Genomic improvement programs for US dairy cattle John












- Slides: 34

Genomic improvement programs for US dairy cattle John B. Cole Animal Genomics and Improvement Laboratory Agricultural Research Service, USDA Beltsville, MD john. cole@ars. usda. gov 2015

U. S. DHI dairy statistics (2011) l 9. 1 million U. S. cows l ~75% bred AI l 47% milk recorded through Dairy Herd Information (DHI) w 4. 4 million cows − 86% Holstein − 8% crossbred − 5% Jersey − <1% Ayrshire, Brown Swiss, Guernsey, Milking Shorthorn, Red & White w 20, 000 herds w 220 cows/herd w 10, 300 kg/cow CRV, Arnhem, The Netherlands, 14 April 2015 (2) Cole

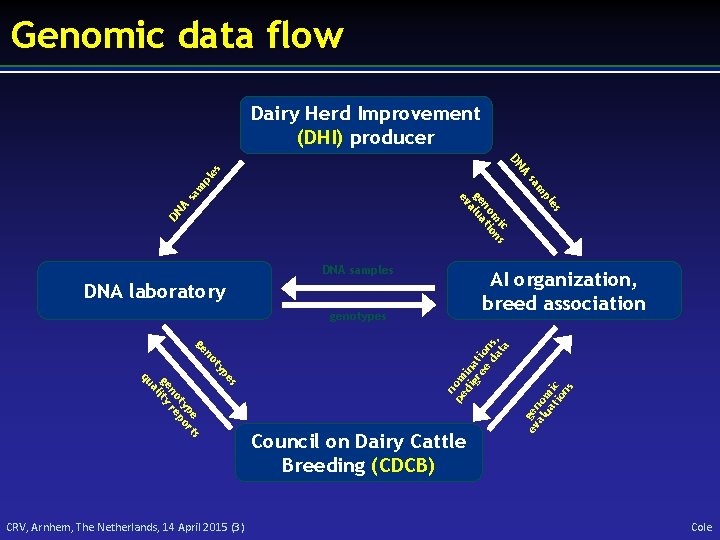
Genomic data flow Dairy Herd Improvement (DHI) producer s pe ty no ge s pe rt ty po no re ge lity a qu n pe om di in gr at ee io da ns, ta genotypes Council on Dairy Cattle Breeding (CDCB) ev gen al om ua tio ic ns sa A DN es AI organization, breed association DNA laboratory CRV, Arnhem, The Netherlands, 14 April 2015 (3) pl m m sa pl A es DN ic s m on no ti ge alua ev DNA samples Cole

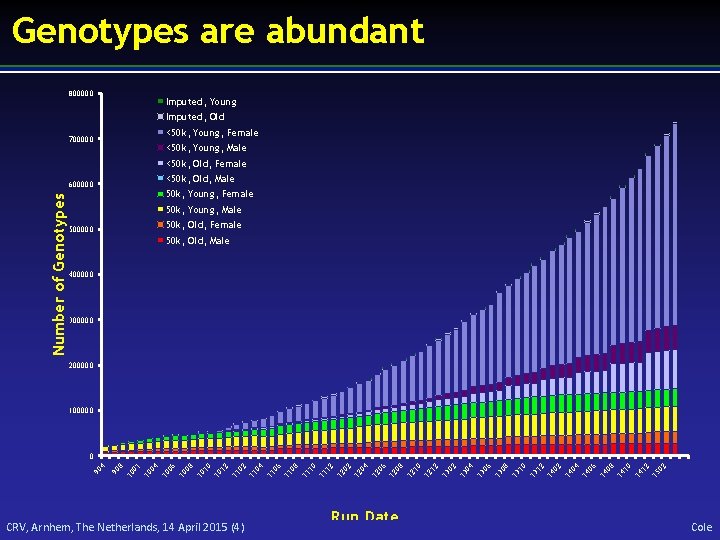
Genotypes are abundant 800000 Imputed, Young Imputed, Old 700000 <50 k, Young, Female <50 k, Young, Male <50 k, Old, Female Number of Genotypes 600000 <50 k, Old, Male 50 k, Young, Female 50 k, Young, Male 500000 50 k, Old, Female 50 k, Old, Male 400000 300000 200000 100000 90 4 90 8 10 01 10 04 10 06 10 08 10 10 10 12 11 04 11 06 11 08 11 10 11 12 12 04 12 06 12 08 12 10 12 12 13 04 13 06 13 08 13 10 13 12 14 04 14 06 14 08 14 10 14 12 15 02 0 CRV, Arnhem, The Netherlands, 14 April 2015 (4) Run Date Cole

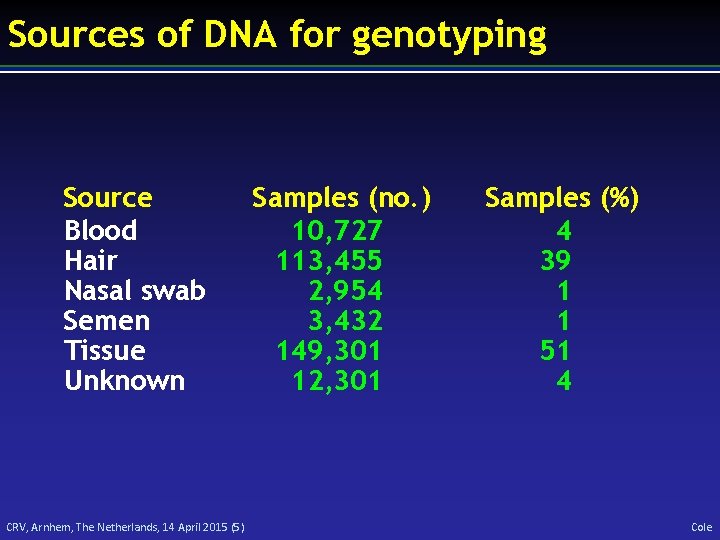
Sources of DNA for genotyping Source Blood Hair Nasal swab Semen Tissue Unknown CRV, Arnhem, The Netherlands, 14 April 2015 (5) Samples (no. ) 10, 727 113, 455 2, 954 3, 432 149, 301 12, 301 Samples (%) 4 39 1 1 51 4 Cole

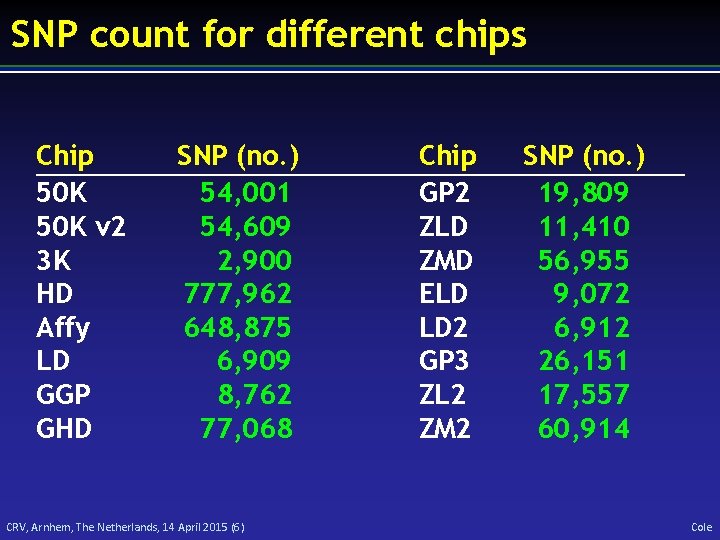
SNP count for different chips Chip 50 K v 2 3 K HD Affy LD GGP GHD SNP (no. ) 54, 001 54, 609 2, 900 777, 962 648, 875 6, 909 8, 762 77, 068 CRV, Arnhem, The Netherlands, 14 April 2015 (6) Chip GP 2 ZLD ZMD ELD LD 2 GP 3 ZL 2 ZM 2 SNP (no. ) 19, 809 11, 410 56, 955 9, 072 6, 912 26, 151 17, 557 60, 914 Cole

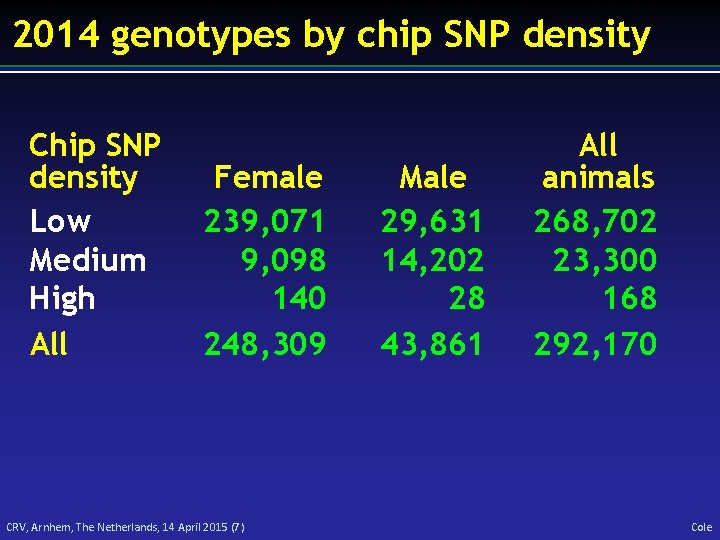
2014 genotypes by chip SNP density Chip SNP density Low Medium High All Female 239, 071 9, 098 140 248, 309 CRV, Arnhem, The Netherlands, 14 April 2015 (7) Male 29, 631 14, 202 28 43, 861 All animals 268, 702 23, 300 168 292, 170 Cole

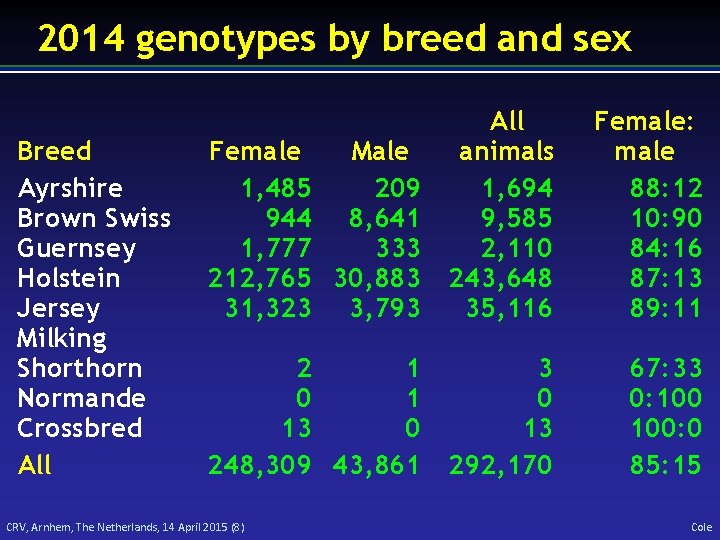
2014 genotypes by breed and sex Breed Ayrshire Brown Swiss Guernsey Holstein Jersey Milking Shorthorn Normande Crossbred All Female Male 1, 485 209 944 8, 641 1, 777 333 212, 765 30, 883 31, 323 3, 793 All animals 1, 694 9, 585 2, 110 243, 648 35, 116 Female: male 88: 12 10: 90 84: 16 87: 13 89: 11 2 1 0 1 13 0 248, 309 43, 861 3 0 13 292, 170 67: 33 0: 100: 0 85: 15 CRV, Arnhem, The Netherlands, 14 April 2015 (8) Cole

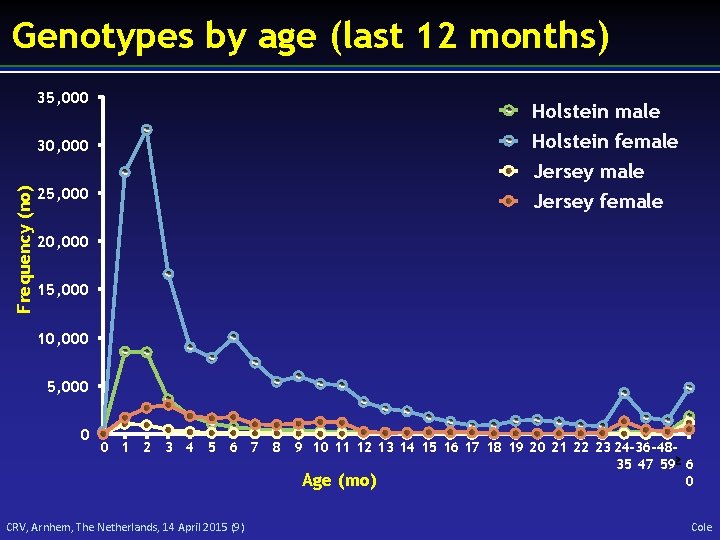
Genotypes by age (last 12 months) 35, 000 Holstein male Holstein female 30, 000 Frequency (no) Jersey male 25, 000 Jersey female 20, 000 15, 000 10, 000 5, 000 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 -36 -4835 47 59 6 0 Age (mo) CRV, Arnhem, The Netherlands, 14 April 2015 (9) Cole

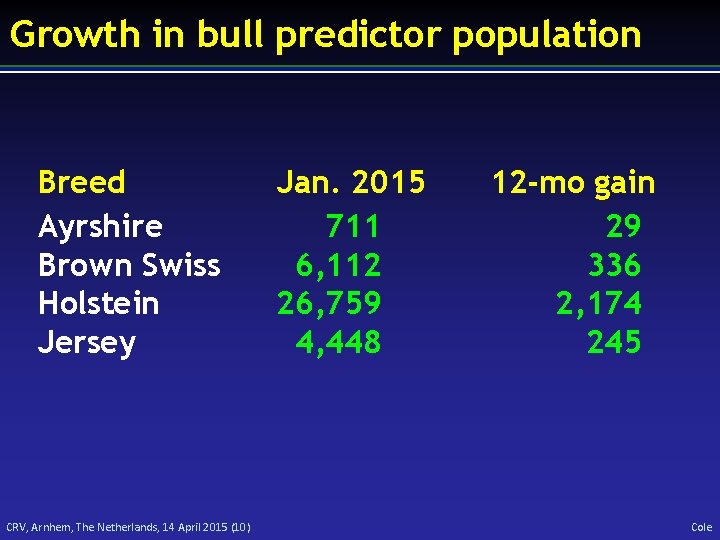
Growth in bull predictor population Breed Ayrshire Brown Swiss Holstein Jersey CRV, Arnhem, The Netherlands, 14 April 2015 (10) Jan. 2015 711 6, 112 26, 759 4, 448 12 -mo gain 29 336 2, 174 245 Cole

Growth in US predictor population Bulls Cows 1, 2 12 -mo Breed Jan. 2015 gain Ayrshire 711 29 69 40 Brown Swiss 6, 112 336 1, 138 350 Holstein 26, 759 2, 174 109, 714 51, 950 Jersey 4, 448 245 26, 012 10, 601 1 Predictor 2 Counts cows must have domestic records. include 3 k genotypes, which are not included in the predictor population. CRV, Arnhem, The Netherlands, 14 April 2015 (11) Cole

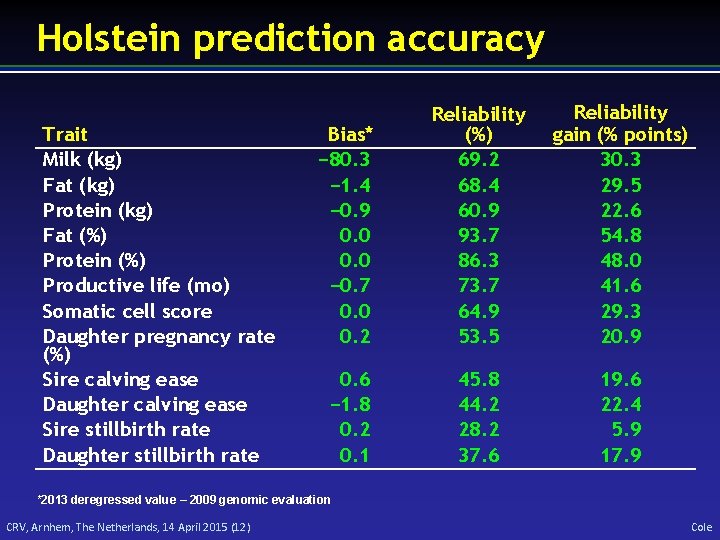
Holstein prediction accuracy Trait Milk (kg) Fat (kg) Protein (kg) Fat (%) Protein (%) Productive life (mo) Somatic cell score Daughter pregnancy rate (%) Sire calving ease Daughter calving ease Sire stillbirth rate Daughter stillbirth rate Bias* − 80. 3 − 1. 4 − 0. 9 0. 0 − 0. 7 0. 0 0. 2 Reliability (%) 69. 2 68. 4 60. 9 93. 7 86. 3 73. 7 64. 9 53. 5 Reliability gain (% points) 30. 3 29. 5 22. 6 54. 8 48. 0 41. 6 29. 3 20. 9 0. 6 − 1. 8 0. 2 0. 1 45. 8 44. 2 28. 2 37. 6 19. 6 22. 4 5. 9 17. 9 *2013 deregressed value – 2009 genomic evaluation CRV, Arnhem, The Netherlands, 14 April 2015 (12) Cole

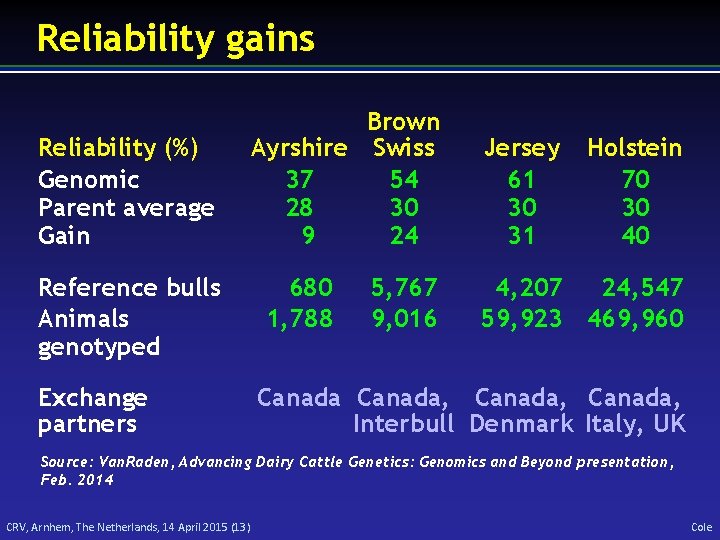
Reliability gains Reliability (%) Genomic Parent average Gain Reference bulls Animals genotyped Exchange partners Brown Ayrshire Swiss 37 54 28 30 9 24 680 1, 788 5, 767 9, 016 Jersey 61 30 31 Holstein 70 30 40 4, 207 24, 547 59, 923 469, 960 Canada, Interbull Denmark Italy, UK Source: Van. Raden, Advancing Dairy Cattle Genetics: Genomics and Beyond presentation, Feb. 2014 CRV, Arnhem, The Netherlands, 14 April 2015 (13) Cole

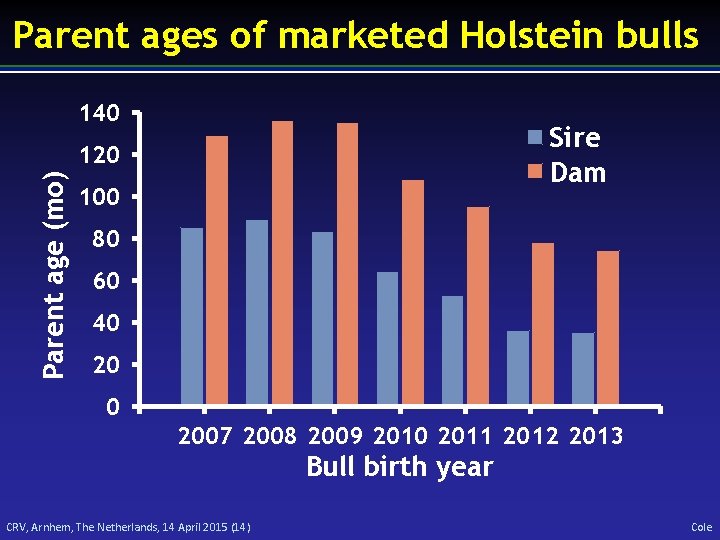
Parent ages of marketed Holstein bulls 140 Sire Dam Parent age (mo) 120 100 80 60 40 2007 2008 2009 2010 2011 2012 2013 Bull birth year CRV, Arnhem, The Netherlands, 14 April 2015 (14) Cole

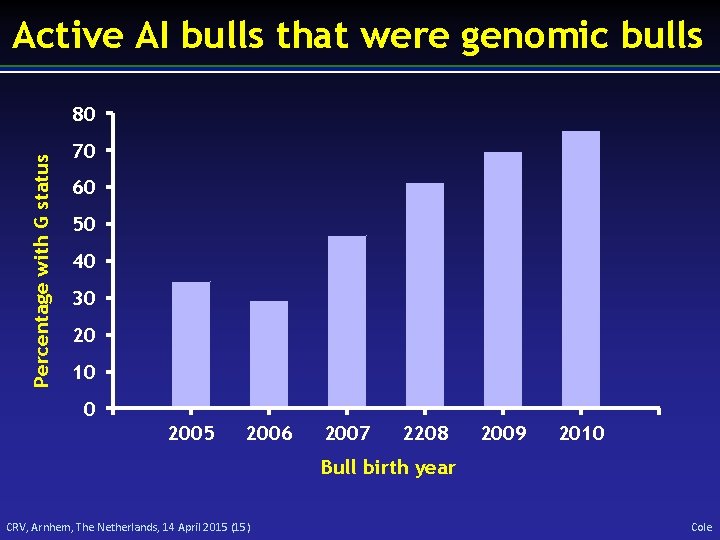
Active AI bulls that were genomic bulls Percentage with G status 80 70 60 50 40 30 20 10 0 2005 2006 2007 2208 2009 2010 Bull birth year CRV, Arnhem, The Netherlands, 14 April 2015 (15) Cole

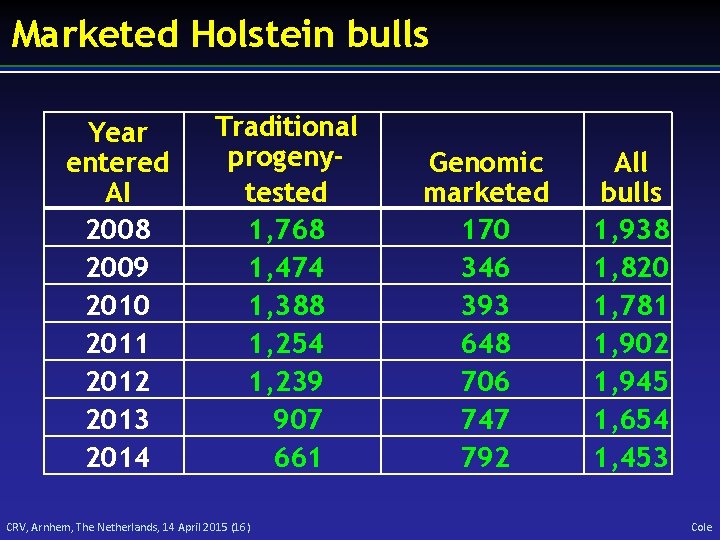
Marketed Holstein bulls Year entered AI 2008 2009 2010 2011 2012 2013 2014 Traditional progenytested 1, 768 1, 474 1, 388 1, 254 1, 239 907 661 CRV, Arnhem, The Netherlands, 14 April 2015 (16) Genomic marketed 170 346 393 648 706 747 792 All bulls 1, 938 1, 820 1, 781 1, 902 1, 945 1, 654 1, 453 Cole

Genetic merit of marketed Holstein bulls 800 Average gain: $85. 60/year Average net merit ($) 700 600 500 400 Average gain: $52. 00/year 300 200 100 0 -100 Average gain: $19. 77/year 00 01 02 03 04 05 06 07 08 09 10 11 12 13 14 Year entered AI CRV, Arnhem, The Netherlands, 14 April 2015 (17) Cole

Stability of genomic evaluations l l l 642 Holstein bulls w Dec. 2012 NM$ compared with Dec. 2014 NM$ w First traditional evaluation in Aug. 2014 w 50 daughters by Dec. 2014 Top 100 bulls in 2012 w Average rank change of 9. 6 w Maximum drop of 119 w Maximum rise of 56 All 642 bulls w Correlation of 0. 94 between 2012 and 2014 w Regression of 0. 92 CRV, Arnhem, The Netherlands, 14 April 2015 (18) Cole

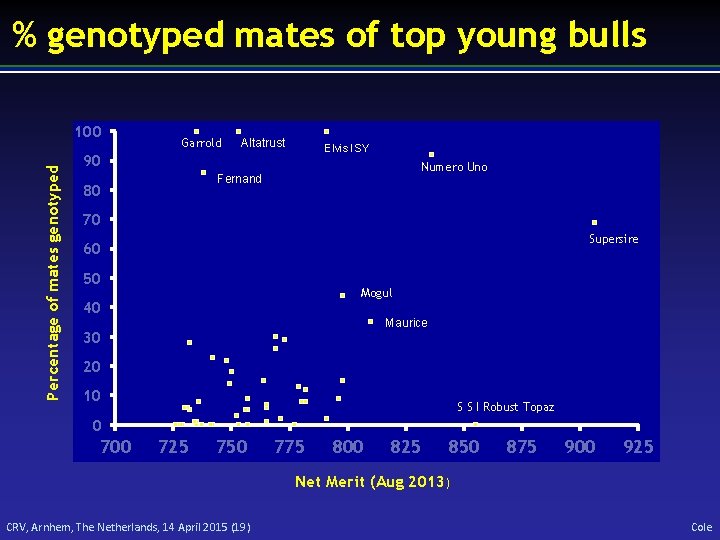
% genotyped mates of top young bulls Percentage of mates genotyped 100 Garrold Altatrust Elvis ISY 90 Numero Uno Fernand 80 70 Supersire 60 50 Mogul 40 Maurice 30 20 10 S S I Robust Topaz 0 700 725 750 775 800 825 850 875 900 925 Net Merit (Aug 2013) CRV, Arnhem, The Netherlands, 14 April 2015 (19) Cole

Haplotypes affecting fertility l l Rapid discovery of new recessive defects w Large numbers of genotyped animals w Affordable DNA sequencing Determination of haplotype location w Significant number of homozygous animals expected, but none observed w Narrow suspect region with fine mapping w Use sequence data to find causative mutation CRV, Arnhem, The Netherlands, 14 April 2015 (20) Cole

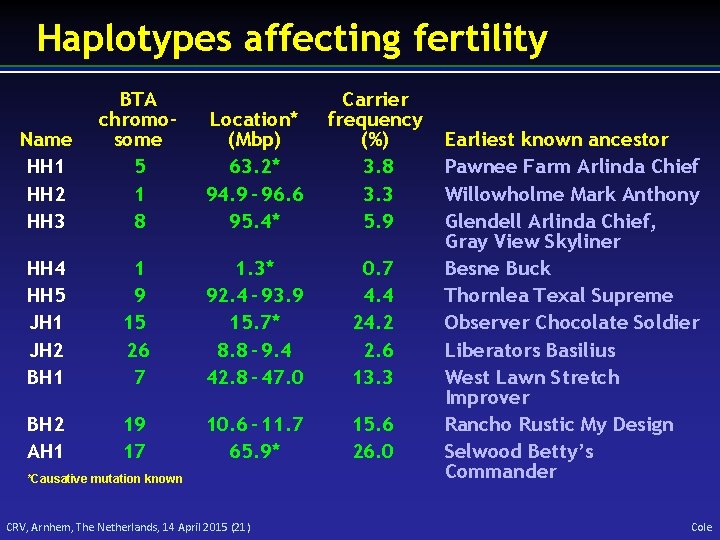
Haplotypes affecting fertility Name HH 1 HH 2 HH 3 BTA chromosome 5 1 8 Location* (Mbp) 63. 2* 94. 9 – 96. 6 95. 4* Carrier frequency (%) 3. 8 3. 3 5. 9 HH 4 HH 5 JH 1 JH 2 BH 1 1 9 15 26 7 1. 3* 92. 4 – 93. 9 15. 7* 8. 8 – 9. 4 42. 8 – 47. 0 0. 7 4. 4 24. 2 2. 6 13. 3 BH 2 AH 1 19 17 10. 6 – 11. 7 65. 9* 15. 6 26. 0 *Causative mutation known CRV, Arnhem, The Netherlands, 14 April 2015 (21) Earliest known ancestor Pawnee Farm Arlinda Chief Willowholme Mark Anthony Glendell Arlinda Chief, Gray View Skyliner Besne Buck Thornlea Texal Supreme Observer Chocolate Soldier Liberators Basilius West Lawn Stretch Improver Rancho Rustic My Design Selwood Betty’s Commander Cole

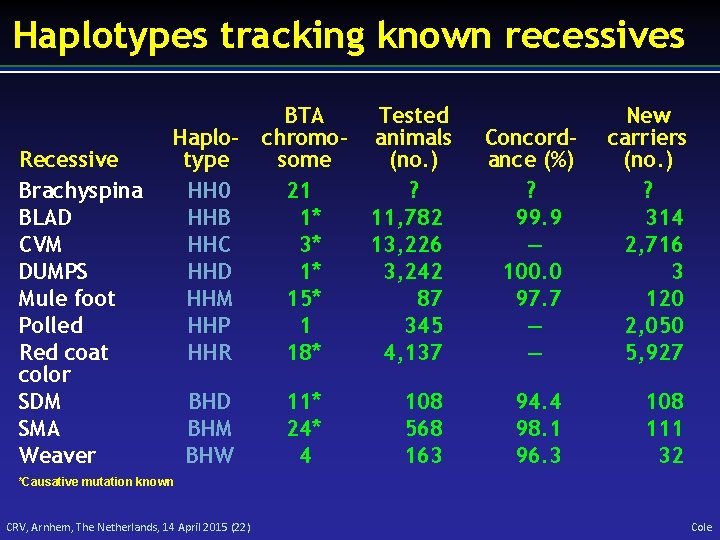
Haplotypes tracking known recessives Recessive Brachyspina BLAD CVM DUMPS Mule foot Polled Red coat color SDM SMA Weaver Haplotype HH 0 HHB HHC HHD HHM HHP HHR BTA chromosome 21 1* 3* 1* 15* 1 18* BHD BHM BHW 11* 24* 4 Tested animals (no. ) ? 11, 782 13, 226 3, 242 87 345 4, 137 108 568 163 Concordance (%) ? 99. 9 — 100. 0 97. 7 — — 94. 4 98. 1 96. 3 New carriers (no. ) ? 314 2, 716 3 120 2, 050 5, 927 108 111 32 *Causative mutation known CRV, Arnhem, The Netherlands, 14 April 2015 (22) Cole

Weekly evaluations l l l Released to nominators, breed associations, and dairy records processing centers at 8 am each Tuesday Calculations restricted to genotypes that first became usable during the previous week Computing time minimized by not calculating reliability or inbreeding CRV, Arnhem, The Netherlands, 14 April 2015 (23) Cole

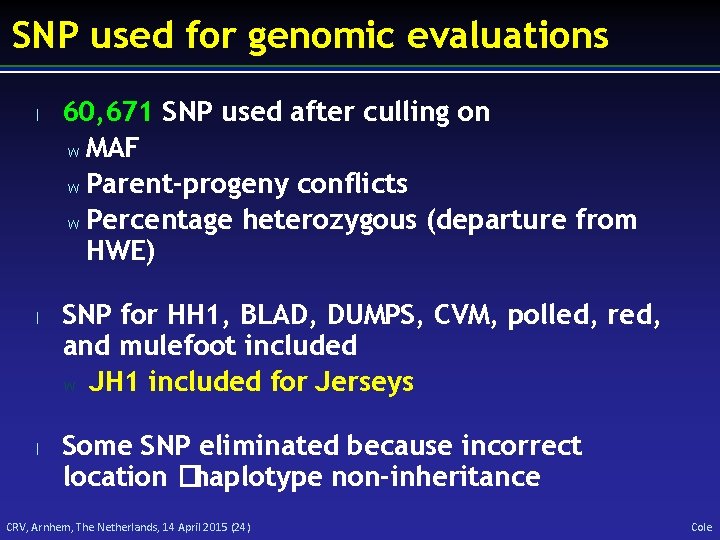
SNP used for genomic evaluations l l l 60, 671 SNP used after culling on w MAF w Parent-progeny conflicts w Percentage heterozygous (departure from HWE) SNP for HH 1, BLAD, DUMPS, CVM, polled, red, and mulefoot included w JH 1 included for Jerseys Some SNP eliminated because incorrect location �haplotype non-inheritance CRV, Arnhem, The Netherlands, 14 April 2015 (24) Cole

Some novel phenotypes studied recently ● Claw health (Van der Linde et al. , 2010) ● Dairy cattle health ● Embryonic development ● Immune response ● Methane production ● Milk fatty acid composition ● Persistency of lactation ● Rectal temperature ● Residual feed intake CRV, Arnhem, The Netherlands, 14 April 2015 (25) (Parker Gaddis et al. , 2013) (Cochran et al. , 2013) (Thompson-Crispi et al. , 2013) (de Haas et al. , 2011) (Soyeurt et al. , 2011) (Cole et al. , 2009) (Dikmen et al. , 2013) (Connor et al. , 2013) Cole

Evaluation methods for traits l Animal model (linear) w w w w l Heritability Yield (milk, fat, protein) Type (AY, BS, GU, JE) Productive life Somatic cell score Daughter pregnancy rate Heifer conception rate Cow conception rate 25 – 40% 7 – 54% 8. 5% 12% 4% 1% 1. 6% Sire–maternal grandsire model (threshold) w w Service sire calving ease Daughter calving ease Service sire stillbirth rate Daughter stillbirth rate CRV, Arnhem, The Netherlands, 14 April 2015 (26) 8. 6% 3. 0% 6. 5% Cole

Holstein daughter pregnancy rate (%) Breeding value (%) 8. 0 Cows 6. 0 4. 0 0. 0 -2. 0 1960 � 0. 1% /yr Sires 2. 0 Phenotypic base = 22. 6% 1970 1980 1990 2000 2010 Birth year CRV, Arnhem, The Netherlands, 14 April 2015 (27) Cole

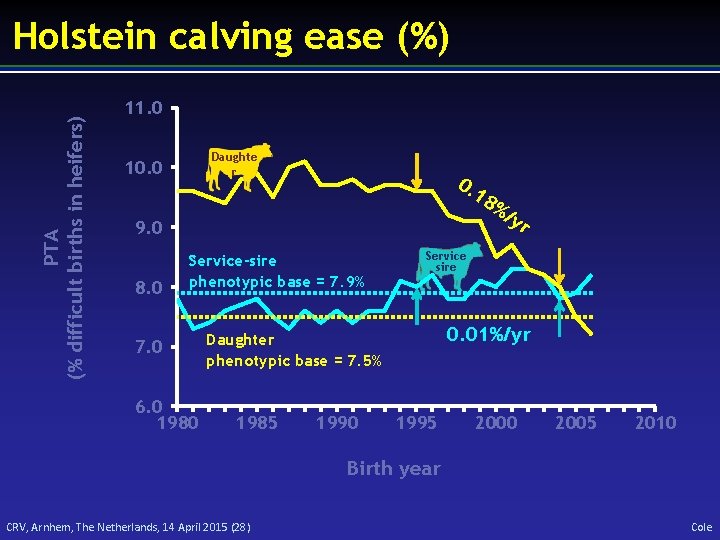
PTA (% difficult births in heifers) Holstein calving ease (%) 11. 0 Daughte r 10. 0 0. 18 9. 0 8. 0 Service-sire phenotypic base = 7. 9% 7. 0 6. 0 1980 1990 yr Service sire 0. 01%/yr Daughter phenotypic base = 7. 5% 1985 %/ 1995 2000 2005 2010 Birth year CRV, Arnhem, The Netherlands, 14 April 2015 (28) Cole

What do US dairy farmers want? National workshop in Tempe, AZ in February Producers, industry, academia, and government Farmers want new tools Additional traits (novel phenotypes) Better management tools Foot health and feed efficiency were of greatest interest CRV, Arnhem, The Netherlands, 14 April 2015 (29) Cole

What can farmers do with novel traits? Put them into a selection index Apply selection for a long time Correlated traits are helpful There are no shortcuts Collect phenotypes on many daughters Repeated records of limited value Genomics can increase accuracy CRV, Arnhem, The Netherlands, 14 April 2015 (30) Cole

What can DRPCs do with novel traits? Short-term – Benchmarking tools for herd management Medium-term – Custom indices for herd management Additional types of data will be helpful Long-term – Genetic evaluations Lots of data needed, which will take time CRV, Arnhem, The Netherlands, 14 April 2015 (31) Cole

International challenges National datasets are siloed Recording standards differ between countries ICAR standards help here Farmers are concerned about the security of their data Many populations are small Low accuracies Small markets CRV, Arnhem, The Netherlands, 14 April 2015 (32) Cole

Conclusions l l Genomic research is ongoing w Detect causative genetic variants w Find more haplotypes affecting fertility w Improve accuracy through more SNPs, more predictor animals, and more traits Genetic trend is favorable for some important, low-heritability traits w More traits are desirable w Data availability remains a challenge for new phenotypes CRV, Arnhem, The Netherlands, 14 April 2015 (33) Cole

Questions? CRV, Arnhem, The Netherlands, 14 April 2015 (34) Cole