Genetic improvement programs for US dairy cattle John

























- Slides: 58

Genetic improvement programs for US dairy cattle John B. Cole Animal Genomics and Improvement Laboratory Agricultural Research Service, USDA Beltsville, MD john. cole@ars. usda. gov

Small dairy farm in western Maryland. Photo courtesy of ARS. Typical US dairies Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (2) Top: Large freestall barn in the state of Florida. Bottom: 7, 000 G (~26, 500 l) milk tankers. Photo courtesy of North Florida Holsteins. Cole

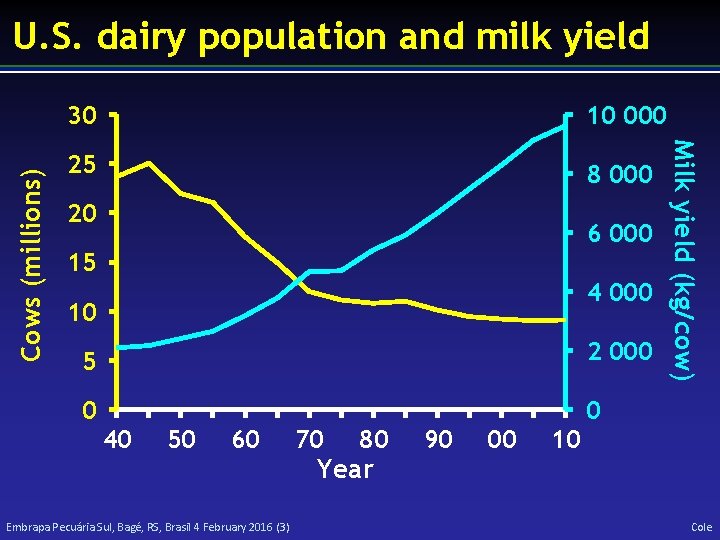
30 10 000 25 8 000 20 6 000 15 4 000 10 5 2 000 0 0 40 50 60 Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (3) 70 80 Year 90 00 Milk yield (kg/cow) Cows (millions) U. S. dairy population and milk yield 10 Cole

U. S. DHI dairy statistics (2011) l 9. 1 million U. S. cows l ~75% bred AI l 47% milk recorded through Dairy Herd Information (DHI) w 4. 4 million cows − 86% Holstein − 8% crossbred − 5% Jersey − <1% Ayrshire, Brown Swiss, Guernsey, Milking Shorthorn, Red & White w 20, 000 herds w 220 cows/herd w 10, 300 kg/cow Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (4) Cole

Genetic evaluation advances Year 1862 1895 1926 1962 1973 1974 1977 1989 1994 Advance Gain, % USDA established USDA begins collecting dairy records Daughter-dam comparison 100 Herdmate comparison 50 Records in progress 10 Modified contemporary comparison 5 Protein evaluated 4 Animal model 4 Net merit, productive life, and somatic cell 50 score 2008 Genomic selection >50 Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (5) Cole

Animal model l 1989 to present l Introduced by Wiggans and Van. Raden l Advantages w w w Information from all relatives Adjustment for genetic merit of mates Uniform procedures for males and females Best prediction (BLUP) Crossbreds included (2007) Genomic information added (2008) Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (6) Cole

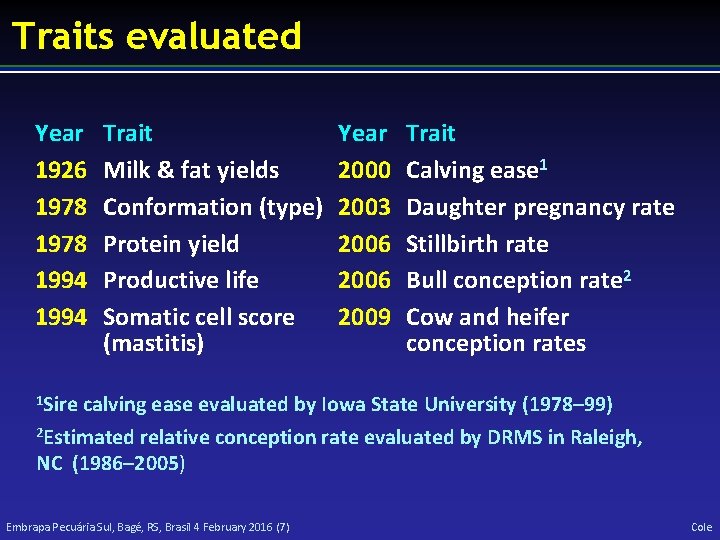
Traits evaluated Year 1926 1978 1994 Trait Milk & fat yields Conformation (type) Protein yield Productive life Somatic cell score (mastitis) Year 2000 2003 2006 2009 Trait Calving ease 1 Daughter pregnancy rate Stillbirth rate Bull conception rate 2 Cow and heifer conception rates 1 Sire calving ease evaluated by Iowa State University (1978– 99) 2 Estimated relative conception rate evaluated by DRMS in Raleigh, NC (1986– 2005) Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (7) Cole

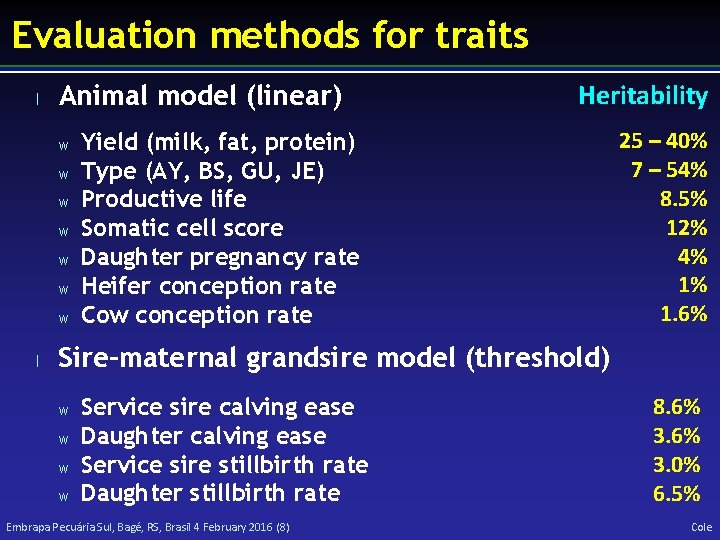
Evaluation methods for traits l Animal model (linear) w w w w l Heritability Yield (milk, fat, protein) Type (AY, BS, GU, JE) Productive life Somatic cell score Daughter pregnancy rate Heifer conception rate Cow conception rate 25 – 40% 7 – 54% 8. 5% 12% 4% 1% 1. 6% Sire–maternal grandsire model (threshold) w w Service sire calving ease Daughter calving ease Service sire stillbirth rate Daughter stillbirth rate Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (8) 8. 6% 3. 0% 6. 5% Cole

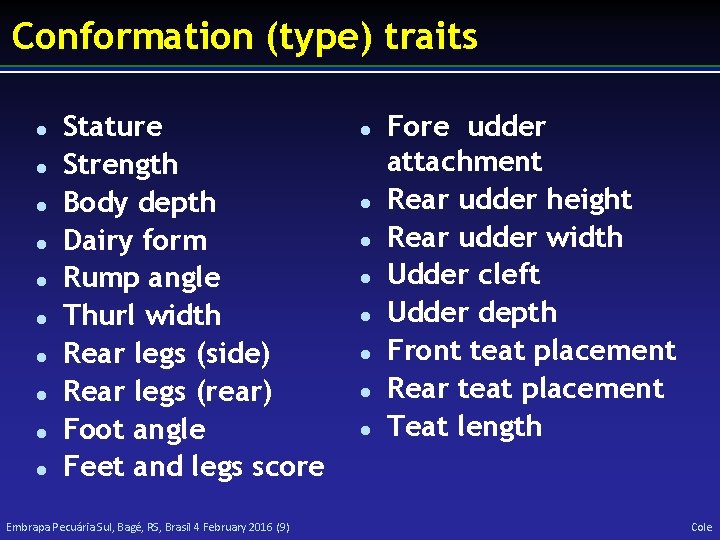
Conformation (type) traits l l l l l Stature Strength Body depth Dairy form Rump angle Thurl width Rear legs (side) Rear legs (rear) Foot angle Feet and legs score Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (9) l l l l Fore udder attachment Rear udder height Rear udder width Udder cleft Udder depth Front teat placement Rear teat placement Teat length Cole

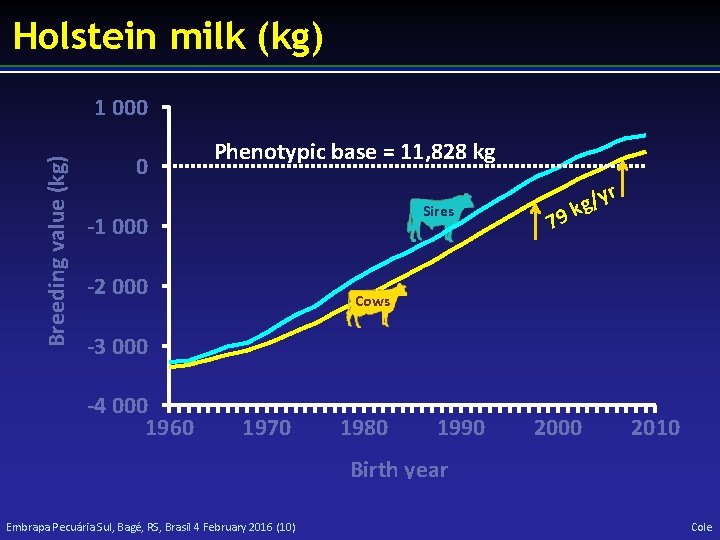
Holstein milk (kg) Breeding value (kg) 1 000 0 Phenotypic base = 11, 828 kg Sires -1 000 -2 000 79 r y / kg Cows -3 000 -4 000 1960 1970 1980 1990 2000 2010 Birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (10) Cole

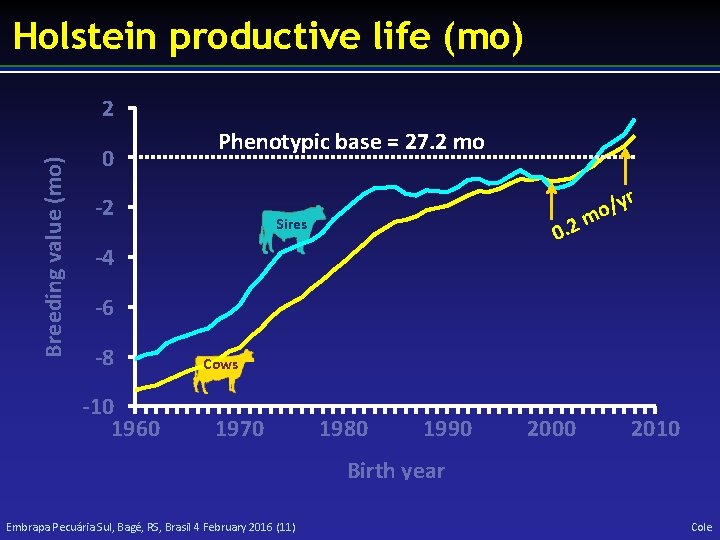
Holstein productive life (mo) Breeding value (mo) 2 0 Phenotypic base = 27. 2 mo -2 r y / o m 2. 0 Sires -4 -6 -8 -10 1960 Cows 1970 1980 1990 2000 2010 Birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (11) Cole

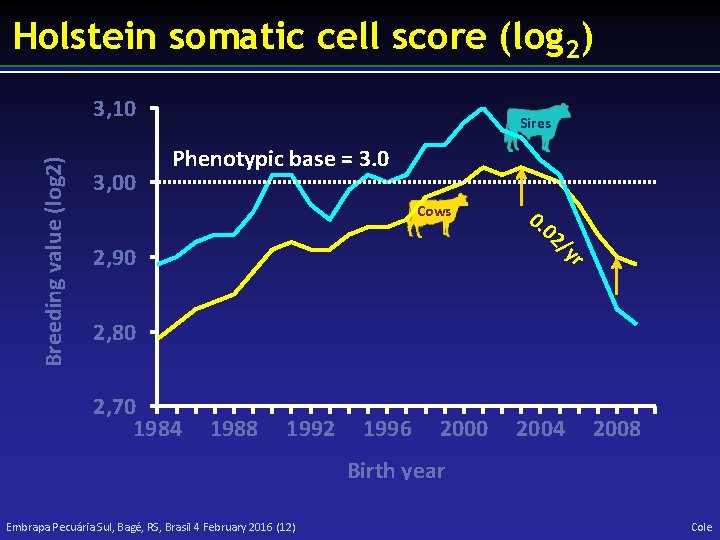
Holstein somatic cell score (log 2) Breeding value (log 2) 3, 10 3, 00 Sires Phenotypic base = 3. 0 Cows 0. 02 /y 2, 90 r 2, 80 2, 70 1984 1988 1992 1996 2000 2004 2008 Birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (12) Cole

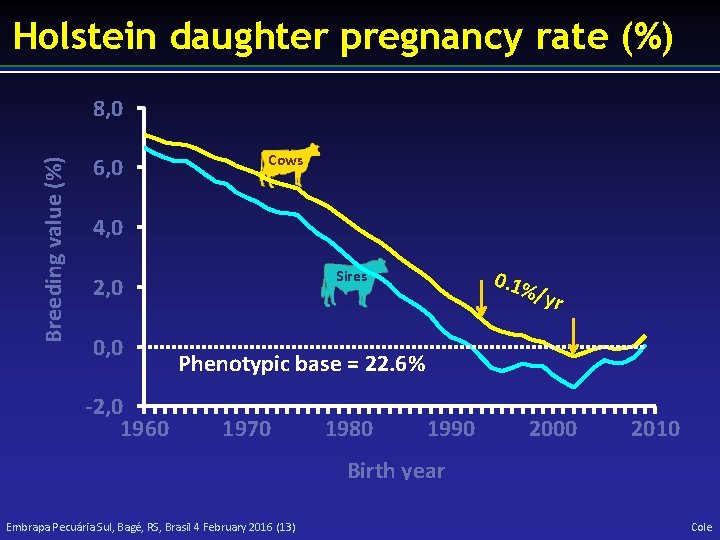
Holstein daughter pregnancy rate (%) Breeding value (%) 8, 0 6, 0 Cows 4, 0 0, 0 -2, 0 1960 0. 1% Sires 2, 0 /yr Phenotypic base = 22. 6% 1970 1980 1990 2000 2010 Birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (13) Cole

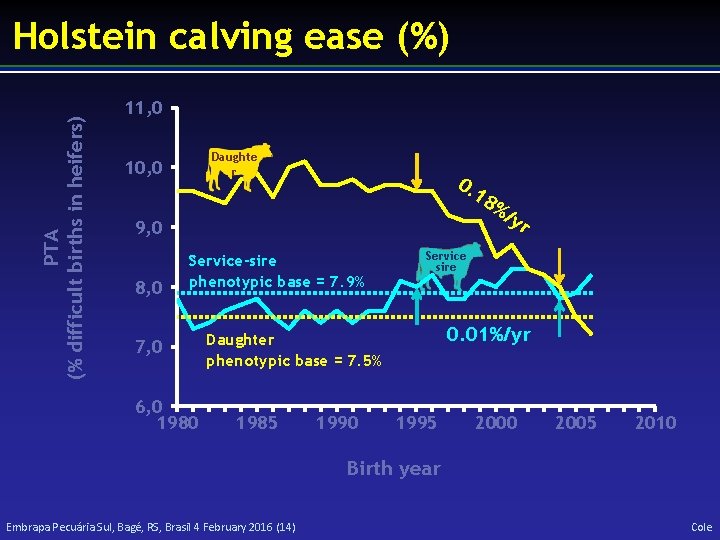
PTA (% difficult births in heifers) Holstein calving ease (%) 11, 0 Daughte r 10, 0 0. 18 9, 0 8, 0 Service-sire phenotypic base = 7. 9% 7, 0 6, 0 1980 1990 yr Service sire 0. 01%/yr Daughter phenotypic base = 7. 5% 1985 %/ 1995 2000 2005 2010 Birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (14) Cole

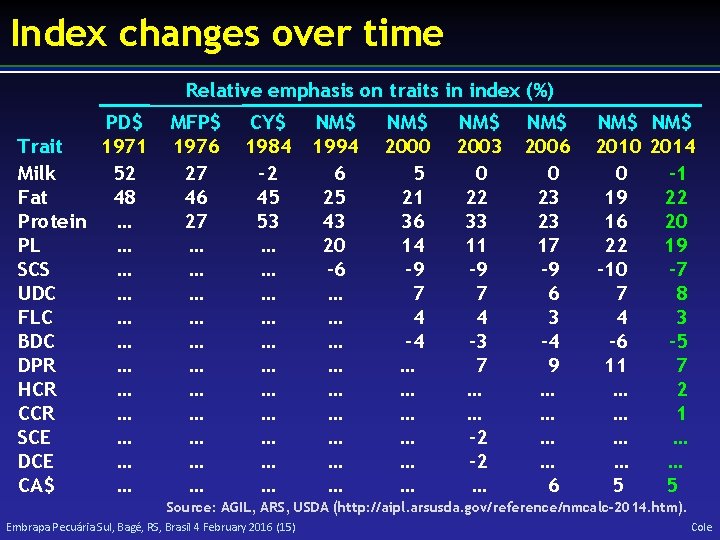
Index changes over time Relative emphasis on traits in index (%) PD$ Trait 1971 Milk 52 Fat 48 Protein … PL … SCS … UDC … FLC … BDC … DPR … HCR … CCR … SCE … DCE … CA$ … MFP$ 1976 27 46 27 … … … CY$ 1984 – 2 45 53 … … … NM$ 1994 6 25 43 20 – 6 … … … … … NM$ 2000 5 21 36 14 – 9 7 4 – 4 … … … NM$ 2003 0 22 33 11 – 9 7 4 – 3 7 … … – 2 … NM$ 2006 0 23 23 17 – 9 6 3 – 4 9 … … 6 NM$ 2010 2014 0 -1 19 22 16 20 22 19 – 10 -7 7 8 4 3 – 6 -5 11 7 … 2 … 1 … … 5 5 Source: AGIL, ARS, USDA (http: //aipl. arsusda. gov/reference/nmcalc-2014. htm). Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (15) Cole

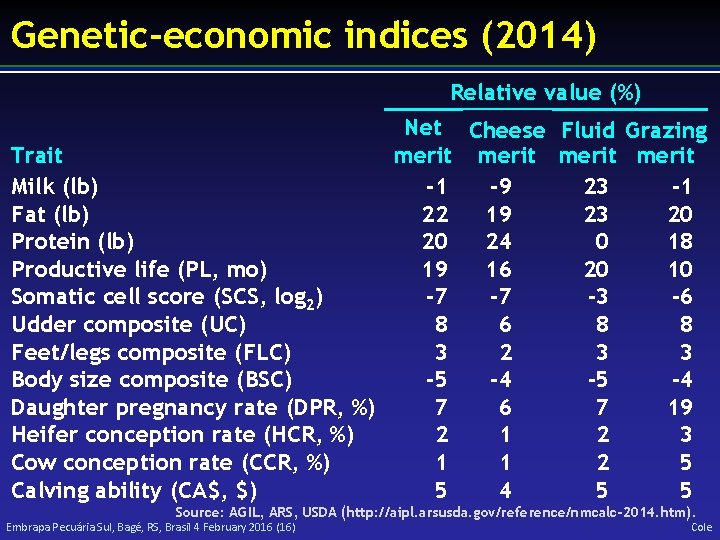
Genetic-economic indices (2014) Relative value (%) Net Cheese Fluid Grazing merit Trait Milk (lb) -1 -9 23 -1 Fat (lb) 22 19 23 20 Protein (lb) 20 24 0 18 Productive life (PL, mo) 19 16 20 10 Somatic cell score (SCS, log 2) -7 -7 -3 -6 Udder composite (UC) 8 6 8 8 Feet/legs composite (FLC) 3 2 3 3 Body size composite (BSC) -5 -4 Daughter pregnancy rate (DPR, %) 7 6 7 19 Heifer conception rate (HCR, %) 2 1 2 3 Cow conception rate (CCR, %) 1 1 2 5 Calving ability (CA$, $) 5 4 5 5 Source: AGIL, ARS, USDA (http: //aipl. arsusda. gov/reference/nmcalc-2014. htm). Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (16) Cole

Traditional evaluation summary l Evaluation procedures have improved l Fitness traits have been added l l l Effective selection has produced substantial annual genetic improvement Indices enable selection for overall economic merit Fertility evaluations prevent continued decline Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (17) Cole

Traditional dairy breeds in the US The six traditional US dairy breeds. Photo courtesy of Bonnie Mohr. Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (18) Cole

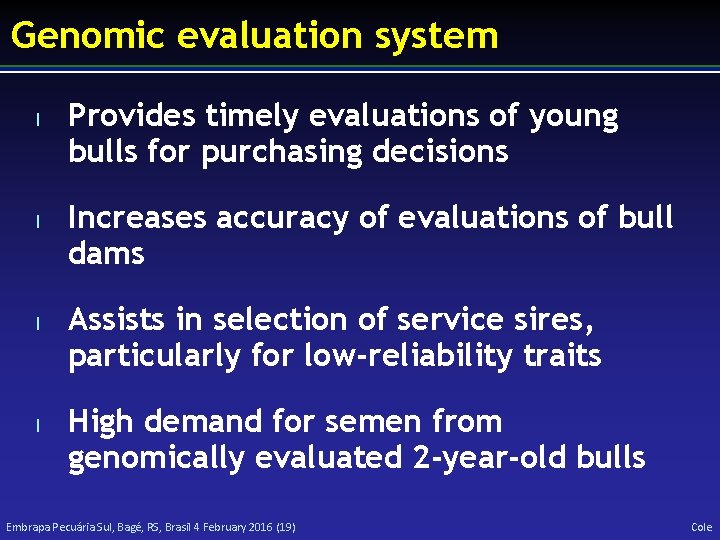
Genomic evaluation system l l Provides timely evaluations of young bulls for purchasing decisions Increases accuracy of evaluations of bull dams Assists in selection of service sires, particularly for low-reliability traits High demand for semen from genomically evaluated 2 -year-old bulls Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (19) Cole

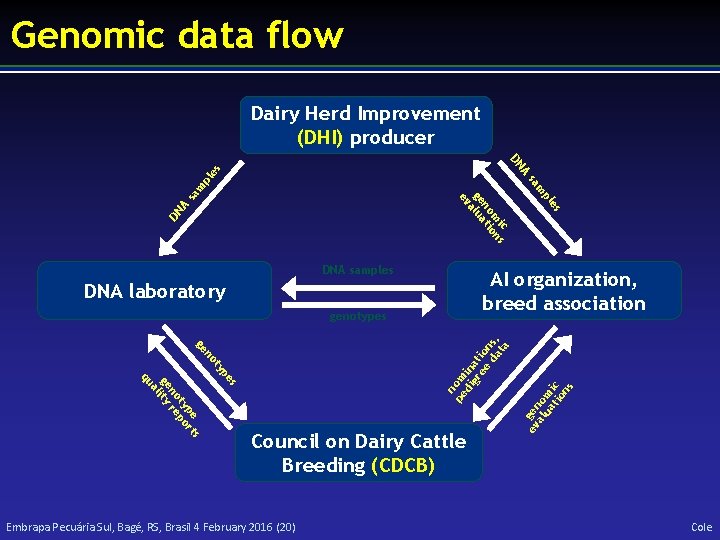
Genomic data flow Dairy Herd Improvement (DHI) producer s s pe rt ty po no re ge lity a qu pe ty no ge n pe om di in gr at ee io da ns, ta genotypes Council on Dairy Cattle Breeding (CDCB) ev gen al om ua tio ic ns sa A DN es AI organization, breed association DNA laboratory Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (20) pl m m sa pl A es DN ic s m on no ti ge alua ev DNA samples Cole

Collaboration with industry l l Council on Dairy Cattle Breeding (CDCB) responsible for receiving data and for computing and delivering US genetic evaluations for dairy cattle AIP responsible for research and development to improve the evaluation system CDCB (Bowie) and AIP (Beltsville) are located near one another Dr. João Dürr is CDCB’s CEO Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (21) Cole

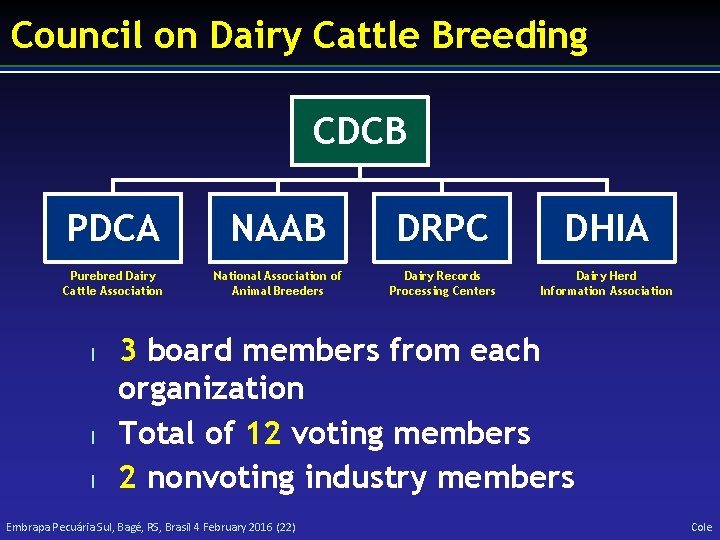
Council on Dairy Cattle Breeding CDCB PDCA NAAB DRPC DHIA Purebred Dairy Cattle Association National Association of Animal Breeders Dairy Records Processing Centers Dairy Herd Information Association l l l 3 board members from each organization Total of 12 voting members 2 nonvoting industry members Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (22) Cole

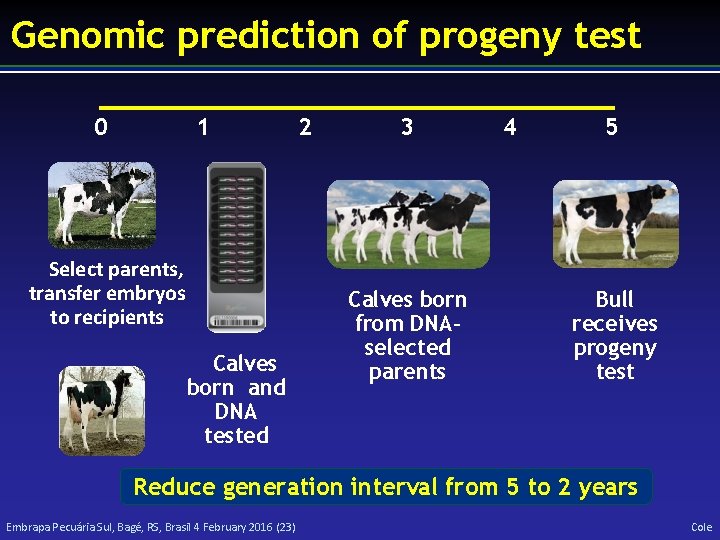
Genomic prediction of progeny test 0 1 Select parents, transfer embryos to recipients Calves born and DNA tested 2 3 Calves born from DNAselected parents 4 5 Bull receives progeny test Reduce generation interval from 5 to 2 years Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (23) Cole

Evaluation flow l l Animal nominated for genomic evaluation by breed association or AI organization Hair or other DNA source sent to genotyping lab DNA extracted and placed on chip for 3 -day genotyping process Genotypes sent from genotyping lab to AIPL for accuracy review Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (24) Cole

Laboratory quality control l l Each SNP evaluated for w Call rate w Portion heterozygous w Parent-progeny conflicts Clustering investigated if SNP exceeds limits Number of failing SNPs indicates genotype quality Target of <10 SNPs in each category Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (25) Cole

Evaluation flow (continued) l Genotype calls modified as necessary l Genotypes loaded into database l Nominators receive reports of parentage and other conflicts l Pedigree or animal assignments corrected l Genotypes extracted and imputed to 45 K l SNP effects estimated Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (26) Cole

Imputation l l Based on splitting genotype into individual chromosomes (maternal and paternal contributions) Missing SNPs assigned by tracking inheritance from ancestors and descendants Imputed dams increase predictor population Genotypes from all chips merged by imputing SNPs not present Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (27) Cole

Evaluation flow (continued) l Final evaluations calculated l Evaluations released to dairy industry w w Download from CDCB FTP site with separate files for each nominator Monthly release for new animals All genomic evaluations updated 3 times each year with traditional evaluations Weekly evaluations are now available Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (28) Cole

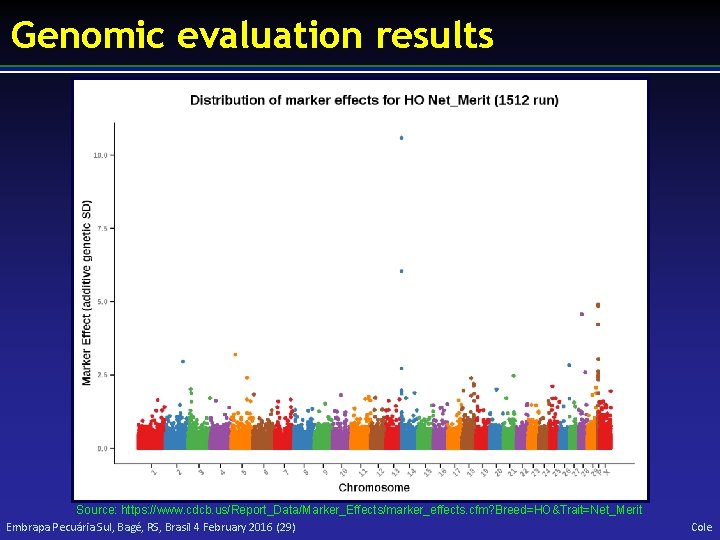
Genomic evaluation results Source: https: //www. cdcb. us/Report_Data/Marker_Effects/marker_effects. cfm? Breed=HO&Trait=Net_Merit Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (29) Cole

Information sources for evaluations l l l Traditional evaluations of genotyped bulls and cows used to estimate SNP effects Combined final evaluation w Sum of SNP effects for an animal’s alleles w Polygenetic effect w Traditional evaluation Pedigree data used and validated by genotypes Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (30) Cole

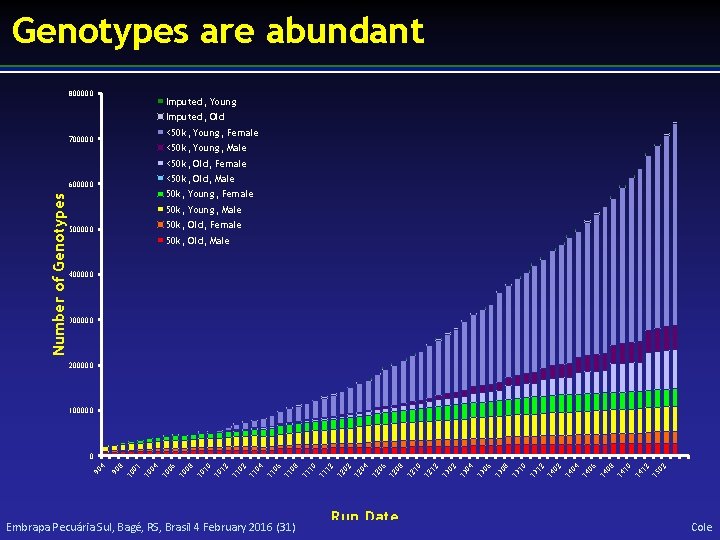
Genotypes are abundant 800000 Imputed, Young Imputed, Old 700000 <50 k, Young, Female <50 k, Young, Male <50 k, Old, Female Number of Genotypes 600000 <50 k, Old, Male 50 k, Young, Female 50 k, Young, Male 500000 50 k, Old, Female 50 k, Old, Male 400000 300000 200000 100000 90 4 90 8 10 01 10 04 10 06 10 08 10 10 10 12 11 04 11 06 11 08 11 10 11 12 12 04 12 06 12 08 12 10 12 12 13 04 13 06 13 08 13 10 13 12 14 04 14 06 14 08 14 10 14 12 15 02 0 Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (31) Run Date Cole

SNP used for genomic evaluations l l l 61, 013 SNP used after culling on w MAF w Parent-progeny conflicts w Percentage heterozygous (departure from HWE) SNP for HH 1, BLAD, DUMPS, CVM, polled, red, and mulefoot included w JH 1 included for Jerseys Some SNP eliminated because incorrect location �haplotype non-inheritance Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (32) Cole

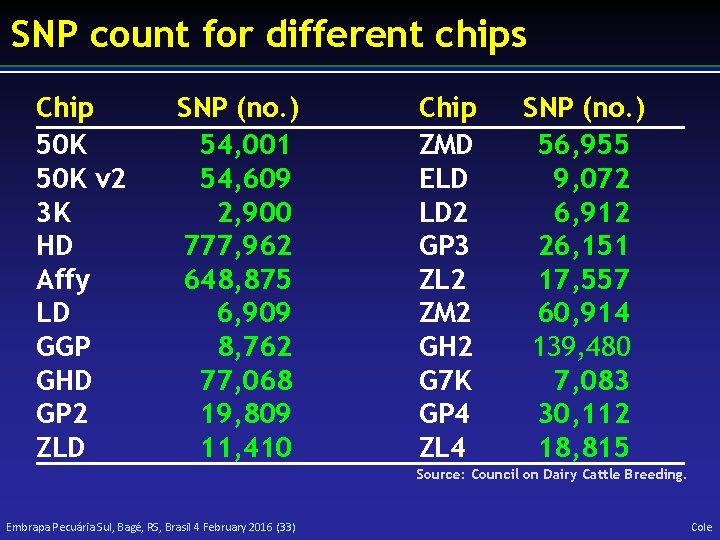
SNP count for different chips Chip 50 K v 2 3 K HD Affy LD GGP GHD GP 2 ZLD SNP (no. ) 54, 001 54, 609 2, 900 777, 962 648, 875 6, 909 8, 762 77, 068 19, 809 11, 410 Chip ZMD ELD LD 2 GP 3 ZL 2 ZM 2 GH 2 G 7 K GP 4 ZL 4 SNP (no. ) 56, 955 9, 072 6, 912 26, 151 17, 557 60, 914 139, 480 7, 083 30, 112 18, 815 Source: Council on Dairy Cattle Breeding. Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (33) Cole

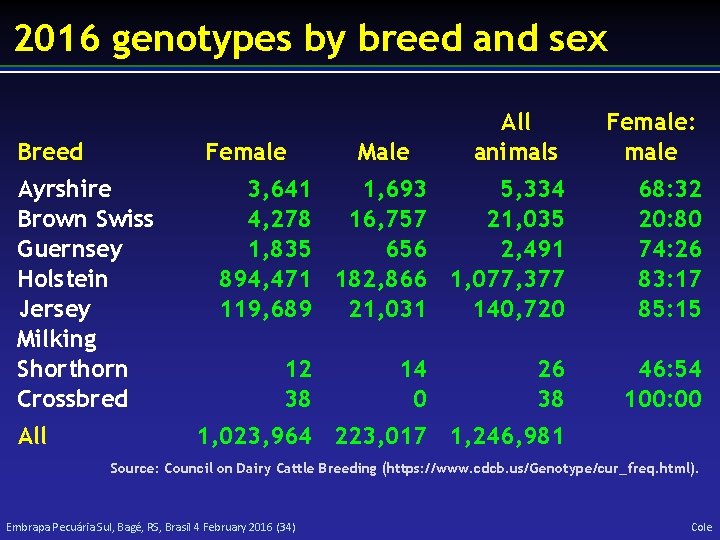
2016 genotypes by breed and sex Breed Female: male 3, 641 1, 693 5, 334 4, 278 16, 757 21, 035 1, 835 656 2, 491 894, 471 182, 866 1, 077, 377 119, 689 21, 031 140, 720 68: 32 20: 80 74: 26 83: 17 85: 15 Female Ayrshire Brown Swiss Guernsey Holstein Jersey Milking Shorthorn Crossbred All animals 12 38 Male 14 0 26 38 46: 54 100: 00 1, 023, 964 223, 017 1, 246, 981 Source: Council on Dairy Cattle Breeding (https: //www. cdcb. us/Genotype/cur_freq. html). Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (34) Cole

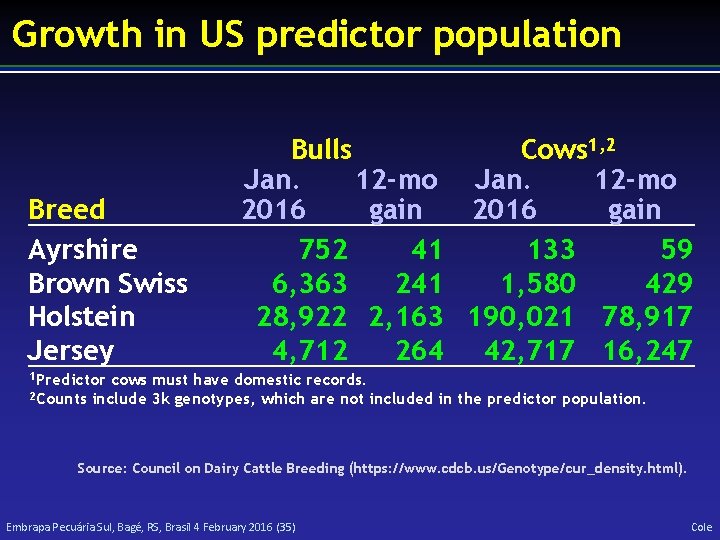
Growth in US predictor population Breed Ayrshire Brown Swiss Holstein Jersey Bulls Cows 1, 2 Jan. 12 -mo 2016 gain 752 41 133 59 6, 363 241 1, 580 429 28, 922 2, 163 190, 021 78, 917 4, 712 264 42, 717 16, 247 1 Predictor 2 Counts cows must have domestic records. include 3 k genotypes, which are not included in the predictor population. Source: Council on Dairy Cattle Breeding (https: //www. cdcb. us/Genotype/cur_density. html). Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (35) Cole

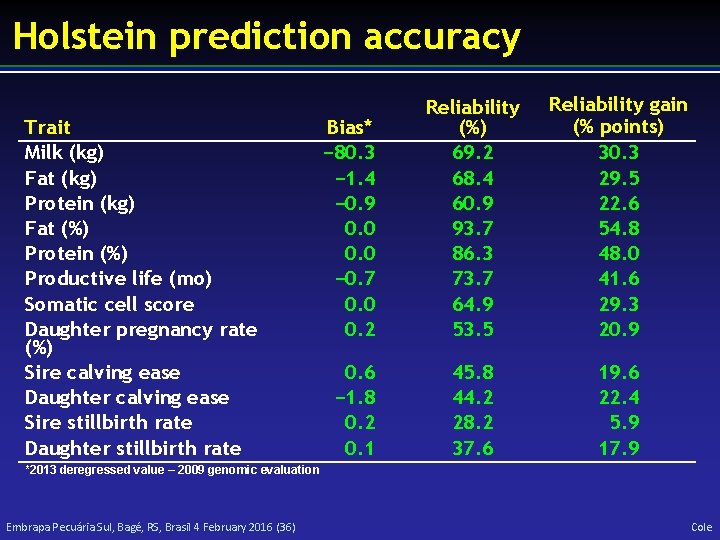
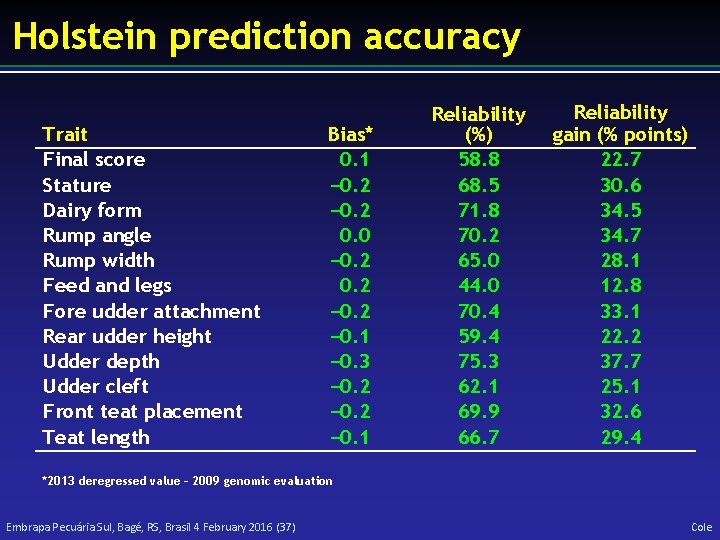
Holstein prediction accuracy Trait Milk (kg) Fat (kg) Protein (kg) Fat (%) Protein (%) Productive life (mo) Somatic cell score Daughter pregnancy rate (%) Sire calving ease Daughter calving ease Sire stillbirth rate Daughter stillbirth rate Bias* − 80. 3 − 1. 4 − 0. 9 0. 0 − 0. 7 0. 0 0. 2 Reliability (%) 69. 2 68. 4 60. 9 93. 7 86. 3 73. 7 64. 9 53. 5 Reliability gain (% points) 30. 3 29. 5 22. 6 54. 8 48. 0 41. 6 29. 3 20. 9 0. 6 − 1. 8 0. 2 0. 1 45. 8 44. 2 28. 2 37. 6 19. 6 22. 4 5. 9 17. 9 *2013 deregressed value – 2009 genomic evaluation Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (36) Cole

Holstein prediction accuracy Trait Final score Stature Dairy form Rump angle Rump width Feed and legs Fore udder attachment Rear udder height Udder depth Udder cleft Front teat placement Teat length Bias* 0. 1 − 0. 2 0. 0 − 0. 2 − 0. 1 − 0. 3 − 0. 2 − 0. 1 Reliability (%) 58. 8 68. 5 71. 8 70. 2 65. 0 44. 0 70. 4 59. 4 75. 3 62. 1 69. 9 66. 7 Reliability gain (% points) 22. 7 30. 6 34. 5 34. 7 28. 1 12. 8 33. 1 22. 2 37. 7 25. 1 32. 6 29. 4 *2013 deregressed value – 2009 genomic evaluation Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (37) Cole

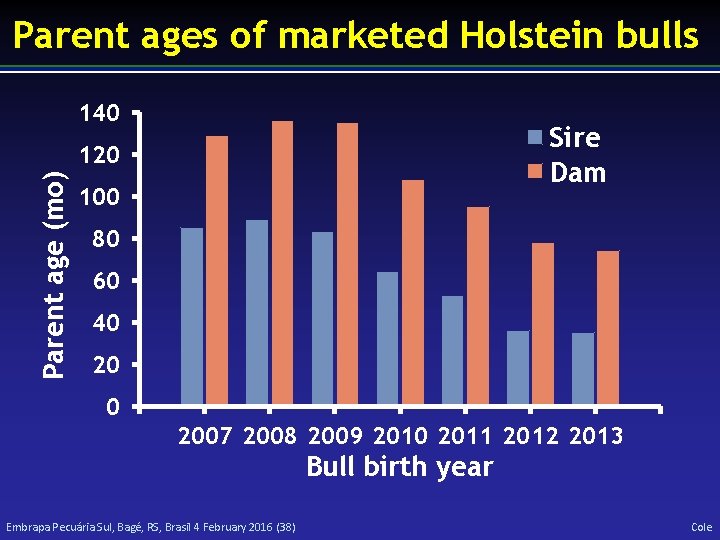
Parent ages of marketed Holstein bulls 140 Sire Dam Parent age (mo) 120 100 80 60 40 2007 2008 2009 2010 2011 2012 2013 Bull birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (38) Cole

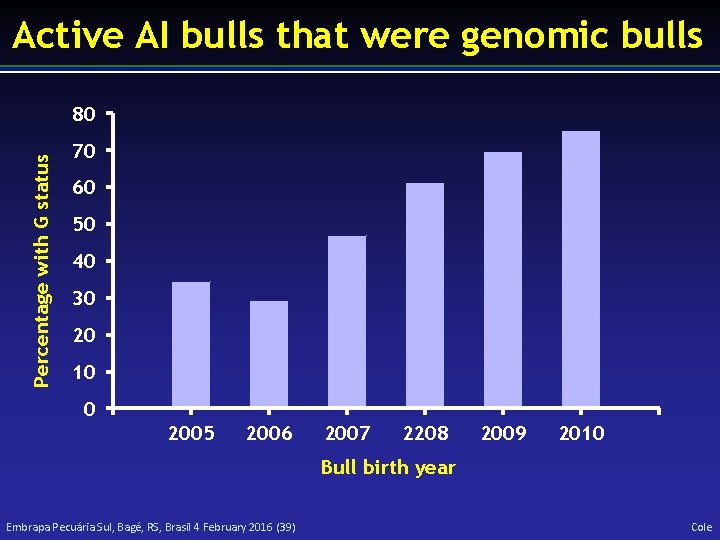
Active AI bulls that were genomic bulls Percentage with G status 80 70 60 50 40 30 20 10 0 2005 2006 2007 2208 2009 2010 Bull birth year Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (39) Cole

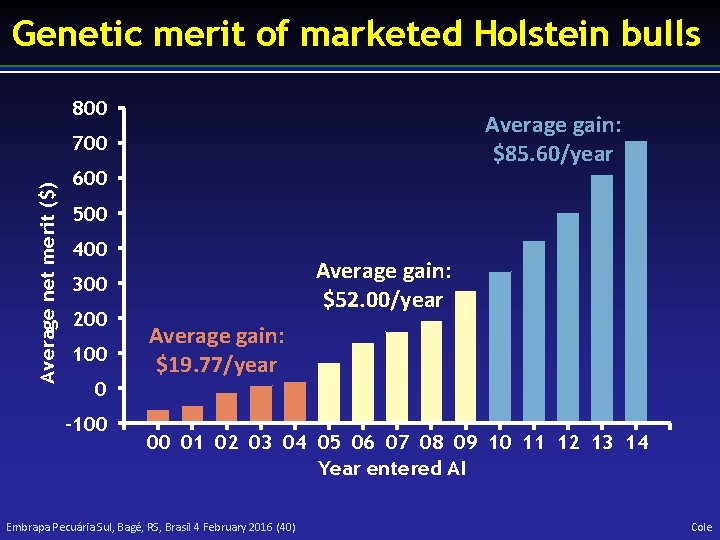
Genetic merit of marketed Holstein bulls 800 Average gain: $85. 60/year Average net merit ($) 700 600 500 400 Average gain: $52. 00/year 300 200 100 0 -100 Average gain: $19. 77/year 00 01 02 03 04 05 06 07 08 09 10 11 12 13 14 Year entered AI Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (40) Cole

Genetic choices l Before genomics: w w l Proven bulls with daughter records (PTA) Young bulls with parent average (PA) After genomics: w w Young animals with DNA test (GPTA) Reliability of GPTA ~70% compared to PA ~35% and PTA ~85% for Holstein NM$ Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (41) Cole

Stability of genomic evaluations l l l 642 Holstein bulls w Dec. 2012 NM$ compared with Dec. 2014 NM$ w First traditional evaluation in Aug. 2014 w 50 daughters by Dec. 2014 Top 100 bulls in 2012 w Average rank change of 9. 6 w Maximum drop of 119 w Maximum rise of 56 All 642 bulls w Correlation of 0. 94 between 2012 and 2014 w Regression of 0. 92 Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (42) Cole

Genomics is not the only innovation The Swedish Agricultural University dairy research center has a state-of-the-art facility equipped with the latest De. Laval technology. Photos courtesy of Albert de Vries. Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (43) Cole

Application to more traits l l l Animal’s genotype good for all traits Traditional evaluations required for accurate estimates of SNP effects Traditional evaluations not currently available for heat tolerance or feed efficiency Research populations could provide data for traits that are expensive to measure Will resulting evaluations work in target population? Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (44) Cole

Some new traits studied recently ● Claw health (Van der Linde et al. , 2010) ● Dairy cattle health ● Embryonic development ● Immune response ● Methane production ● Milk fatty acid composition ● Persistency of lactation ● Rectal temperature ● Residual feed intake Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (45) (Parker Gaddis et al. , 2013) (Cochran et al. , 2013) (Thompson-Crispi et al. , 2013) (de Haas et al. , 2011) (Soyeurt et al. , 2011) (Cole et al. , 2009) (Dikmen et al. , 2013) (Connor et al. , 2013) Cole

Parentage validation and discovery l l l Parent-progeny conflicts detected w Animal checked against all other genotypes w Reported to breeds and requesters w Correct sire usually detected Maternal grandsire (MGS) checking w SNP at a time checking w Haplotype checking more accurate Breeds moving to accept SNPs in place of microsatellites Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (46) Cole

Haplotypes affecting fertility l l Rapid discovery of new recessive defects w Large numbers of genotyped animals w Affordable DNA sequencing Determination of haplotype location w Significant number of homozygous animals expected, but none observed w Narrow suspect region with fine mapping w Use sequence data to find causative mutation Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (47) Cole

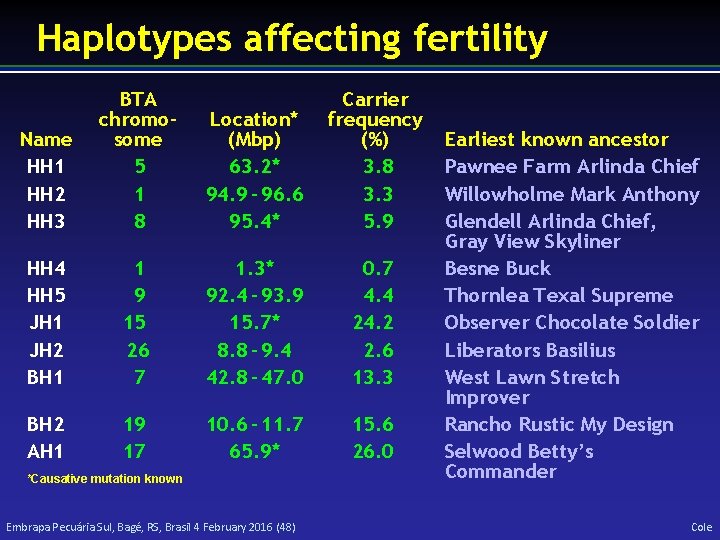
Haplotypes affecting fertility Name HH 1 HH 2 HH 3 BTA chromosome 5 1 8 Location* (Mbp) 63. 2* 94. 9 – 96. 6 95. 4* Carrier frequency (%) 3. 8 3. 3 5. 9 HH 4 HH 5 JH 1 JH 2 BH 1 1 9 15 26 7 1. 3* 92. 4 – 93. 9 15. 7* 8. 8 – 9. 4 42. 8 – 47. 0 0. 7 4. 4 24. 2 2. 6 13. 3 BH 2 AH 1 19 17 10. 6 – 11. 7 65. 9* 15. 6 26. 0 *Causative mutation known Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (48) Earliest known ancestor Pawnee Farm Arlinda Chief Willowholme Mark Anthony Glendell Arlinda Chief, Gray View Skyliner Besne Buck Thornlea Texal Supreme Observer Chocolate Soldier Liberators Basilius West Lawn Stretch Improver Rancho Rustic My Design Selwood Betty’s Commander Cole

Haplotypes tracking known recessives Recessive Brachyspina BLAD CVM DUMPS Mule foot Polled Red coat color SDM SMA Weaver Haplotype HH 0 HHB HHC HHD HHM HHP HHR BTA chromosome 21 1* 3* 1* 15* 1 18* BHD BHM BHW 11* 24* 4 Tested animals (no. ) ? 11, 782 13, 226 3, 242 87 345 4, 137 108 568 163 Concordance (%) ? 99. 9 — 100. 0 97. 7 — — 94. 4 98. 1 96. 3 New carriers (no. ) ? 314 2, 716 3 120 2, 050 5, 927 108 111 32 *Causative mutation known Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (49) Cole

Impact on breeders l l Haplotype and gene tests in selection and mating programs Trend towards a small number of elite breeders that are investing heavily in genomics About 30% of young males genotyped directly by breeders since April 2013 Prices for top genomic heifers can be very high (e. g. , $265, 000 or R$1, 060, 000) Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (50) Cole

Impact on dairy producers l General w Reduced generation interval w Increased rate of genetic gain w More inbreeding/homozygosity? Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (51) Cole

Impact on dairy producers l (continued) Sires w w Higher average genetic merit of available bulls More rapid increase in genetic merit for all traits Larger choice of bulls in terms of traits and semen price Greater use of young bulls Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (52) Cole

Why genomics works for dairy cattle l Extensive historical data available l Well-developed genetic evaluation program l Widespread use of AI sires l Progeny-test programs l l High-value animals worth the cost of genotyping Long generation interval that can be reduced substantially by genomics Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (53) Cole

Key issues for the dairy industry l l l Inbreeding and genetic diversity (including across breeds) Sequencing, new genes, and mutations Novel traits, resource populations (feed efficiency, health, milk properties) Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (54) Cole

Conclusions l l l Genomic evaluation has dramatically changed dairy cattle breeding Rate of gain is increasing primarily because of a large reduction in generation interval Genomic research is ongoing w Detect causative genetic variants w Find more haplotypes affecting fertility w Improve accuracy through more SNPs, more predictor animals, and more traits Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (55) Cole

Acknowledgments l l Appropriated project 1265 -31000 -096 -00, "Improving Genetic Predictions in Dairy Animals Using Phenotypic and Genomic Information", ARS, USDA CNPq “Science Without Borders” project 301025/2014 -2 Kristen Gaddis, Dan Null, Paul Van. Raden, and George Wiggans Council on Dairy Cattle Breeding Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (56) Cole

Disclaimer Mention of trade names or commercial products in this presentation is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (57) Cole

Questions? Embrapa Pecuária Sul, Bagé, RS, Brasil 4 February 2016 (58) Cole