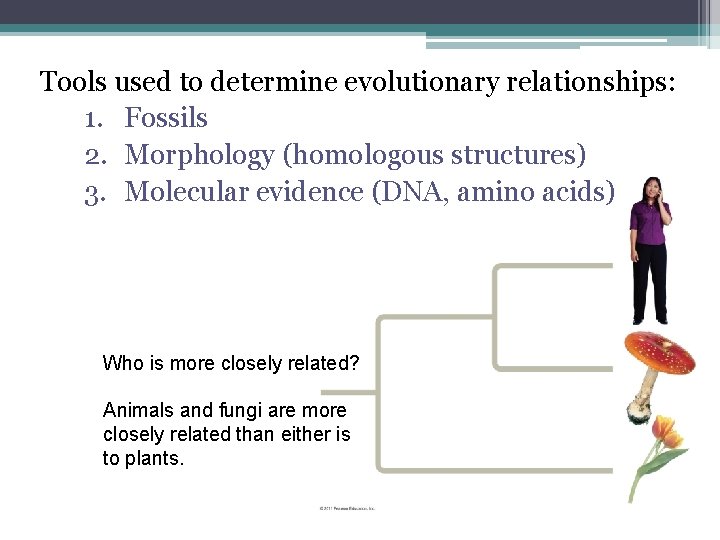
Tools used to determine evolutionary relationships: 1. Fossils 2. Morphology (homologous structures) 3. Molecular evidence (DNA, amino acids) Who is more closely related? Animals and fungi are more closely related than either is to plants.

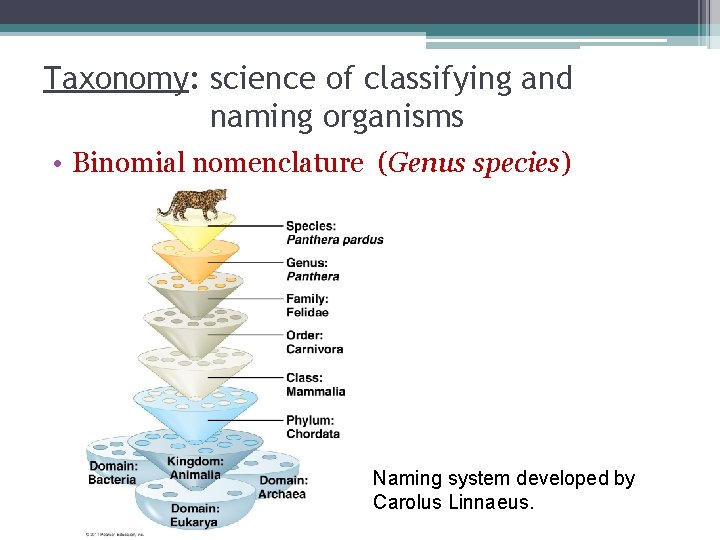
Taxonomy: science of classifying and naming organisms • Binomial nomenclature (Genus species) Naming system developed by Carolus Linnaeus.

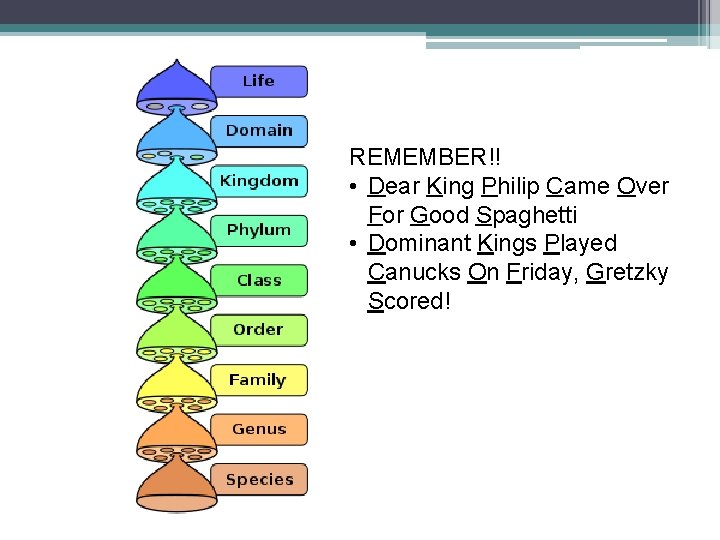
REMEMBER!! • Dear King Philip Came Over For Good Spaghetti • Dominant Kings Played Canucks On Friday, Gretzky Scored!

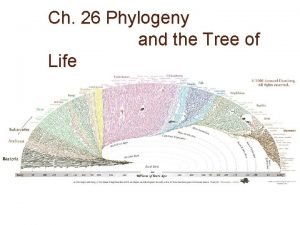
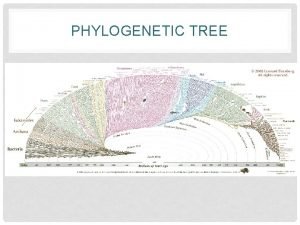
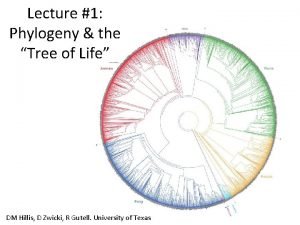
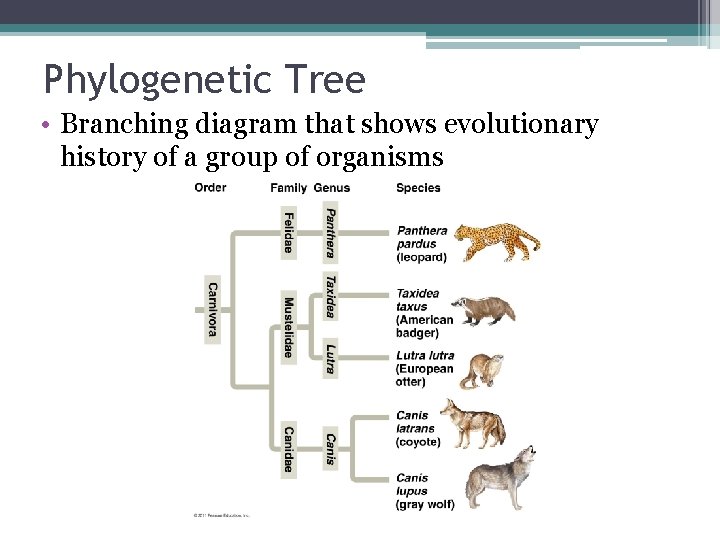
Phylogenetic Tree • Branching diagram that shows evolutionary history of a group of organisms

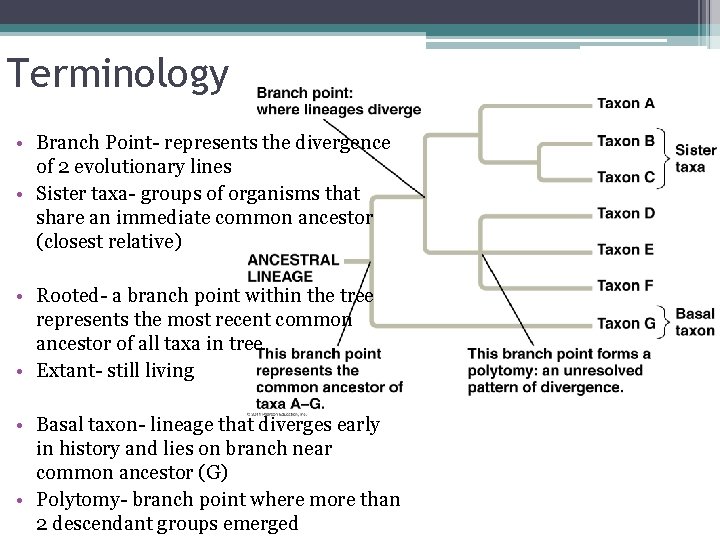
Terminology • Branch Point- represents the divergence of 2 evolutionary lines • Sister taxa- groups of organisms that share an immediate common ancestor (closest relative) • Rooted- a branch point within the tree represents the most recent common ancestor of all taxa in tree • Extant- still living • Basal taxon- lineage that diverges early in history and lies on branch near common ancestor (G) • Polytomy- branch point where more than 2 descendant groups emerged

3 points about Phylogenetic Trees 1) Intended to show patterns of descent not phenotypic similarities Ex) Crocodiles most closely related to birds than lizards but look more like lizards

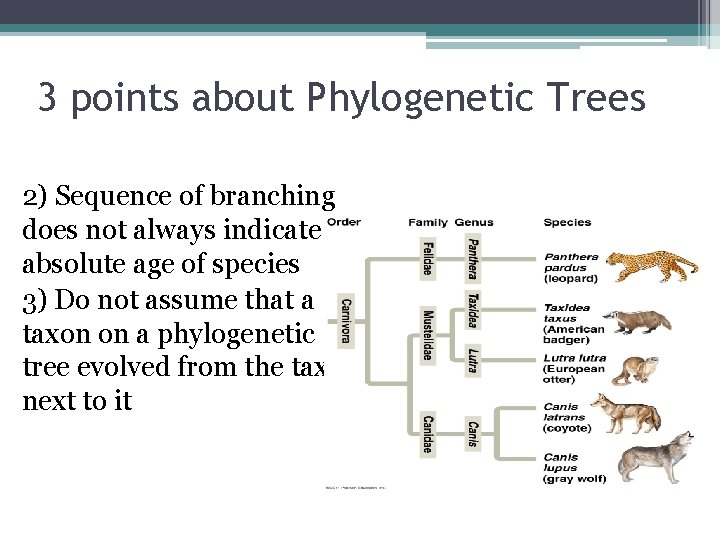
3 points about Phylogenetic Trees 2) Sequence of branching does not always indicate absolute age of species 3) Do not assume that a taxon on a phylogenetic tree evolved from the taxon next to it

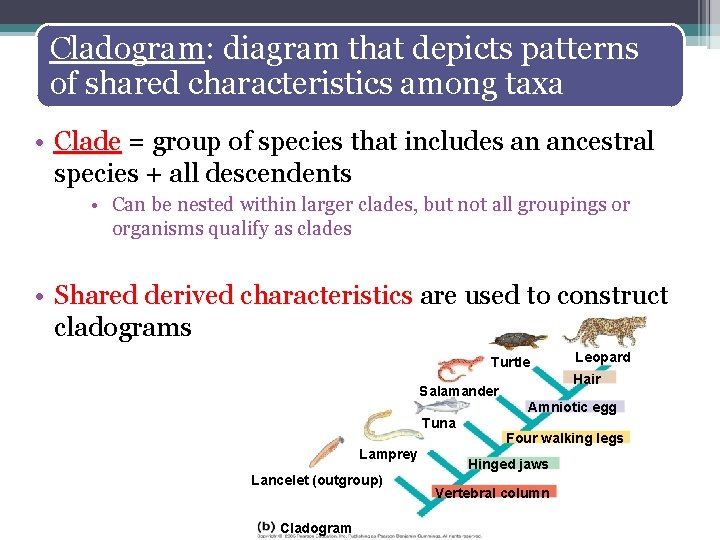
Cladogram: diagram that depicts patterns of shared characteristics among taxa • Clade = group of species that includes an ancestral species + all descendents • Can be nested within larger clades, but not all groupings or organisms qualify as clades • Shared derived characteristics are used to construct cladograms Turtle Leopard Hair Salamander Amniotic egg Tuna Lamprey Lancelet (outgroup) Cladogram Four walking legs Hinged jaws Vertebral column

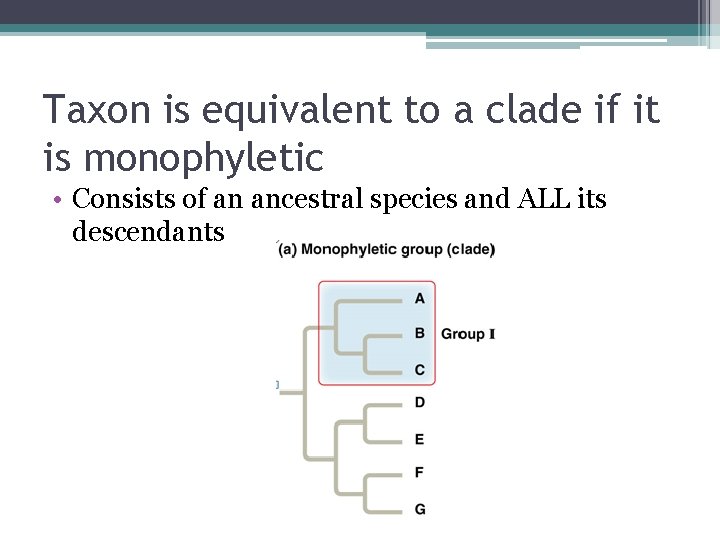
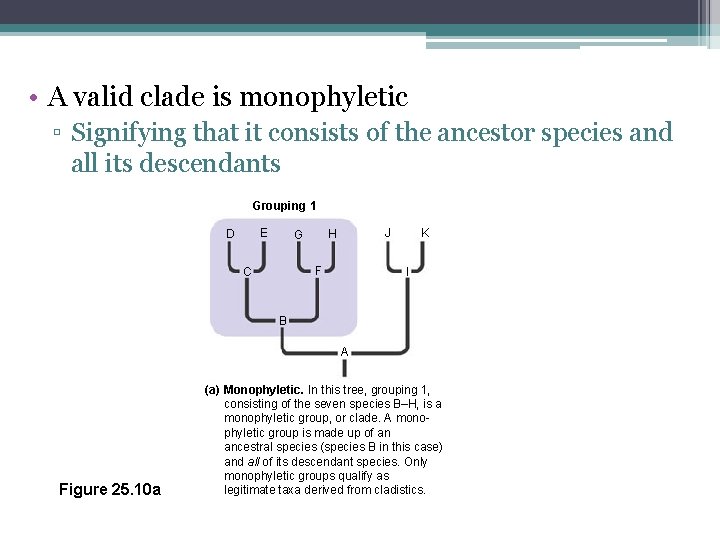
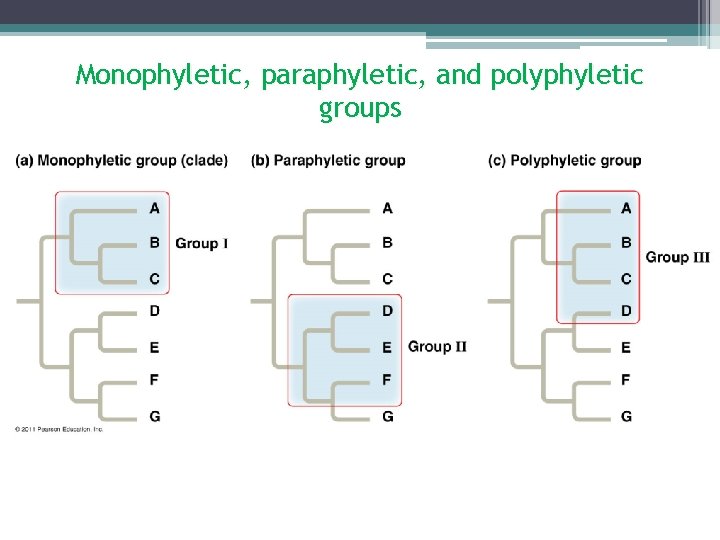
Taxon is equivalent to a clade if it is monophyletic • Consists of an ancestral species and ALL its descendants

• A valid clade is monophyletic ▫ Signifying that it consists of the ancestor species and all its descendants Grouping 1 E D J H G F C K I B A Figure 25. 10 a (a) Monophyletic. In this tree, grouping 1, consisting of the seven species B–H, is a monophyletic group, or clade. A monophyletic group is made up of an ancestral species (species B in this case) and all of its descendant species. Only monophyletic groups qualify as legitimate taxa derived from cladistics.

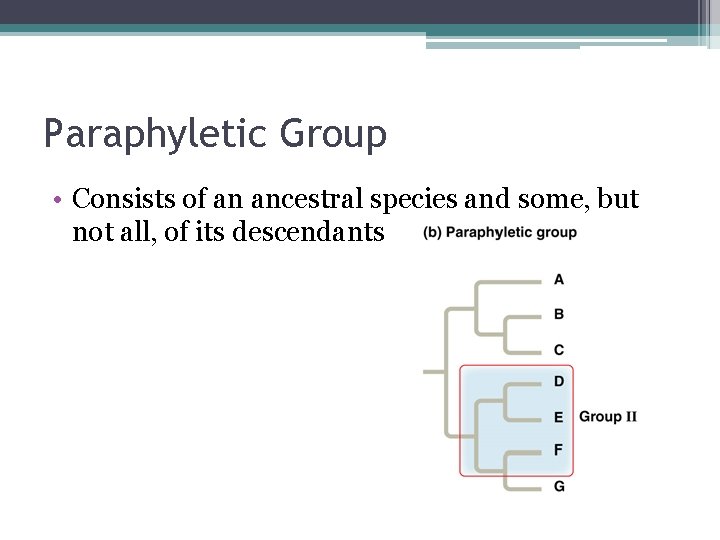
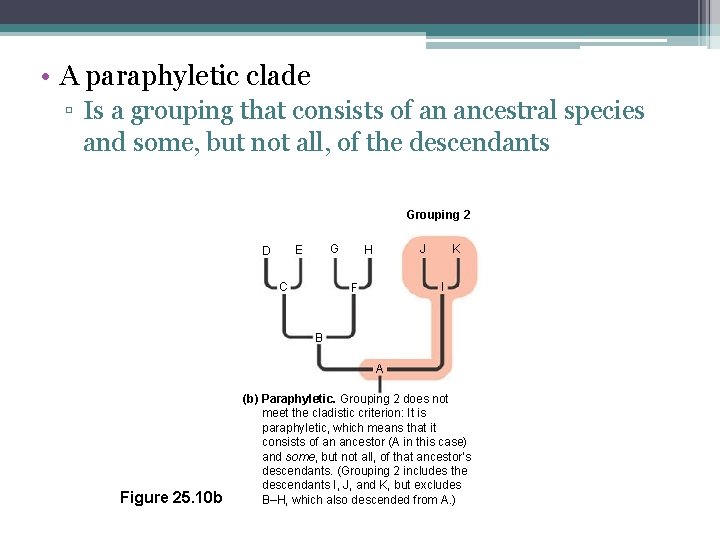
Paraphyletic Group • Consists of an ancestral species and some, but not all, of its descendants

• A paraphyletic clade ▫ Is a grouping that consists of an ancestral species and some, but not all, of the descendants Grouping 2 G E D C J H K I F B A Figure 25. 10 b (b) Paraphyletic. Grouping 2 does not meet the cladistic criterion: It is paraphyletic, which means that it consists of an ancestor (A in this case) and some, but not all, of that ancestor’s descendants. (Grouping 2 includes the descendants I, J, and K, but excludes B–H, which also descended from A. )

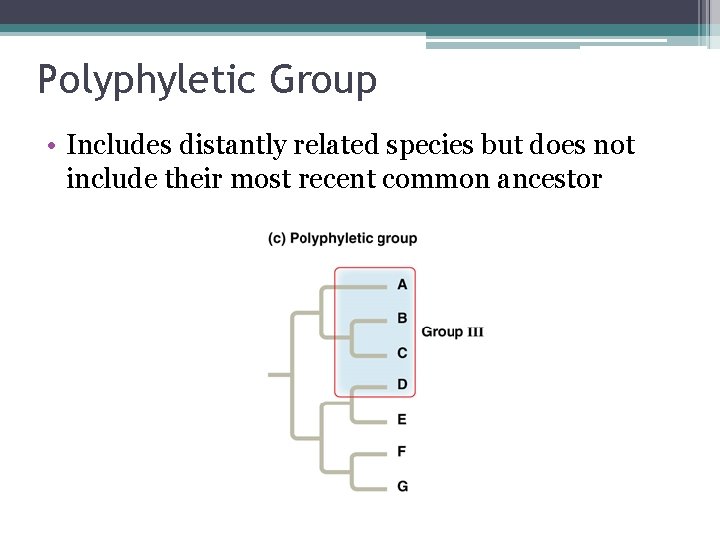
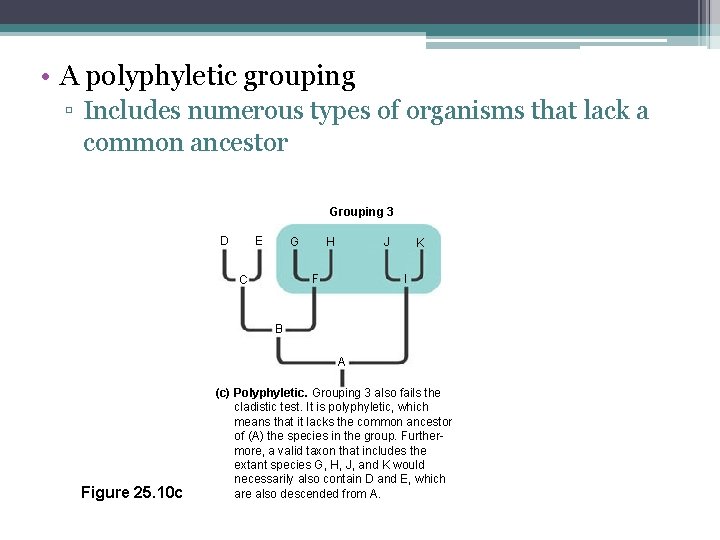
Polyphyletic Group • Includes distantly related species but does not include their most recent common ancestor

• A polyphyletic grouping ▫ Includes numerous types of organisms that lack a common ancestor Grouping 3 D E G J H I F C K B A Figure 25. 10 c (c) Polyphyletic. Grouping 3 also fails the cladistic test. It is polyphyletic, which means that it lacks the common ancestor of (A) the species in the group. Furthermore, a valid taxon that includes the extant species G, H, J, and K would necessarily also contain D and E, which are also descended from A.

Monophyletic, paraphyletic, and polyphyletic groups

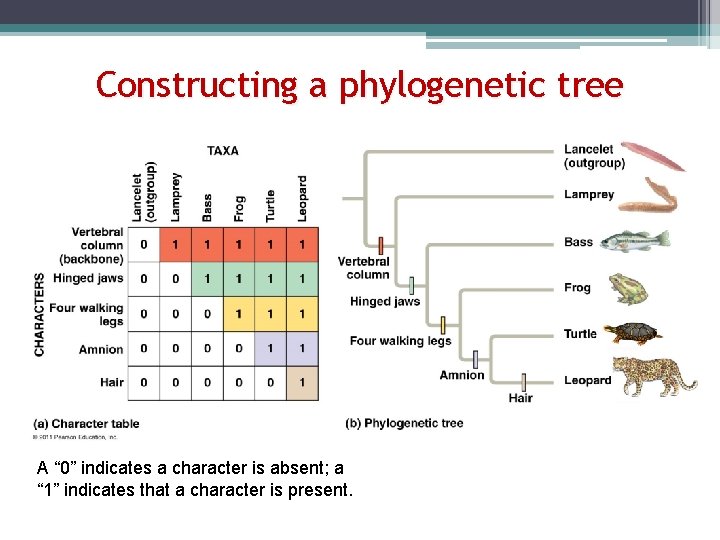
Constructing a phylogenetic tree A “ 0” indicates a character is absent; a “ 1” indicates that a character is present.

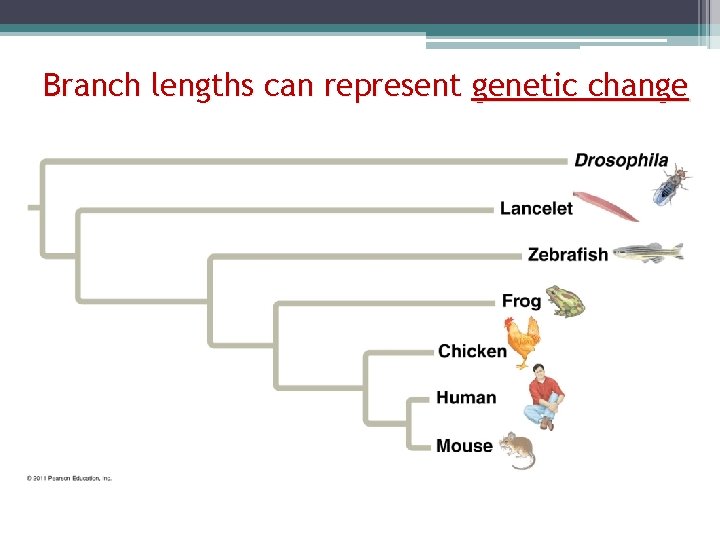
Branch lengths can represent genetic change

Branch lengths can indicate time

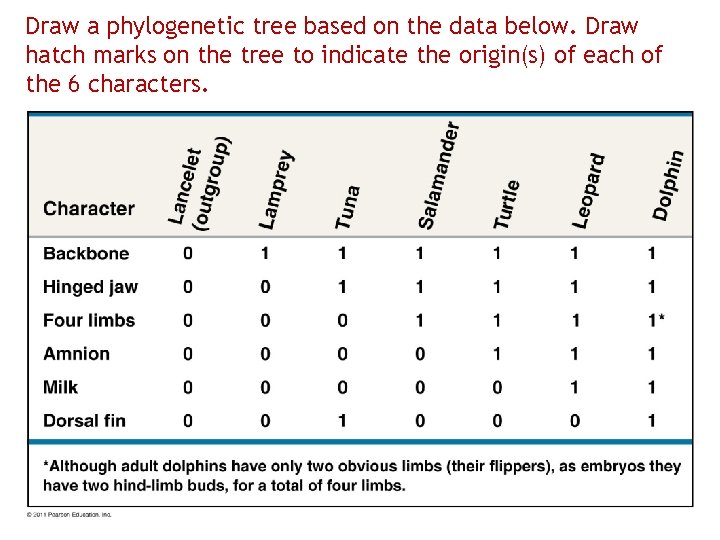
Draw a phylogenetic tree based on the data below. Draw hatch marks on the tree to indicate the origin(s) of each of the 6 characters.

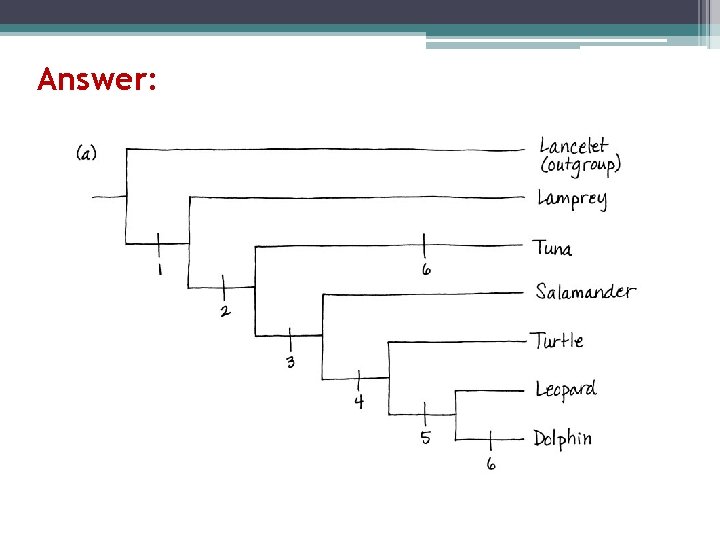
Answer:

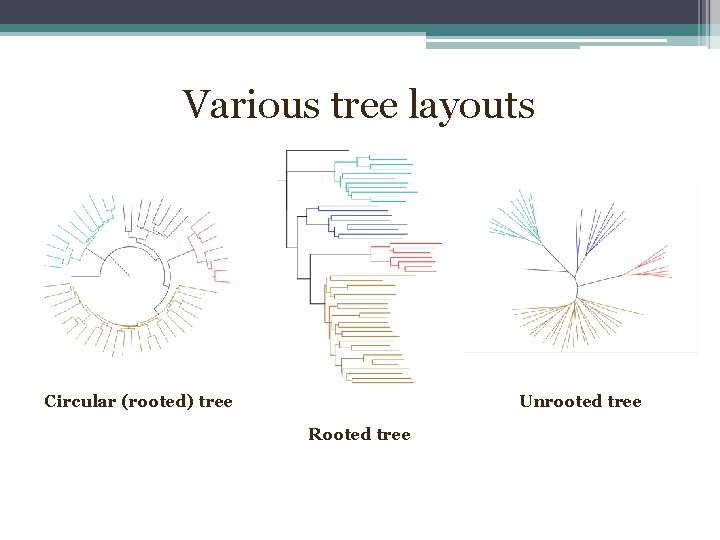
Various tree layouts Circular (rooted) tree Unrooted tree Rooted tree

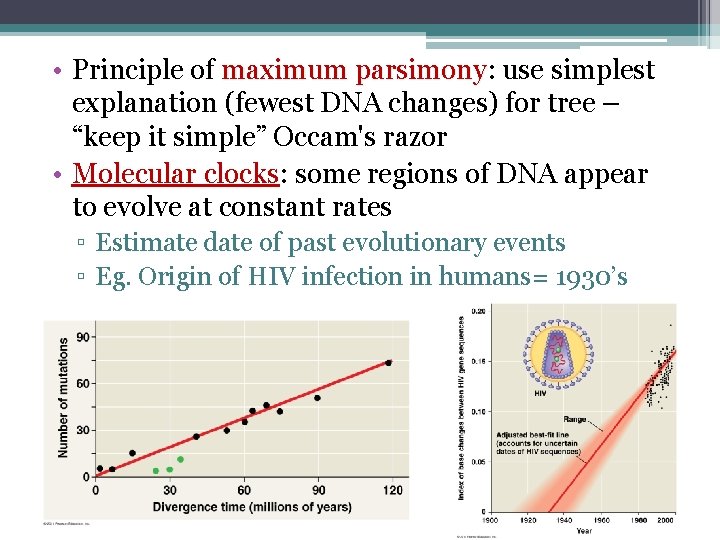
• Principle of maximum parsimony: parsimony use simplest explanation (fewest DNA changes) for tree – “keep it simple” Occam's razor • Molecular clocks: some regions of DNA appear to evolve at constant rates ▫ Estimate date of past evolutionary events ▫ Eg. Origin of HIV infection in humans= 1930’s

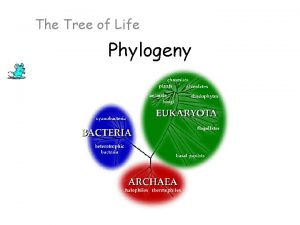
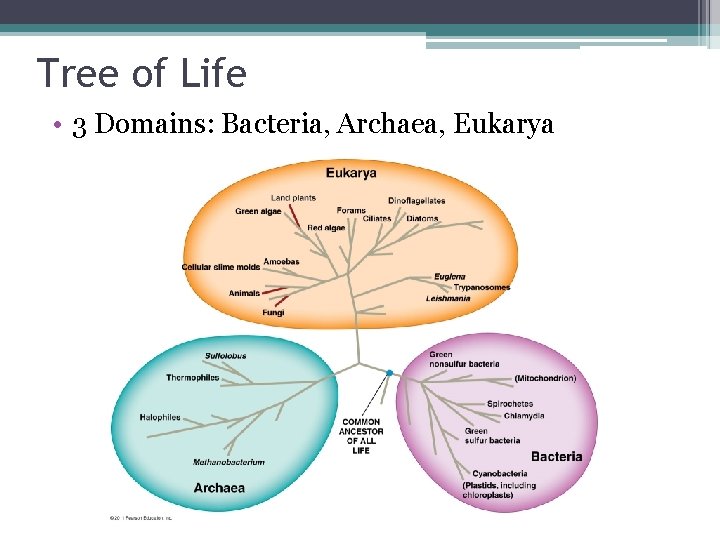
Tree of Life • 3 Domains: Bacteria, Archaea, Eukarya

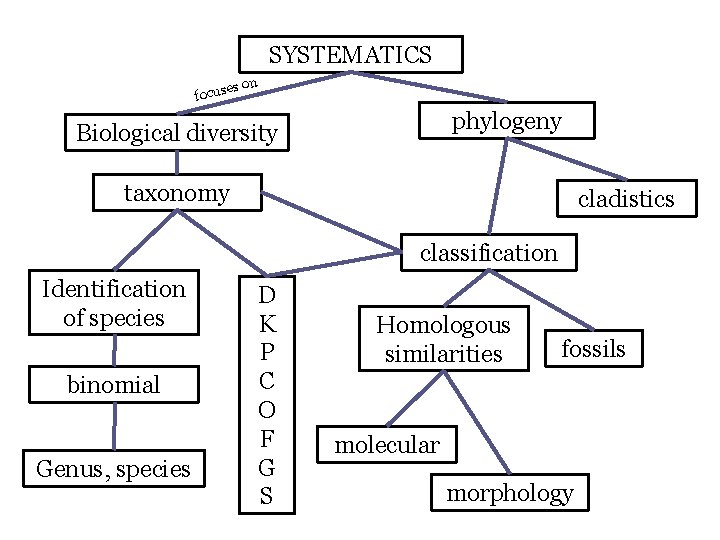
SYSTEMATICS es focus on phylogeny Biological diversity taxonomy cladistics classification Identification of species binomial Genus, species D K P C O F G S Homologous similarities fossils molecular morphology
 Monophyletic group
Monophyletic group Chapter 20 phylogeny and the tree of life
Chapter 20 phylogeny and the tree of life Monophyletic paraphyletic polyphyletic
Monophyletic paraphyletic polyphyletic Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Phylogeny and the tree of life chapter 26
Phylogeny and the tree of life chapter 26 Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Taxa phylogenetic tree
Taxa phylogenetic tree Outgroup in phylogeny
Outgroup in phylogeny What is a sister group in phylogeny
What is a sister group in phylogeny Photos
Photos Crab cladogram
Crab cladogram Biological taxonomy
Biological taxonomy Polytomy
Polytomy Animal kingdom cladogram
Animal kingdom cladogram Phylogeny
Phylogeny Fur
Fur Monophyletic group
Monophyletic group Classification of platyhelminthes
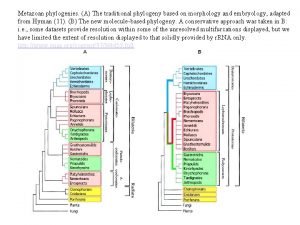
Classification of platyhelminthes Metazoan phylogeny
Metazoan phylogeny Anatomy of trout
Anatomy of trout Avian phylogeny
Avian phylogeny Ap biology phylogeny
Ap biology phylogeny Clustal omega alignment
Clustal omega alignment Phylogenetic tree
Phylogenetic tree Ontogeny recapitulates phylogeny
Ontogeny recapitulates phylogeny Closed circulatory system
Closed circulatory system Fish phylogeny
Fish phylogeny Chordate
Chordate